Significance
Kaposi's sarcoma-associated herpesvirus (KSHV) and EBV are cancer-causing human herpesviruses. Their smallest capsid proteins (SCPs) were shown to be required for capsid assembly and are potential drug targets for curbing viral infections, but how they work is unclear. By cryoEM and genetic engineering, we determine the structures of KSHV capsids bearing full-length or truncated SCPs and localize regions of SCP that are important for capsid assembly. We show that a long kinked helix of SCP cross-links neighboring subunits of the major capsid protein of hexons to stabilize the capsid. Our results explain how SCP, acting like a cementing protein found in bacterial viruses, facilitates tumor herpesvirus capsid assembly and viral maturation.
Keywords: Kaposi's sarcoma-associated herpesvirus, smallest capsid protein, capsid assembly, capsid stabilization, cementing protein
Abstract
With just one eighth the size of the major capsid protein (MCP), the smallest capsid protein (SCP) of human tumor herpesviruses—Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV)—is vital to capsid assembly, yet its mechanism of action is unknown. Here, by cryoEM of KSHV at 6-Å resolution, we show that SCP forms a crown on each hexon and uses a kinked helix to cross-link neighboring MCP subunits. SCP-null mutation decreased viral titer by 1,000 times and impaired but did not fully abolish capsid assembly, indicating an important but nonessential role of SCP. By truncating the C-terminal half of SCP and performing cryoEM reconstruction, we demonstrate that SCP’s N-terminal half is responsible for the observed structure and function whereas the C-terminal half is flexible and dispensable. Serial truncations further highlight the critical importance of the N-terminal 10 aa, and cryoEM reconstruction of the one with six residues truncated localizes the N terminus of SCP in the cryoEM density map and enables us to construct a pseudoatomic model of SCP. Fitting of this SCP model and a homology model for the MCP upper domain into the cryoEM map reveals that SCP binds MCP largely via hydrophobic interactions and the kinked helix of SCP bridges over neighboring MCPs to form noncovalent cross-links. These data support a mechanistic model that tumor herpesvirus SCP reinforces the capsid for genome packaging, thus acting as a cementing protein similar to those found in many bacteriophages.
Herpesviridae is a large family of dsDNA viruses containing several widespread human pathogens. It is classified into three subfamilies, namely alpha-, beta-, and gammaherpesviruses (1). Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a member of the oncogenic gammaherpesvirus subfamily (2). All herpesviruses share the same architecture with a genome-containing capsid surrounded by a poorly defined tegument layer, which in turn is enclosed in a lipid envelope. The capsids of different herpesviruses are similar in composition and structure (3–5), each composed of at least five capsid proteins (encoding genes of KSHV given in parentheses): the major capsid protein (MCP, ORF25) in hexameric and pentameric capsomers, triplex proteins 1 and 2 (ORF62 and ORF26, respectively) forming heterotrimers in a 1:2 ratio to connect the capsomers, the smallest capsid protein (SCP, ORF65) previously shown to decorate tips of hexons (6–9), and the portal protein (ORF43) forming a dodecameric structure for DNA genome packaging at one of the 12 icosahedral vertices (10).
Assembly pathway of herpesvirus capsid resembles what has been established for dsDNA bacteriophages such as T4, λ, and P22 (11, 12): a spherical, porous intermediate named “procapsid” is first assembled around a core of scaffold proteins; the scaffold proteins are then cut by a maturational protease and extruded through the large holes of the procapsid shell; concurrently, DNA genome is packaged through the portal; the procapsid undergoes dramatic conformational changes and matures into a more stable, angularized icosahedral capsid (12).
The assembly of herpesvirus capsid was proposed to be a spontaneous process, as the capsid of HSV-1 (herpes simplex virus 1, an alphaherpesvirus) could be assembled in a cell-free system (13, 14). The minimum set of capsid proteins required for HSV-1 capsid assembly was determined to include the MCP, the two triplex proteins, the scaffold protein, and the maturational protease. The SCP of HSV-1 was dispensable for capsid assembly (13) and virus propagation (15). However, SCPs of KSHV and Epstein-Barr virus (EBV), the two known human pathogens of the gammaherpesvirus subfamily, have been demonstrated to be vital for capsid assembly in vitro (16, 17) and important for virus propagation (18). How SCP of tumor herpesviruses affects capsid assembly is poorly understood because of the lack of high-resolution capsid structures of these viruses.
Here we use cryo electron microscopy (cryoEM) combined with BAC mutagenesis to determine structures of KSHV virion capsids bearing full-length or truncated SCP. The results provide molecular and mechanistic insights into the role of this 170-residue small protein in KSHV capsid assembly and viral propagation.
Materials and Methods
Construction of KSHV Mutant BACs.
KSHV BAC16 genome was modified according to a previously described method (19). Briefly, for KSHV-SCPnull, WT sequence (ATC CCG CCT TTG AAT TCC ACC CAT CCT CCT CAG; nucleotides 112,532–112,564; GQ994935.1) in the ORF65 coding region was replaced by a mutant sequence (ATC CCG CCT Tag atc tca ACC CAT CCT CCT CAG) to introduce a stop codon that can abolish ORF65 protein translation and a Bgl II restriction site that would facilitate the screening of the desired mutation. For KSHV SCP truncation mutants, shortened sequence with the corresponding amino acids removed was used to replace the WT sequence by homologous recombination. The restriction patterns of BAC plasmids were verified by comparing them with BAC16 to ensure their overall integrity without gross changes other than the expected ones. Fragments with the mutations in the middle were PCR-amplified from the BAC plasmids and sequenced to confirm that all mutations were correct. All mutant BAC plasmids were further introduced into iSLK-puro cells, followed by selection with 1,200 µg/mL hygromycin B, 1 µg/mL puromycin, and 250 µg/mL G418 for 1 mo to generate cell lines latently infected by the KSHV mutants.
Titration of Infectious KSHV Virus.
To determine the concentration of infectious KSHV virions released from iSLK cells harboring KSHV WT or ORF65 mutant genome, the supernatants were collected from iSLK cell cultures 3 d after induction with 1 mM sodium butyrate plus 1 µg/mL doxycycline, centrifuged at 10,000 × g for 15 min to remove cellular debris, serially diluted in DMEM with 10% (vol/vol) FBS, and then used to infect 293T cells by spinoculation (3,000 × g for 1 h at 30 °C). Because the KSHV BAC16 virus harbors a GFP expression cassette driven by the cellular EF1_promoter, 293T cells will express GFP when the viral genome has been delivered into the cells, providing a means to measure the amount of infectious viruses present in the original supernatant solution. Three days after infection, GFP-positive cell clusters containing two or more cells were counted under a fluorescence microscope to determine the titers of KSHV viruses. Infectious units are expressed as the number of GFP-positive cell clusters in each well at the time of analysis.
Western Blotting and Antibodies.
Cell lysates were resolved by SDS/PAGE, transferred onto PVDF membrane, and further probed with rabbit polyclonal antibody against SCP (20) or mouse monoclonal antibody to actin (Abcam).
Transmission EM.
The iSLK-WT, iSLK-SCPnull, or iSLK-SCPN86 cells were treated with 1 µg/mL doxycycline plus 1 mM sodium butyrate for 3 d to induce viral lytic replication. Then, cells were collected and subjected to plastic embedding and transmission EM by the method previously described (21). Briefly, cells were washed with PBS solution, fixed with 2% (vol/vol) glutaraldehyde in PBS solution for 1 h, postfixed in 1% OsO4 for 1 h, en bloc stained in 2% (wt/vol) uranyl acetate for 1 h, dehydrated in an ascending ethanol series, and embedded in Spurr resin. Approximately 75-nm sections were stained with saturated aqueous uranyl acetate and lead citrate and examined with an FEI Tecnai F20 electron microscope operated at 200 kV.
Virion Purification.
The iSLK cell lines harboring WT or mutant KSHV BAC16 were cultured in DMEM with 10% FBS, 1 µg/mL puromycin, 250 µg/mL G418, and 1,200 µg/mL hygromycin to 80% confluence, and then treated with 1 mM sodium butyrate and 1 µg/mL doxycycline in fresh DMEM plus 10% FBS for 3–5 d to induce viral lytic replication. WT and mutant KSHV virions were purified from supernatants of the culture, following the procedure developed previously for purification of HCMV virions (4, 22).
CryoEM and 3D Reconstruction.
Viral sample was applied to holey carbon-coated grids, plunge-frozen in liquid ethane, and imaged at liquid nitrogen temperature in an FEI Titan Krios electron microscope. We first imaged the KSHV-WT sample on Kodak SO-163 films at a magnification of 59,000×, and the films were digitalized with a Nikon Coolscan 9000ED scanner at 1.1 Å per pixel and used to obtain an 8-Å-resolution map. To improve the image quality, we subsequently imaged the KSHV-WT sample on a Gatan K2 direct electron detector at super-resolution mode with a nominal magnification of 14,000×, giving a pixel size of 1.03 Å per pixel. The KSHV-SCPN86 and KSHV-SCPΔN6 mutants were imaged on the Gatan K2 detector under the same conditions. For the KSHV-SCPnull mutant, images were recorded on a Gatan UltraScan 4k × 4k CCD camera at a nominal magnification of 47,000×, giving a pixel size of 1.89 Å per pixel. An accumulated electron dose of 25 e−/Å2 was used in all cases. For all K2 datasets, each image was fractioned into 26 frames, and all frames were aligned and averaged for drift correction with a method described elsewhere (23).
Defocus value of each image was determined with CTFFIND3 (24) to range from 0.5 to 2.5 µm underfocus. Particles were picked with Ethan (25) and preprocessed with EMAN (26). Center and orientation parameters of each particle were determined and refined iteratively with the common-line–based method by using the IMIRS software package (27, 28). Three-dimensional reconstructions were carried out with the GPU program eLite3D (29). Resolution was determined based on the 0.143 Fourier shell correlation criterion (30). Visualization of the density maps was carried out with Chimera (31).
Pseudoatomic Model Building.
The pseudoatomic model of KSHV SCP was built ab initio with Coot (32). The pseudoatomic homology model of KSHV MCPud was first built with MODELER (33) by using the atomic model of HSV-1 MCPud (PDB ID code 1NO7) (34) as template, and then manually adjusted in Coot by matching the secondary structures resolved in the cryoEM map as described previously (35).
Results
KSHV SCP Cross-Links Neighboring MCPs in the Hexon.
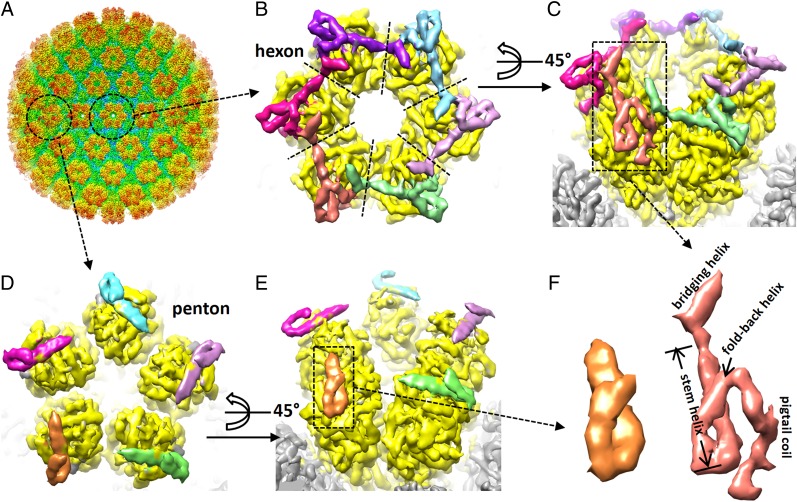
Sample availability has been a major hurdle for structural studies of human tumor herpesviruses. We purified infectious KSHV virion to high concentration with the newly developed BAC16 plasmid and the iSLK-puro cell line (36, 37). From ∼5,000 particle images recorded on a direct electron detector with superresolution mode, we reconstructed the structure of KSHV virion capsid to 6-Å resolution (Fig. 1A and Movie S1). Taking advantage of the known crystallographic structure of HSV-1 MCP upper domain [MCPud; Protein Data Bank (PDB) ID code 1NO7] (34) and the simple fold of KSHV SCP (it is merely 170 residues long), we were able to identify the SCP–MCP boundary, segment SCP monomers (Fig. 1 B–E), and trace the SCP backbone (Fig. 1F), although identification of the N and C termini was problematic.
Fig. 1.
The cryoEM structure of KSHV virion capsid reveals that SCP cross-links hexon MCPs. (A) The cryoEM reconstruction of KSHV virion capsid at 6-Å resolution. The density map is radially colored. Note that the tegument proteins surrounding the pentons identified previously are at low density level (50) and thus not visible at the contour level optimal for displaying the capsid proteins. (B and C) Zoom-in views of a hexon in the KSHV virion capsid structure. For clarity, the six MCP subunits are all colored in yellow, the six SCP molecules are colored differently, and all other densities of the capsid are gray (C) or hidden. The dashed lines in B roughly mark boundaries of neighboring MCP subunits. Note that each hexon SCP binds two neighboring MCPs to cross-link them. (D and E) Zoom-in views of a penton in the KSHV virion capsid structure. The coloring scheme is similar to that in B and C. (F) SCP monomers segmented out from the penton (Left) or from the hexon (Right). Secondary structural elements identified are labeled in the hexon SCP structure.
SCP densities were identified in hexons and pentons (Fig. 1 B and D), in contrast to the previous notion that KSHV SCP decorates hexons but not pentons (6). In the hexon, six copies of SCP form a crown-like rim of the hexon (Fig. 1 B and C and Movie S1). Each hexon SCP density consists of a “stem helix” binding in a surface groove of the underlying MCP, flanked by a “bridging helix” extended over the neighboring MCP, and a “fold-back helix” and “pigtail coil” on the other side (Fig. 1F). With their bridging helices, the six SCP molecules in a hexon cross-link and fasten the six MCP subunits (Fig. 1 B and C). In the penton, five copies of SCP bind the MCP with their stem helices in a way similar to that in the hexon, but no cross-linking is observed for neighboring MCP/SCP subunits (Fig. 1 D and E). Actually, no bridging helix and pigtail coil densities are visualized for penton SCP (Fig. 1F), suggesting structural flexibility of these segments. It is likely that, because of an enlarged distance and/or relative rotation (34) between neighboring MCP subunits in the penton compared with the hexon (compare Fig. 1D with Fig. 1B), the bridging helix of penton SCP can no longer reach to and interact with the neighboring SCP and MCP, rendering the bridging helices and pigtail coils of all penton SCPs flexible.
Structure-Guided Mutagenesis Establishes SCP’s Auxiliary Role in Capsid Assembly.
The aforementioned cross-linking of hexon MCPs by SCPs suggests that SCP may function by strengthening the integrity of hexons in the capsid. In this regard, previous studies with recombinant capsid proteins in a baculovirus expression system have suggested an essential role of SCP in KSHV and EBV in vitro capsid assembly (16, 17), and the “essential” region of SCP was further mapped to the N-terminal half by mutagenesis scan with the in vitro capsid assembly system (17, 38). To assess whether SCP is also essential for in vivo viral assembly and to better understand its mechanism of action, we performed conventional transmission EM of mutant virus-infected cells and cryoEM of purified viruses bearing SCP-null mutation or SCP C-terminal truncation.
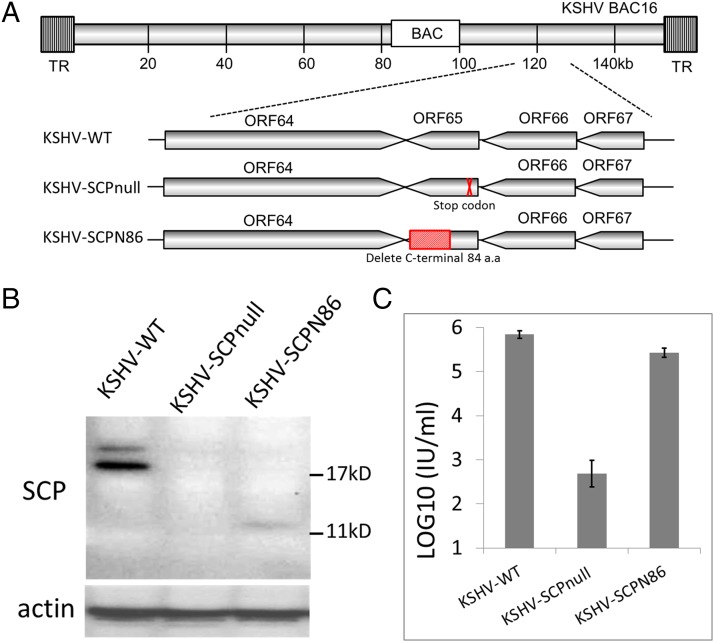
We constructed an SCP-null mutant by introducing a stop codon at the 5′ end of ORF65 in the KSHV-BAC16 plasmid (KSHV-SCPnull). The SCP C-terminal truncation mutant was generated by truncating SCP at residue number 86 (KSHV-SCPN86) to delete the C-terminal 84 aa (Fig. 2A). Absence of SCP expression in iSLK cells harboring KSHV-SCPnull genome (iSLK-SCPnull) and expression of truncated SCP in cells harboring KSHV-SCPN86 genome (iSLK-SCPN86) were confirmed by Western blotting with anti-SCP antibodies (Fig. 2B). We then tested the role of SCP in KSHV lytic replication by measuring progeny virus production of the two mutants. As shown in Fig. 2C, compared with that of the WT (KSHV-WT), the viral titer of KSHV-SCPnull mutant decreased approximately 1,000 fold, which is much more significant than the previously reported 30-fold decrease of total viral particles released, which was estimated by real-time PCR of extracellular viral genome (18). By contrast, the KSHV-SCPN86 mutant had only a moderate decrease in viral titer (approximately threefold), suggesting that the C-terminal half of KSHV SCP plays only a minor role in viral propagation.
Fig. 2.
Construction of KSHV SCP-null and SCP C-terminal truncation mutants. (A) Schematic illustration of KSHV BAC16 plasmids at the region encoding SCP (ORF65). Single stop codon was introduced into the 5′ end of ORF65 to generate the KSHV-SCPnull mutant; the coding sequence for the SCP C-terminal 84 aa was removed to generate the truncated mutant KSHV-SCPN86. (B) Expression of SCP proteins during KSHV reactivation. iSLK cells harboring KSHV-WT or mutants (KSHV-SCPnull or KSHV-SCPN86) were induced with sodium butyrate plus doxycycline for 3 d, and expression of SCP proteins were analyzed by Western blotting with anti-SCP antibodies. (C) Virus production from KSHV-WT or mutants. Supernatants from the cells were collected, and titers of infectious virus were determined on 293T cells.
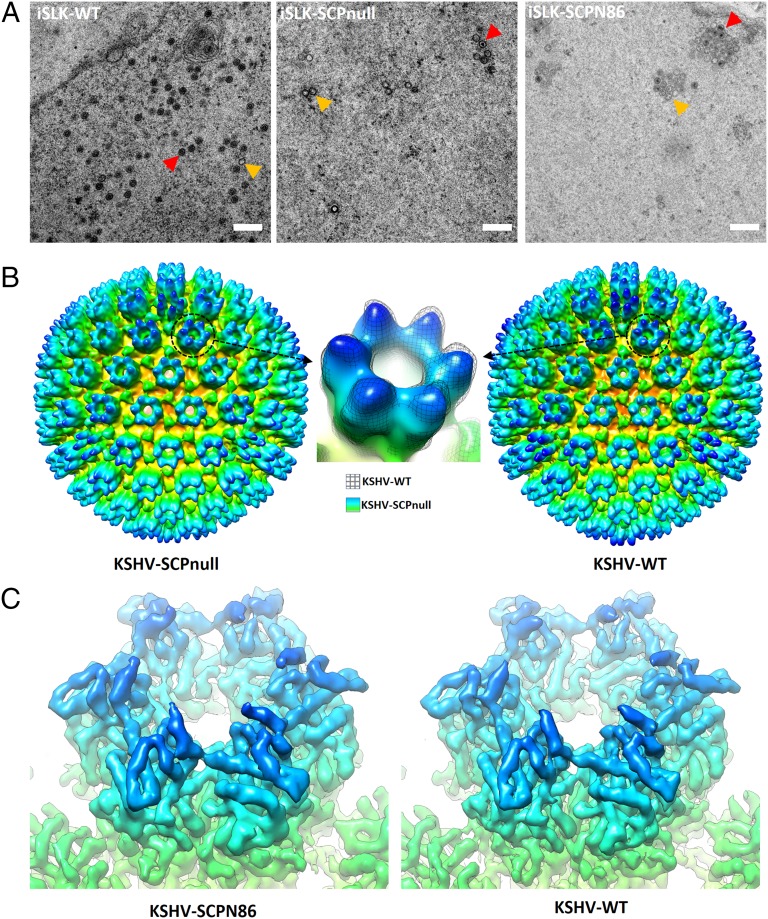
To verify whether the observed decrease of viral titer in absence of SCP expression is indeed a result of deficiency of capsid assembly, we examined thin sections of iSLK-WT, iSLK-SCPnull, or iSLK-SCPN86 cells 3 d after induction of KSHV lytic replication (Fig. 3A). As expected, although capsid-like particles were occasionally observed in nuclei of some iSLK-SCPnull cells, the number of cells that contained assembled capsids, and the number of capsids per cell, if any, were much lower than those of iSLK-WT cells (Table S1). Moreover, a higher percentage of empty capsids and a lower percentage of DNA-filled capsids were observed in iSLK-SCPnull cells compared with iSLK-WT cells. Consistently, we also observed by cryoEM that the percentage of DNA-filled C-capsids in purified virion samples was much lower for KSHV-SCPnull mutant (∼15%) compared with that for the KSHV-WT (more than 90%). We also compared the number of assembled capsids in iSLK-SCPN86 cell nuclei with that in iSLK-WT cells. Indeed, these numbers were comparable, consistent with viral titer measurements (Fig. 3A and Table S1). These ultrastructural observations confirm that the effects of SCP mutations on viral growth can be correlated to capsid assembly.
Fig. 3.
Comparative structural studies of KSHV SCP-null and SCP C-terminal truncation mutants. (A) Capsid assembly in the nuclei of KSHV-WT or mutant infected cells during viral lytic replication. The iSLK cells harboring KSHV-WT or mutants (KSHV-SCPnull or KSHV-SCPN86) were induced with sodium butyrate plus doxycycline for 3 d, and viral capsid assembly in the nuclei was examined with plastic embedding, ultrathin sectioning, and transmission EM. Red arrow marks DNA-filled C-capsid; yellow arrow marks empty A- or B-capsid. (Scale bar, 500 nm.) (B) Comparing the cryoEM reconstruction of KSHV-SCPnull mutant with that of the KSHV-WT at 30-Å resolution. The WT map was reconstructed from randomly selected particles in the same number as for the mutant. The two structures are very similar except that a little bit more densities are present on tip of capsomers in the WT, which is attributable to SCP (Inset), suggesting correct assembly of the KSHV-SCPnull capsids. (C) Comparison of the cryoEM reconstruction of KSHV-SCPN86 mutant with that of the KSHV-WT at 7-Å resolution. Zoom-in view of one hexon instead of the entire capsid structure is shown to emphasize that the C-terminal half truncated SCP has the same density as the full-length SCP, suggesting the flexible nature of KSHV SCP C-terminal half in the WT virus.
We then compared cryoEM reconstructions of purified KSHV-SCPnull particles and KSHV-SCPN86 particles with that of the WT. A structure of KSHV-SCPnull capsid at ∼30-Å resolution was obtained from 137 particle images. This structure resembles that of the WT filtered to the same resolution (Fig. 3B). Together with the fact that DNA-packaged virions were observed in cryoEM images of KSHV-SCPnull, this result confirms that KSHV capsid is correctly assembled and capable of encapsidating viral genome in the absence of SCP. Therefore, in contrast to the suggested essential role of SCP in the in vitro assembly studies, KSHV SCP is not essential for capsid assembly in vivo, but rather dramatically enhances the efficiency of capsid assembly. It is also possible that capsids were also assembled in the absence of SCP in previous in vitro studies but might have eluded detection as a result of their scarcity (16, 38). Our structure and in vivo mutagenesis studies support the notion that SCP is nonessential and plays a secondary or auxiliary role. We also obtained a 7-Å-resolution structure of KSHV-SCPN86 capsid from ∼1,500 particle images (Fig. 3C). Other than revealing less density, the structure of this mutant virus containing the C-terminal-half truncated SCP is nearly identical to that of the WT at secondary structure level; particularly, the truncated form of SCP retains the same density as the full-length SCP (Fig. 3C). It indicates that the SCP density visualized in KSHV-WT is contributed only by residues within the N-terminal half, and that the C-terminal half is flexible.
Taken together with our structural data, these observations suggest that hexon cross-linking by SCP plays a role in stabilizing assembled capsids and consequently enhancing viral genome containment, as discussed later.
Structural and Functional Mapping of the SCP N Terminus.
To better understand how the SCP interacts with the MCP and with each other to function, it is necessary to map the SCP density to its sequence and build a model for the SCP. However, the 6-Å resolution of the density map does not provide enough structural details, such as side-chain densities, to allow us to register the protein sequence directly. Particularly, we could not determine unambiguously which end of the SCP density corresponds to the N terminus of the SCP.
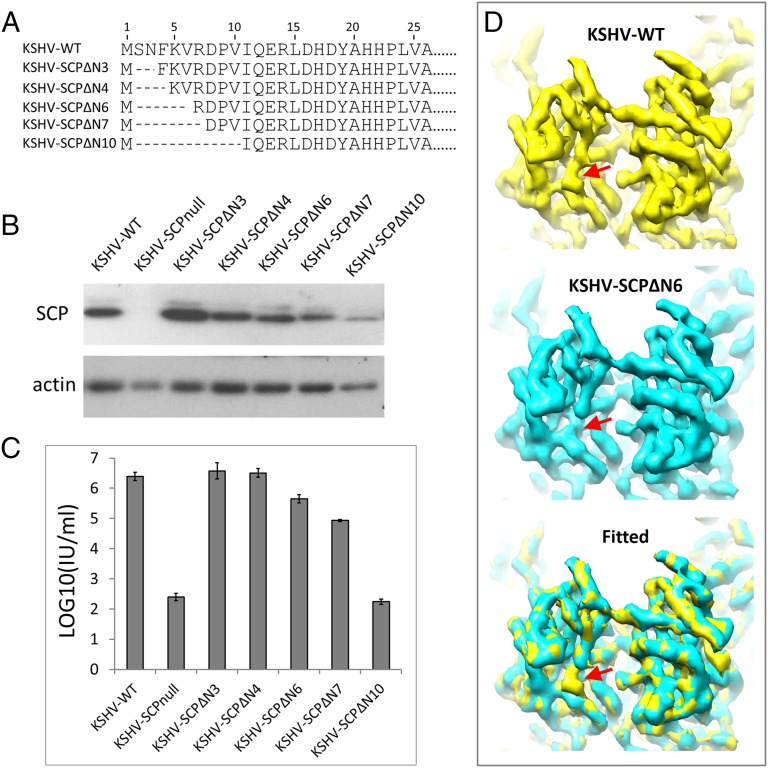
To localize the SCP N terminus and to test the function of SCP N-terminal residues in the context of KSHV replication, we constructed a set of mutant viruses with their SCP truncated in the N terminus for 3, 4, 6, 7, and 10 residues, respectively (e.g., KSHV-SCPΔN3, KSHV-SCPΔN4, and so on; Fig. 4A). Successful SCP expressions were confirmed by Western blotting with anti-SCP antibodies (Fig. 4B). Measurement of progeny virus production indicated that removal of 3 or 4 aa from KSHV SCP N terminus did not affect viral propagation, but removal of 6 or 7 aa exerted significant impact, with virion production decreased 81.6% or 95.7%, respectively, and removal of the N-terminal 10 aa reduced viral production to the level same as SCP-null mutation (Fig. 4C).
Fig. 4.
Structural and functional mapping of KSHV SCP N-terminal residues. (A) Partial protein sequences of WT SCP and the N-terminal truncated mutants. (B) Expression of SCP in iSLK cells harboring BAC plasmids of KSHV-WT or the SCP N-terminal truncated mutants during KSHV reactivation. (C) Virus production from KSHV-WT or the mutants during KSHV reactivation. (D) Comparison of cryoEM reconstructions of KSHV-SCPΔN6 mutant and the WT. It is obvious that a small trunk of density (red arrows) is missing in the cryoEM map of the mutant, which corresponds to the truncated N-terminal six residues of SCP.
We then selected the KSHV-SCPΔN6 mutant for cryoEM study because it showed attenuated viral titer but still yielded an adequate amount of virions for cryoEM reconstruction. A 7-Å-resolution virion capsid structure was obtained from ∼1,200 particles. As expected, fitting the structure with that of the WT clearly showed the absence of a short segment of density in the KSHV-SCPΔN6 mutant at the end of the pigtail coil (Fig. 4D). This result unambiguously mapped the N terminus of SCP to the pigtail coil.
Pseudoatomic Models of SCP and MCPud and SCP–MCP Interactions.
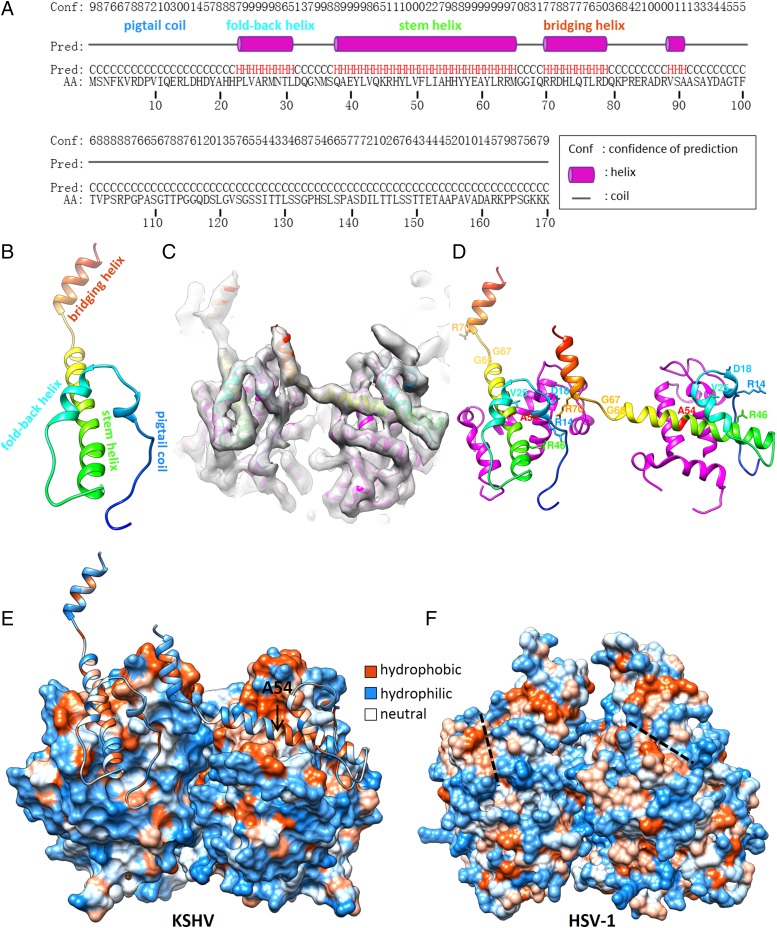
With the N terminus established, we then correlated the SCP density with the secondary structure prediction (Fig. 5A) and built a pseudoatomic model for KSHV SCP (Fig. 5B). Taking advantage of the known crystallographic structure of HSV-1 MCPud, and the fact that it is highly structurally conserved with KSHV MCPud, we also built a homology model for KSHV MCPud (Fig. 5C and Fig. S1).
Fig. 5.
Pseudoatomic models and interactions of KSHV SCP and MCPud. (A) Secondary structure prediction of KSHV SCP sequence (51). The predicted secondary structures correlate well with the observed structure of SCP in the density map, and their assignments to the real structure are marked. (B) Pseudoatomic model of KSHV SCP. The model is rainbow-colored from N′ (blue) to C′ (red). Key structural segments as named in Fig. 1F are also marked here. (C) Fitting the pseudoatomic model of SCP (rainbow-colored) and the homology model of MCPud (magenta) into the cryoEM density map of KSHV-WT hexon (translucent gray). (D) KSHV SCP–MCP and SCP–SCP interactions revealed by the pseudoatomic models. The orientation and coloring are the same as in B. For clarity, only part of the MCPud is shown, including the three helices forming the SCP-binding groove and a loop involved in SCP cross-linking. Positions are marked for SCP residues R14, D18, V25, R46, A54, G66, and R70, which were demonstrated previously to be vital for KSHV capsid assembly (38, 39). A stereo-pair version of D is also available as Fig. S2. (E and F) Hydrophobicity surface presentations of KSHV and HSV-1 MCPud. In E, the KSHV SCP model is also shown and colored according to its side-chain hydrophobicity to demonstrate the complementarity between the SCP and MCP interfaces. The position of SCP residue A54 is marked, and is buried in a hydrophobic packet of the MCP groove. In F, the two dashed lines mark the most likely binding area on HSV-1 MCPud for the stem helix of HSV-1 SCP. Note that this SCP-binding groove in HSV-1 is not as prominent as that in KSHV.
The model shows that association of SCP to the capsid is primarily via its stem helix (amino acids 39–65) binding in a groove formed by three helices at top surface of the MCP, namely helices 763–778, 826–840, and 871–883 (Fig. 5D, Fig. S2, and Movie S2). In the hexon, the bridging helix (amino acids 70–82) of SCP is also interacting with loop 837–856 of the neighboring MCP and pigtail coil amino acids 10–20 of another SCP, forming the cross-link (Fig. 5 C and D and Fig. S2). The stem helix and bridging helix of each SCP can also be regarded as one long helix kinked by a flexible glycine pivot point (G66 and G67). Pigtail coil amino acids 1–10 of hexon SCP also makes specific contacts with the surface of underlying MCP (Fig. 5C). As we have demonstrated, very short truncations in this region affected viral replication considerably (Fig. 4C). Therefore, these 10 aa may help to maintain the entire pigtail coil in the right position and right conformation to form the cross-link.
Six residues—R14, D18, V25, R46, G66, and R70—of KSHV SCP were demonstrated previously to be vital for capsid assembly: when any of these residues was mutated to alanine, capsid assembly was “abolished” or reduced (38). Based on our model, R14, D18, and R70 are likely directly involved in forming the cross-link (Fig. 5D and Fig. S2). G66, together with G67, may serve as a pivot point for the bridging helix to have some flexibility relative to the stem helix, as glycine is the smallest and most flexible amino acid. Conceivably, having a somewhat flexible kinked helix, instead of a rigid straight long helix sticking out, might be a good strategy to form the cross-link without causing steric hindrance in earlier stages of capsid assembly. Interestingly, G67 is conserved among SCPs of several gammaherpesviruses (Fig. S3), as discussed later in more detail.
Hydrophobicity analysis of the model suggests that hydrophobic interactions might be an important driving force for SCP binding MCP. Highly hydrophobic patches are identified in the MCP surface groove, which are complementary to several hydrophobic or neutral residues on the groove-binding side of SCP stem helix, especially the segment LVFLIA (amino acids 49–54; Fig. 5E, marked position of A54). In support of this, it had been demonstrated that substitution of A54 with polar residue lysine abolished SCP binding and capsid assembly, whereas substitutions with hydrophobic residues leucine, valine, or even proline did not (39). In addition, L49K and I53K mutations also resulted in partially defective assemblies (39).
Although identities among SCP sequences from alpha-, beta-, and gammaherpesviruses are relatively low, the presence of an MCP-binding stem helix is likely a conserved structural feature among all SCPs (3, 4, 40, 41). However, MCPs from different herpesvirus subfamilies may have evolved different surface properties for SCP binding. Comparing surface presentations of MCPud from KSHV and HSV-1, it is obvious that KSHV MCP has a more prominent groove than that in HSV-1 MCP at the SCP-binding area (Fig. 5 E and F and Movie S2). As seen from the models, helix 763–778 contributing one cliff of the groove in KSHV MCP is replaced by loop 767–781 in HSV-1 (Fig. S1, black arrows), and resulted in a not-so-prominent groove. We propose that evolution may have strengthened the MCP binding of KSHV SCP for its pivotal role in viral propagation or, conversely, weakened evolutionary pressure caused by dispensability of HSV-1 SCP might have led to weakened binding of HSV-1 SCP.
Discussion
Our 6-Å KSHV virion capsid structure shows that SCP forms a crown at the rim of each hexon to cross-link neighboring MCP subunits. SCP-null mutation confirmed an important but nonessential role of SCP. Mutagenesis studies with C- and N-terminal truncations established that the N-terminal half is responsible for SCP to cross-link hexon MCPs. Integration of structural and functional studies has enabled us to construct a pseudoatomic model for SCP and to define the largely hydrophobic interactions between SCP and MCP. Taken together, the present description of SCP–MCP chemical interactions for a herpesvirus, to our knowledge the first of its kind, helps in the understanding of the functional roles of SCP in different subfamilies of herpesviruses.
Within the gammaherpesvirus subfamily, secondary structure predictions of SCPs from four gammaherpesviruses—KSHV, EBV, rhesus monkey rhadinovirus (RRV), and herpesvirus saimiri (HVS)—show similar patterns of three-helix composition in their N-terminal segments (Fig. S3). Besides, multiple-sequence alignment shows that their N-terminal segments are more conserved compared with their C-terminal segments (Fig. S3), consistent with what we have observed here for KSHV SCP, in which the N-terminal half is functionally important but the C-terminal half is dispensable. Indeed, the SCP of EBV was also demonstrated to be important for capsid assembly in vitro, and only its N-terminal half was required and sufficient (17), which is strikingly similar to KSHV. Therefore, it is reasonable to expect that the cross-linking of hexon MCP by a crown of SCP we observed in KSHV is a conserved feature among many, if not all, gammaherpesviruses.
The auxiliary role of SCP in gammaherpesvirus capsid stabilization mirrors that of cementing proteins often found in dsDNA bacteriophages, such as the Soc protein in phage T4 (42, 43) and the gpD protein in phage λ (44), as proposed previously for VP26 of HSV (45). In phage λ, the 11.4-kDa gpD forms a trimer and binds to capsid surface at quasi- and icosahedral threefold axes after capsid maturation; it stabilizes the capsid structure by fastening six gpE (the MCP) subunits from three neighboring capsomers (44). These nonessential auxiliary proteins in phages “cement” adjacent MCPs to stabilize the mature capsid against extremes in pH and temperature as well as other factors in their hostile environment. They also help the thin capsid shell to withstand the pressure exerted by the packaged DNA (42, 44, 46, 47). In gammaherpesvirus, it is conceivable that the cross-linking by hexon SCP would also fasten the MCP subunits in hexons and thus increase the overall stability of the capsid. Indeed, we observed lower percentage of genome-containing C-capsids in SCP-null mutant-infected cells and also in cryoEM images of purified virion compared with WT. It is possible that, in the absence of SCP, some capsids may collapse during and/or after DNA packaging.
Our finding of gammaherpesvirus SCP acting as an auxiliary cementing protein agrees with previous notions of similarities between herpesviruses and dsDNA bacteriophages (12, 45, 48). It has been well documented that these two groups of viruses use strategies of capsid assembly that bear many similarities, including the use of scaffold proteins, the formation of procapsids, and the incorporation of a portal complex at one unique capsid vertex for translocation of genome (12). Moreover, herpesvirus MCP contains an HK97-like fold in the capsid floor domain (48). All these similarities support a common ancestry for herpesviruses and tailed dsDNA bacteriophages as proposed before (48).
Across the three subfamilies of Herpesviridae, SCP is the least conserved among all capsid proteins in terms of the identities and sizes of their sequences. Nonetheless, our notion of SCP acting as a cementing protein can be extrapolated to all herpesviruses. For alphaherpesviruses whose SCP is dispensable, the effect of SCP on capsid stability may have been somehow weakened over the course of evolution, possibly because of strengthened interactions among other capsid components. For betaherpesviruses, other auxiliary proteins, such as the capsid-associated tegument protein pp150 in human cytomegalovirus (HCMV), may work together with SCP to cement capsid proteins. Indeed, HCMV SCP is required for the association of pp150 to the capsid (4). In this scenario, the extensive cementing network formed by SCP and pp150 in cytomegalovirus may provide additional strength that is needed for the encapsidation of its genome, which is the largest among all herpesviruses (49).
Supplementary Material
Acknowledgments
We thank Dr. Jae U. Jung and Dr. Don Ganem for providing us the KSHV BAC16 plasmid and the iSLK-puro cell line used for viral culture and mutagenesis. This research was supported in part by National Institutes of Health (NIH) Grants AI046420/AI094386, CA091791, DE023591, and GM071940, and a University of California, Los Angeles (UCLA) Clinical and Translational Science Institute (CTSI) Core Voucher Award. We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA, NIH Instrumentation Grants 1S10RR23057 and 1S10OD018111, and National Science Foundation Grant DBI-1338135.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinate has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3J9A), and EMDataBank, www.emdatabank.org (accession nos. EMD-6038, EMD-6212, EMD-6213, and EMD-6214).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420317112/-/DCSupplemental.
References
- 1.Pellet P, Roizman B. The family Herpesviridae: A brief introduction. In: Knipe D, Howley P, editors. Fields Virology. 5th Ed. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 2479–2500. [Google Scholar]
- 2.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3.Zhou ZH, et al. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288(5467):877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 4.Dai X, et al. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog. 2013;9(8):e1003525. doi: 10.1371/journal.ppat.1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu XK, et al. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J Virol. 2003;77(24):13182–13193. doi: 10.1128/JVI.77.24.13182-13193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo P, Yu X, Atanasov I, Chandran B, Zhou ZH. Three-dimensional localization of pORF65 in Kaposi’s sarcoma-associated herpesvirus capsid. J Virol. 2003;77(7):4291–4297. doi: 10.1128/JVI.77.7.4291-4297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X, et al. Three-dimensional localization of the smallest capsid protein in the human cytomegalovirus capsid. J Virol. 2005;79(2):1327–1332. doi: 10.1128/JVI.79.2.1327-1332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou ZH, et al. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2(11):1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 9.Trus BL, et al. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: Structural authenticity and localization of VP26. J Virol. 1995;69(11):7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rochat RH, et al. Seeing the portal in herpes simplex virus type 1 B capsids. J Virol. 2011;85(4):1871–1874. doi: 10.1128/JVI.01663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter E. Virus assembly. In: Knipe D, Howley P, editors. Fields Virology. 5th Ed. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 141–168. [Google Scholar]
- 12.Brown JC, Newcomb WW. Herpesvirus capsid assembly: Insights from structural analysis. Curr Opin Virol. 2011;1(2):142–149. doi: 10.1016/j.coviro.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newcomb WW, Homa FL, Thomsen DR, Ye Z, Brown JC. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68(9):6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newcomb WW, et al. Assembly of the herpes simplex virus capsid: Characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263(3):432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 15.Desai P, DeLuca NA, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247(1):115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 16.Perkins EM, et al. Small capsid protein pORF65 is essential for assembly of Kaposi’s sarcoma-associated herpesvirus capsids. J Virol. 2008;82(14):7201–7211. doi: 10.1128/JVI.00423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henson BW, Perkins EM, Cothran JE, Desai P. Self-assembly of Epstein-Barr virus capsids. J Virol. 2009;83(8):3877–3890. doi: 10.1128/JVI.01733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sathish N, Yuan Y. Functional characterization of Kaposi’s sarcoma-associated herpesvirus small capsid protein by bacterial artificial chromosome-based mutagenesis. Virology. 2010;407(2):306–318. doi: 10.1016/j.virol.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Gong D, et al. Kaposi’s sarcoma-associated herpesvirus ORF18 and ORF30 are essential for late gene expression during lytic replication. J Virol. 2014;88(19):11369–11382. doi: 10.1128/JVI.00793-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin SF, et al. Identification, expression, and immunogenicity of Kaposi’s sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71(4):3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng L, Ryazantsev S, Sun R, Zhou ZH. Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure. 2010;18(1):47–58. doi: 10.1016/j.str.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai X, Zhou ZH. Purification of Herpesvirus virions and capsids. Bio-protocol. 2014;4(15):e1193. doi: 10.21769/bioprotoc.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 25.Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. Local average intensity-based method for identifying spherical particles in electron micrographs. J Struct Biol. 2000;131(2):126–134. doi: 10.1006/jsbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- 26.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 27.Liang Y, Ke EY, Zhou ZH. IMIRS: A high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137(3):292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, et al. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J Struct Biol. 2008;161(1):64–73. doi: 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Zhang X, Zhou ZH. Low cost, high performance GPU computing solution for atomic resolution cryoEM single-particle reconstruction. J Struct Biol. 2010;172(3):400–406. doi: 10.1016/j.jsb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. EMBO J. 2003;22(4):757–765. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou ZH, et al. Four levels of hierarchical organization, including noncovalent chainmail, brace the mature tumor herpesvirus capsid against pressurization. Structure. 2014;22(10):1385–1398. doi: 10.1016/j.str.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brulois KF, et al. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol. 2012;86(18):9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myoung J, Ganem D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J Virol Methods. 2011;174(1-2):12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreitler D, et al. The assembly domain of the small capsid protein of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2012;86(21):11926–11930. doi: 10.1128/JVI.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capuano CM, et al. A hydrophobic domain within the small capsid protein of Kaposi’s sarcoma-associated herpesvirus is required for assembly. J Gen Virol. 2014;95(pt 8):1755–1769. doi: 10.1099/vir.0.064303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker ML, et al. Ab initio modeling of the herpesvirus VP26 core domain assessed by CryoEM density. PLOS Comput Biol. 2006;2(10):e146. doi: 10.1371/journal.pcbi.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homa FL, et al. Structure of the pseudorabies virus capsid: Comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol. 2013;425(18):3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin L, Fokine A, O’Donnell E, Rao VB, Rossmann MG. Structure of the small outer capsid protein, Soc: A clamp for stabilizing capsids of T4-like phages. J Mol Biol. 2010;395(4):728–741. doi: 10.1016/j.jmb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao VB, Black LW. Structure and assembly of bacteriophage T4 head. Virol J. 2010;7:356. doi: 10.1186/1743-422X-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lander GC, et al. Bacteriophage lambda stabilization by auxiliary protein gpD: Timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16(9):1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rixon FJ, Chiu W. Studying large viruses. Adv Protein Chem. 2003;64:379–408. doi: 10.1016/s0065-3233(03)01011-8. [DOI] [PubMed] [Google Scholar]
- 46.Booy FP, et al. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64(5):1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauer DW, Huffman JB, Homa FL, Evilevitch A. Herpes virus genome, the pressure is on. J Am Chem Soc. 2013;135(30):11216–11221. doi: 10.1021/ja404008r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79(23):14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, et al. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J Struct Biol. 2011;174(3):451–460. doi: 10.1016/j.jsb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai X, Gong D, Wu TT, Sun R, Zhou ZH. Organization of capsid-associated tegument components in Kaposi’s sarcoma-associated herpesvirus. J Virol. 2014;88(21):12694–12702. doi: 10.1128/JVI.01509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.