Significance
Polo-like kinases (Plks) are a conserved family of enzymes that function as master regulators for the process of cell division. Among their duties, Plks control the assembly of centrosomes, tiny organelles that facilitate mitotic spindle assembly and maintain the fidelity of chromosome inheritance. Plks are overexpressed in cancer, and therefore it is critical to unravel the normal regulation of these kinases. Here, we studied Plk4 regulation whose activity controls centrosome number. We showed that, as do other Plks, Plk4 autoinhibits its kinase activity. However, Plk4 is unique in its ability to relieve autoinhibition through a third Polo box domain not present in other Plk family members. Moreover, autoinhibition controls Plk4 oligomerization, which ultimately governs its stability and thus centrosome duplication.
Keywords: autoinhibition, centriole, centrosome, Polo box, Polo-like kinase 4
Abstract
Polo-like kinase 4 (Plk4) is a master regulator of centriole duplication, and its hyperactivity induces centriole amplification. Homodimeric Plk4 has been shown to be ubiquitinated as a result of autophosphorylation, thus promoting its own degradation and preventing centriole amplification. Unlike other Plks, Plk4 contains three rather than two Polo box domains, and the function of its third Polo box (PB3) is unclear. Here, we performed a functional analysis of Plk4’s structural domains. Like other Plks, Plk4 possesses a previously unidentified autoinhibitory mechanism mediated by a linker (L1) near the kinase domain. Thus, autoinhibition is a conserved feature of Plks. In the case of Plk4, autoinhibition is relieved after homodimerization and is accomplished by PB3 and by autophosphorylation of L1. In contrast, autophosphorylation of the second linker promotes separation of the Plk4 homodimer. Therefore, autoinhibition delays the multiple consequences of activation until Plk4 dimerizes. These findings reveal a complex mechanism of Plk4 regulation and activation which govern the process of centriole duplication.
The Polo-like kinase (Plk) family of proteins functions as master regulators of cell-cycle progression, cell division, centrosome maturation, and centriole duplication (1–3). All four members (Plk1–4) of this serine/threonine protein kinase family share sequence similarity and domain structure with the founding member, Drosophila Polo kinase (the homolog of human Plk1) (4). Plks are highly expressed in proliferating cells and are overexpressed in a variety of cancers where they have the potential to promote chromosomal instability and tumorigenesis (5–9). Previous studies have shown that Plk kinase activity can be limited to brief periods within the cell cycle through mechanisms involving the transcription, localization, degradation, and autoinhibition of the kinase (3, 10–13). New regulatory mechanisms of Plks continue to be identified (14–16), making it clear that our understanding of Plk regulation is incomplete.
All Plks contain an N-terminal kinase domain followed by one or more Polo box (PB) motifs separated by linkers of varying length (4). PBs are ∼100-aa multifunctional domains that serve as hubs of protein interaction and are important for dimerization, substrate binding, intracellular targeting, and autoinhibition of kinase activity (3, 4, 12, 13, 17). Plk1–3 contain two PBs, whereas Plk4 contains three distinct PBs (18). Of all Plk members, Plk1 regulation is the best understood, in part because the recent crystallization of the kinase domain in complex with its PB-linker elements (16) has revealed insights into its mechanism of autoinhibition. The two PBs of Plk1 form an intramolecular dimer joined by two linkers (19) and together make extensive contact with the kinase domain (16, 20, 21). This interaction rigidifies the hinge region of the kinase domain, thereby decreasing the flexibility of the ATP cleft and likely crippling nucleotide hydrolysis (16). Inhibition is relieved either by phosphopeptide binding to the PB dimer or by phosphorylation within the kinase domain (22–26) which disrupts the kinase domain–PB linker interaction (16). In addition, full Plk1 activity requires phosphorylation of its activation loop (AL) by Aurora A (27, 28), but this phosphorylation is hindered by the interdomain linker that connects the kinase domain to the PB dimer (16). Thus, Plk1 normally is inactive because of autoinhibition and requires multiple cell-cycle–dependent inputs to achieve full mitotic activation.
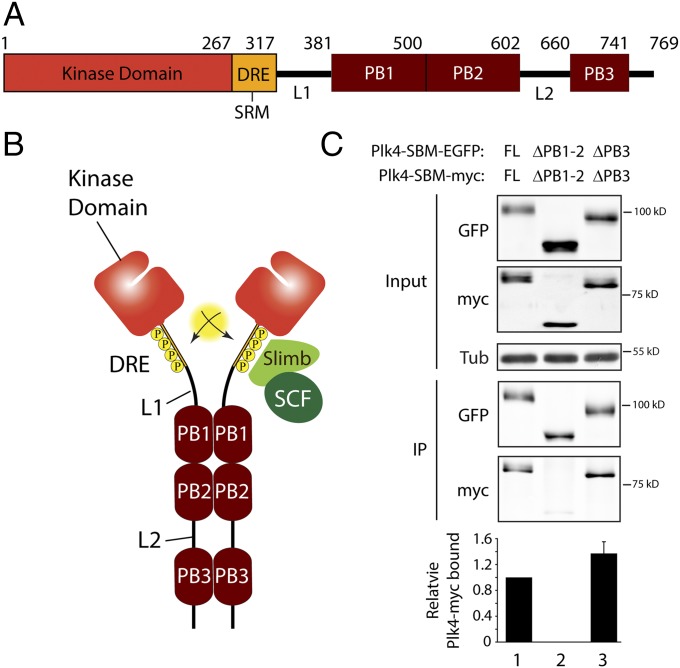
Plk4 is the master regulator of centriole duplication, and its hyperactivation drives centriole amplification (29–34), a phenomenon observed in cancer (35). Plk4 is distinct from its monomeric relatives because it forms a homodimer and contains an additional PB, PB3 (Fig. 1A) (18, 36). Crystal structure analysis suggests that Plk4 homodimerization is mediated by protein–protein interactions between the PB domains (18, 37), and its stability is regulated primarily through ubiquitin (Ubi)-mediated proteolysis (10, 11, 38). Plk4 generates its own phosphodegron that ultimately results in its degradation (36, 39–41). Following assembly into a homodimer, Plk4 extensively trans-autophosphorylates its downstream regulatory element (DRE) containing the phylogenetically conserved supernumerary limb (Slimb)-recognition motif (36, 39–41). This phosphorylation recruits the SCFSlimb/β-TrCP [the Skp1/Cullin/F-box E3 ubiquitin ligase complex containing Slimb (in Drosophila) or β-transducin repeat-containing protein (β-TrCP, in vertebrates)] to ubiquitinate Plk4 (Fig. 1B), although the specific ubiquitinated residues are not known.
Fig. 1.
Tandem Polo boxes PB1 and PB2 are required domains for Plk4 homodimerization. (A) Linear map of the Drosophila Plk4 polypeptide showing functional and structural domains including PB1–3, the DRE [containing the SRM (Slimb recognition motif)], L1, and L2. (B) Model of Plk4 autodestruction. Homodimerization facilitates trans-autophosphorylation, generating an extensive phosphodegron within each DRE. The SCFSlimb/β-TrCP Ubi-ligase binds the phosphodegron and ubiquitinates Plk4, targeting it for proteasomal degradation. (C) Anti-GFP immunoprecipitates (IPs) were prepared from lysates of S2 cells transiently co-overexpressing the indicated inducible nondegradable SBM EGFP and myc-tagged Plk4 constructs. Blots of the input lysates and IPs were probed for α-tubulin, GFP, and myc. Error bars indicate SEM. Amounts of Plk4-EGFP and associated Plk4-myc in the IPs were determined by densitometry of the anti-GFP and myc immunoblots and then normalizing the measurements with the amounts of SBM-Plk4 present in the IPs. The plotted values are relative to the coimmunoprecipitation in lane 1.
To understand how the PBs and linker regions regulate Plk4, we studied their impact on Plk4 activity and stability in cultured Drosophila S2 cells. Our analyses reveal that PBs not only are crucial for Plk4 homodimerization and ubiquitination but also relieve autoinhibition caused by linker 1 (L1). Relief of autoinhibition is mediated by downstream PB3, demonstrating a previously unidentified role for this third PB and supporting a multistep model for Plk4 activation. Thus, autoinhibition is a conserved regulatory mechanism of the Plk family and, in the case of Plk4, controls oligomerization.
Results
PBs Involved in Plk4 Dimerization.
Structures of purified fly PB1–PB2 and mouse PB3 have been solved, and although each PB is unique, they all adopt a classic PB-fold and form stable homodimers in vitro (18, 37). In the case of the purified PB1–PB2 cassette, homodimerization is mediated by contacts at both the PB1–PB1 and PB2–PB2 interfaces. These findings have led to a model in which all three PBs mediate Plk4 homodimerization (Fig. 1B), but this model has not been tested in cells.
To determine which PBs actually mediate homodimerization, we performed dimerization assays using S2 cells coexpressing different EGFP- and myc-tagged Plk4 constructs. Because Plk4 normally is degraded rapidly, we stabilized expressed Plk4 in transfected cells by introducing the Slimb-binding mutation (SBM, a double-Ala substitution in the DRE) into the different constructs used in this assay (10, 11). Plk4-EGFP protein was immunoprecipitated from clarified cell lysates, and quantitative immunoblotting was used to measure the amounts of associated Plk4-myc. As expected, Plk4-myc binds Plk4-EGFP (Fig. 1C, lane 1). We next evaluated the importance of the PB1–PB2 tandem as a single unit because these PBs are adjacent and are separated by only a short Thr–Pro linker. Plk4 lacking PB1–PB2 (∆PB1–PB2) was unable to self-associate (Fig. 1C, lane 2 and Fig. S1A, lane 2). Thus, PB1–PB2 is critical for Plk4 dimerization. In contrast, PB3 is not required for dimerization, because proteins lacking PB3 associate and, in fact, dimerize to a greater extent than full-length Plk4 (Fig. 1C, lane 3).
The individual contributions of PB1 and PB2 to dimerization were examined by deleting either domain from Plk4-EGFP and then measuring association with full-length (WT) Plk4-myc. Deletion of either PB1 or PB2 dramatically reduced binding, but neither disrupted binding to the extent of ∆PB1–PB2, underscoring the importance of the PB1–PB2 cassette in dimerization (Fig. S1A). The PB1 and PB2 deletions are not equivalent: ∆PB2 bound about three times less WT-Plk4 than ∆PB1. Using a similar assay, we also examined the sufficiency of EGFP- and myc-tagged PB1–PB2 fragments for dimerization. As expected, PB1–PB2-myc associated with PB1–PB2-EGFP but not with PB3-EGFP (Fig. S1B, lanes 1–4). Thus, Plk4 homodimerization is mediated primarily through contacts between PB1–PB2 cassettes.
Ubiquitination Sites in Plk4.
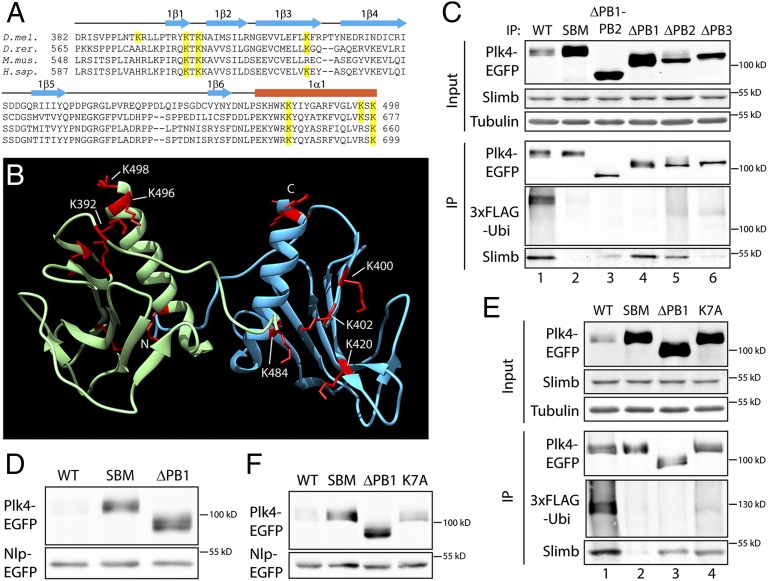
Specific sites of Plk4 ubiquitination are unknown. To identify ubiquitinated residues, we immunoprecipitated Plk4-EGFP expressed in S2 cells and mapped di-glycine (Gly–Gly)–modified Lys residues using tandem MS (MS/MS). Trypsin treatment of ubiquitinated proteins cleaves Ubi near its C terminus, leaving only a Gly–Gly remnant of Ubi covalently bound to a Lys of the modified protein. Therefore, Gly–Gly–modified Lys are diagnostic for ubiquitination sites (42). MS analysis of full-length Plk4 (total coverage was 97% and included all Lys residues) identified several Lys-linked Gly–Gly modifications (Fig. S2 A and B and Table S1). Seven of the modified residues reside in PB1, a region in close proximity to the Slimb-binding DRE, and five of these sites are conserved in humans (Fig. 2A). Within Drosophila PB1, the modified residues cluster in two regions. In the first region, K496 and K498 of the C terminus of the 1α1 helix are spatially clustered with K392 in a nearby loop (Fig. 2B). In the second, the remaining four ubiquitinated Lys (K484 in the N terminus of 1α1, K400 and K420 within β-strands 1 and 3, respectively, and K402 within the 1β1–1β2 linker) have their side chains oriented toward the opposite face of PB1.
Fig. 2.
PB1 contains multiple ubiquitination sites. (A) Plk4 PB1 sequence alignment across four species. Fly PB1 contains nine Lys residues, seven of which were Gly–Gly modified (highlighted in yellow), indicating that these residues were ubiquitinated in vivo. Five of the ubiquitinated residues are conserved in human PB1. (B) Quaternary structure of homodimerized PB1. Ubiquitinated Lys residues are shown in red. (C) Anti-GFP IPs from lysates of S2 cells transiently expressing 3xFLAG-Ubi and the indicated Plk4-EGFP construct. Blots were probed with anti-GFP, anti-FLAG, and anti-Slimb antibodies. Note that ubiquitination is greatly diminished in Plk4 lacking PB1 (lane 4), even though it binds endogenous Slimb. (D) Anti-GFP immunoblots of lysates of S2 cells transiently expressing the indicated inducible Plk4-EGFP constructs were used to examine the relative stabilities of the Plk4 proteins. (Cotransfected Nlp-EGFP was expressed under its endogenous promoter and served as a loading control in gels.) (E) Anti-GFP IPs from S2 cell lysates transiently expressing 3xFLAG-Ubi and the indicated inducible Plk4-EGFP construct. Blots were probed with anti-GFP, anti-FLAG, and anti-Slimb antibodies. Note that although the K7A mutant binds Slimb, its ubiquitination is decreased dramatically. (F) As in D, anti-GFP immunoblots of S2 cell lysates were used to examine the relative stabilities of the indicated inducible Plk4-EGFP constructs.
To test which PBs are important for ubiquitination, we expressed various PB deletion-EGFP mutants together with 3xFLAG-Ubi in S2 cells and then assessed their ubiquitination states. As expected, WT-Plk4 coprecipitated with endogenous Slimb and was robustly ubiquitinated, whereas nondegradable SBM-Plk4 did not bind Slimb, nor was it ubiquitinated (Fig. 2C, lanes 1 and 2). Likewise, EGFP alone was not labeled with FLAG-Ubi (Fig. S2C). As previously reported (18), PB1–PB2 deletion (ΔPB1–PB2) reduces Slimb binding and abolishes incorporation of FLAG-Ubi (Fig. 2C, lane 3). Without PB1–PB2, this mutant fails to dimerize and so probably also fails to become trans-autophosphorylated, a prerequisite for Slimb binding and ubiquitination (40, 41). Supporting this inference is its electrophoretic mobility which is a readout of Plk4 autophosphorylation (11, 41): Plk4–ΔPB1–PB2 migrates as a sharp band on SDS/PAGE, indicating that this mutant is less phosphorylated than WT-Plk4, which migrates as a diffuse phosphorylated species. Likewise, deletion of PB2, which severely impairs dimerization, migrates primarily as a sharp band and displays reduced Slimb binding and ubiquitination (Fig. 2C, lane 5). Deletion of PB1, which has less impact on dimerization than ΔPB2, does not appear to block autophosphorylation. This mutant displayed the diffuse appearance of a phosphorylated species on SDS/PAGE and bound a similar amount of Slimb as WT-Plk4 (Fig. 2C, lane 4). Strikingly, although Plk4–ΔPB1 binds Slimb, its ubiquitination was reduced to an undetectable level (Fig. 2C, lane 4), suggesting that PB1 is a major site of Plk4 ubiquitination. Moreover, Plk4–ΔPB1 total protein levels were dramatically elevated in cells, similar to nondegradable SBM-Plk4 (Fig. 2D).
Next, we tested whether the seven Gly–Gly–modified Lys in PB1 contributed to Plk4 ubiquitination. To block their ubiquitination, all seven residues were mutated to Ala (K7A) in an otherwise WT-Plk4-EGFP background and then were coexpressed with 3xFLAG-Ubi in cells. Immunoprecipitation of K7A revealed that it recruits Slimb, suggesting that it is capable of homodimerization and trans-autophosphorylation, but ubiquitination was reduced almost to the same extent as in Plk4–ΔPB1 (Fig. 2E). [We confirmed that the K7A mutation does not inhibit dimerization using an immunoprecipitation assay (Fig. S2D).]
Reduced ubiquitination of K7A was accompanied by an increase in its protein levels in cells, although Plk4–ΔPB1 was noticeably more stable (Fig. 2F). To test whether K7A is protective against proteasome-mediated degradation, Plk4 expression was induced overnight, and then cycloheximide (CHX) was introduced to block further protein synthesis. Plk4 protein levels then were assayed every 2 h (Fig. S2 E and F). WT-Plk4 is relatively short-lived (55% of WT was eliminated 2 h after CHX addition), and although ∆PB1 showed greater stability, turnover of both ∆PB1 and K7A were suppressed throughout the time course. Furthermore, the K7A mutation did not disrupt centriole targeting, nor did it prevent Plk4 activity because it induced centriole amplification when overexpressed (Fig. S3 A and B). These findings indicate that at least some of the Lys in PB1 are physiologically important targets of Plk4 ubiquitination.
PB3 Regulates Plk4 Kinase Activity.
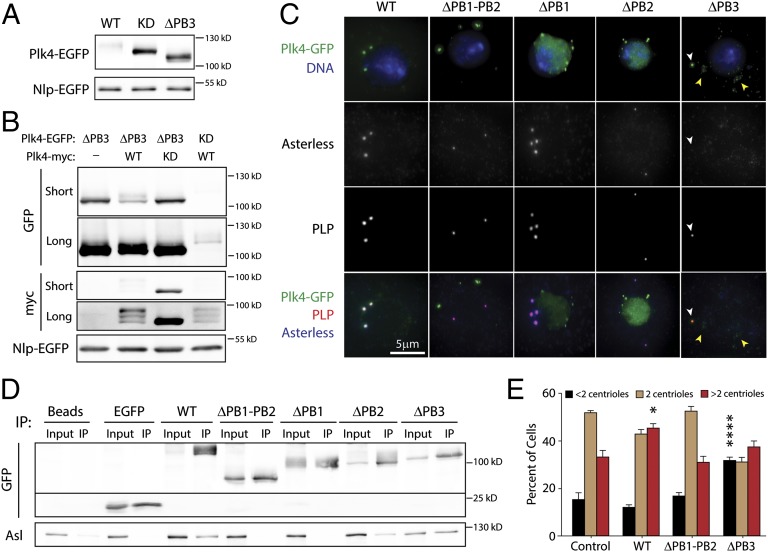
The function of PB3 is unknown. The dimerization-promoting and centriole-targeting functions of PB1–PB2 are not shared with PB3, because we show PB3 is not necessary for Plk4 dimerization and, previously, we found it targets centrioles only weakly when expressed as a GFP fusion (18). Therefore, we performed a functional analysis of PB3 by first expressing a PB3-deletion (ΔPB3) mutant of Plk4-EGFP and examining its protein level in S2 lysates. Surprisingly, deletion of PB3 dramatically stabilized the mutant protein compared with WT-Plk4 (Fig. 3A, lanes 1 and 3). Levels of Plk4–ΔPB3 were comparable to kinase-dead (KD)-Plk4, a mutant which is unable to autophosphorylate and avoids Slimb-mediated degradation (36, 39) (Fig. 3A, lane 2). Measurements of protein stability confirmed that Plk4–ΔPB3 turnover is greatly reduced compared with WT-Plk4 (Fig. S2 E and F). Last, when coexpressed with 3xFLAG-Ubi, immunoprecipitated Plk4–ΔPB3 displayed reduced ubiquitination and Slimb binding (Fig. 2C, lane 6). All these observations are consistent with the hypothesis that kinase activity is impaired in Plk4–ΔPB3, reducing its ability to trans-autophosphorylate and recruit Slimb and thereby increasing its stability.
Fig. 3.
PB3 is required for full kinase activity. (A) Anti-GFP immunoblots of transiently transfected S2 cell lysates demonstrate the similar stabilities of expressed KD and ∆PB3 constructs. (B) Lysates of S2 cells transiently coexpressing the indicated inducible EGFP- and myc-tagged Plk4 constructs were analyzed by immunoblotting to examine the relative protein stabilities. Anti-GFP and -myc immunoblots are shown at short and long exposures. (C) S2 cells coexpressing the indicated Plk4-EGFP construct (green) and the transfection marker Nlp-EGFP (green nuclei) were immunostained for PLP to mark centrioles (red) and Asterless (Asl; blue in bottom row). DNA is shown as blue in top row. Plk4–∆PB3 targets centrioles (white arrowhead) but also forms cytoplasmic punctate aggregates (yellow arrowheads). (D) Anti-GFP immunoprecipitates from lysates of S2 cells transiently expressing the indicated inducible Plk4-EGFP construct (or control GFP) were probed for GFP and endogenous Asl. (E) Transfected S2 cells were induced to express Plk4-EGFP constructs for 3 d, then were immunostained for PLP (a centriole marker), and their centrioles were counted. Nlp-EGFP–transfected cells were used as control. Each bar shows the average percent of cells containing the indicated number of centrioles (n = 300 cells were counted per treatment in each of three experiments). Asterisks mark significant differences (relative to control) for comparisons mentioned in the text. Error bars indicate SEM. Centriole amplification (an increase in the percentage of cells with more than two centrioles) occurs in cells expressing WT-Plk4 (P = 0.01). Even though Plk4–∆PB3 localizes to centrioles, it does not induce centriole amplification but instead significantly increases the percentage of cells with fewer than two centrioles (P < 0.0001).
To test the impact of PB3 on kinase activity further, we coexpressed Plk4–ΔPB3-EGFP with WT- or KD-Plk4-myc in S2 cells and examined the heterodimers for trans-autophosphorylation. As previously described, WT-Plk4-myc bound and trans-autophosphorylated KD-Plk4-EGFP, efficiently promoting degradation of this otherwise stable KD mutant (Fig. 3B, lane 4) (36, 39–41). Likewise, WT-Plk4 bound and converted Plk4–ΔPB3 into a more slowly migrating phosphorylated species and promoted its degradation, compared with cells expressing Plk4–ΔPB3 alone (Fig. 3B, lanes 1 and 2). In contrast, Plk4–ΔPB3 did not promote KD-Plk4 degradation (Fig. 3B, lane 3) even though they heterodimerized and coimmunoprecipitated (Fig. S3C), suggesting that Plk4–ΔPB3 lacks the ability to trans-autophosphorylate.
As an additional test of ΔPB3 mutant kinase activity, we examined its localization and ability to induce centriole amplification in S2 cells. Although WT-Plk4 colocalized with pericentrin-like protein (PLP)- and Asterless (Asl)-labeled centrioles, centriole targeting was disrupted by the deletion of PB1–PB2 or either PB1 or PB2 (Fig. 3C and Fig. S3D). This result is consistent with the critical role PB1–PB2 plays in binding Asl, which then targets Plk4 to centrioles (43–45). In contrast, Plk4–ΔPB3 colocalized with centrioles (Fig. 3C). Further, we note that Plk4–ΔPB3 expression phenocopied two characteristics of KD-Plk4 expression: First, Plk4–ΔPB3 formed numerous small aggregates in the cytoplasm, reminiscent of KD-Plk4 overexpression (Fig. 3C) (41). Second, Asl staining was diminished dramatically on PLP-labeled centrioles in these cells, similar to the effects observed with KD-Plk4 overexpression (Fig. 3C) (41).
Previously, we found that KD-Plk4 expression causes centriole loss by sequestering Asl and preventing its centriole targeting (41). Therefore, we tested the ability of Asl to coimmunoprecipitate with WT-Plk4 and PB-deletion mutants. Consistent with previous work (18), Asl associated with WT-Plk4 but not with ΔPB1–PB2 or ΔPB1 mutants (Fig. 3D). In contrast, deletion of PB3 had no effect on Asl binding (Fig. 3D), suggesting that, like KD-Plk4, Plk4–ΔPB3 may sequester Asl and prevent its centriole localization.
S2 cells can divide in the absence of centrioles, and if centriole duplication is inhibited, then the constant complement of centrioles becomes diluted among the exponentially increasing number of cells, thus increasing the percentage of acentriolar cells. Therefore we expressed Plk4-EGFP constructs in S2 cells for 3 d, then immunostained them for PLP (a centriole protein that coats the outer surface of mature centrioles) (46), and counted the centrioles per cell. Because of their small size, mother and daughter centrioles cannot be distinguished within an engaged pair using standard light microscopy in most fly cells. Nevertheless, the number of PLP spots is an accurate readout of centriole loss (fewer than two spots) and amplification (more than two spots) in these cells (18, 41, 44, 47). As expected, WT-Plk4 expression significantly increased the percentage of cells with centriole amplification (P = 0.01), whereas Plk4–ΔPB1–PB2 had no effect (Fig. 3E). In contrast, Plk4–ΔPB3 expression significantly increased the percentage of cells with fewer than two centrioles (Fig. 3E; P < 0.0001), similar to the effects of KD-Plk4 expression (41). All our functional assays lead to the conclusion that PB3 deletion severely impairs kinase activity, thus revealing a surprising new role for PB3 in stimulating kinase activity.
The Plk4 Phosphorylation Pattern.
The finding that PB3 deletion impairs kinase activity raises the possibility that Plk4 possesses a previously unidentified autoinhibitory mechanism that can be relieved by PB3. In the case of human Plk1, autoinhibition is relieved by phosphorylation of its kinase domain (16, 22, 23, 25–28). Because Plk4 extensively autophosphorylates its DRE to recruit Slimb (39–41), it is plausible that similar autophosphorylation of other domains, in conjunction with PB3, regulates a putative autoinhibitory mechanism. Therefore, it was necessary to characterize the Plk4 phosphorylation pattern better.
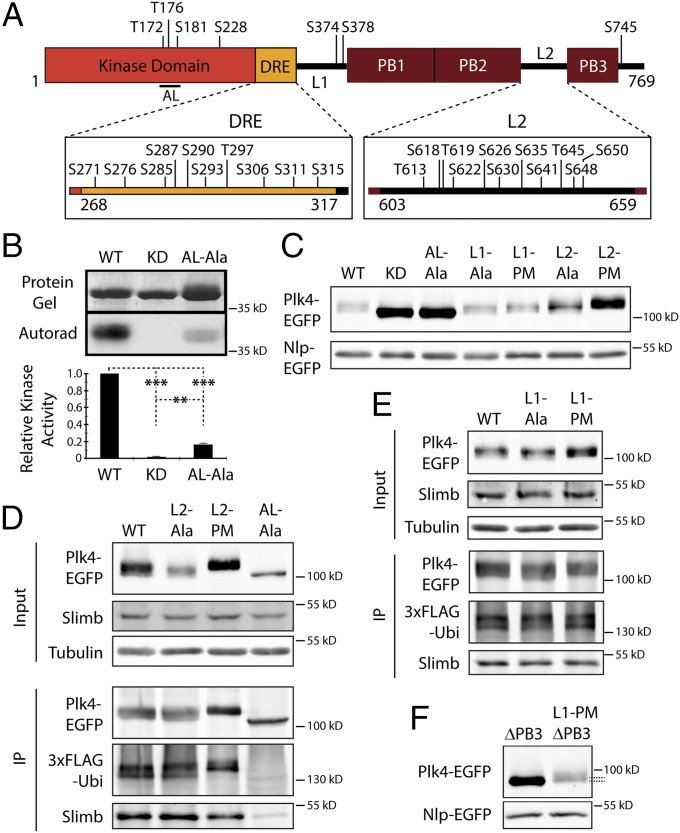
First, we isolated full-length Plk4-EGFP from S2 cells and mapped phosphorylated Ser/Thr residues using MS/MS. From 97% coverage, several clusters of phosphorylated residues were identified throughout the protein (Fig. 4A, Fig. S4 A and B, and Table S2). Many of these residues were confirmed as in vitro autophosphorylation targets by performing MS on samples of purified fly Plk4 (containing the kinase domain) incubated with MgATP and purified proteins containing different downstream regions of Plk4 (Fig. S4 A–E and Table S2). Notably, autophosphorylation sites were not detected within any PB region but instead were restricted primarily to linker and loop regions. Aside from the 10 previously described residues in the DRE (41), the phosphorylation sites include three residues in the AL of the kinase domain, one residue in the C-terminal lobe of the kinase domain, two residues near the end of L1, 11 residues in linker 2 (L2), and one residue in the C-terminal segment (Fig. 4A).
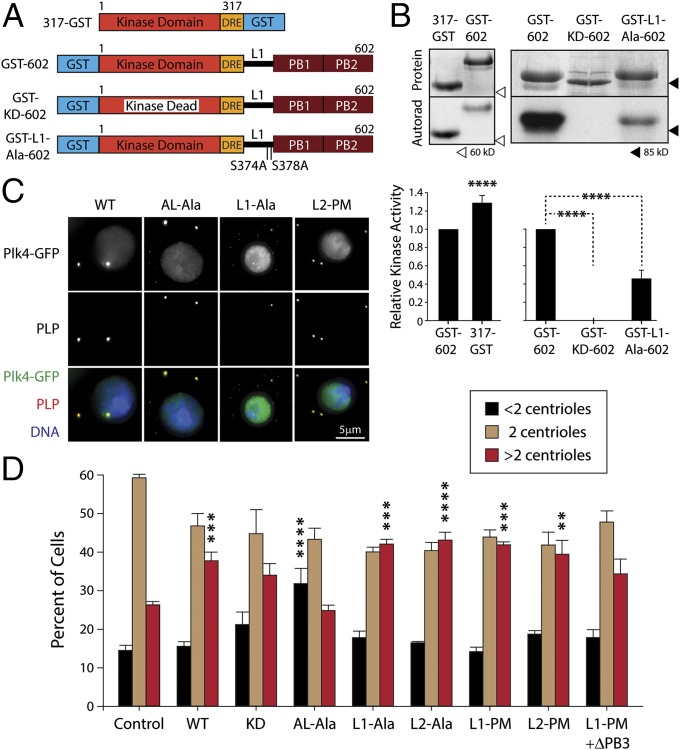
Fig. 4.
Plk4 displays autoinhibition which can be relieved by L1 autophosphorylation. (A) Linear map of Plk4 showing phosphorylated residues identified by MS/MS. Note the high concentration of phosphorylated residues within the AL of the kinase domain, the DRE, and L2. (B) In vitro autophosphorylation of purified His6-tagged Plk4 kinase domain + DRE (amino acids 1–317) is reduced significantly in the triple-Ala AL (AL-Ala) mutant, as is KD-Plk4. (Upper Top) Coomassie-stained SDS/PAGE gel. (Upper Bottom) Corresponding autoradiograph. (Lower) Quantitation of autophosphorylation activity (normalized by protein load and displayed relative to WT kinase activity). The average activity levels are significantly different (WT vs. KD, P < 0.0001; WT vs. AL-Ala, P < 0.0001; KD vs. AL-Ala, P < 0.0001). Error bars indicate SEM. (C) The indicated Plk4 constructs were analyzed by immunoblotting lysates of S2 cells coexpressing the indicated inducible Plk4-EGFP construct and Nlp-EGFP. (D and E) Anti-GFP IPs from lysates of S2 cells transiently coexpressing inducible 3xFLAG-Ubi and the indicated Plk4-EGFP construct. Blots were probed with anti-GFP, anti-FLAG, and anti-Slimb antibodies. (F) Immunoblots of lysates of S2 cells coexpressing the indicated inducible Plk4-EGFP construct and Nlp-EGFP. Note the mobility shift of Plk4–L1-PM–∆PB3 compared with Plk4–∆PB3 (dashed lines), indicating that the addition of L1-PM restored kinase activity to Plk4–∆PB3.
A Mechanism for Plk4 Autoinhibition.
Phosphorylation of the human Plk1 AL by Aurora A stimulates its activity and governs a critical step in Plk1 regulation (20, 22, 23, 27, 28). Our identification of autophosphorylated residues in the Plk4 AL suggested that Plk4 may be regulated in a similar manner and that autoinhibition might function to block autophosphorylation of this loop. We tested the importance of these residues by first mutating all three of the phosphorylated residues of the Plk4 AL to Ala (T172A/T176A/S181A) and then comparing the activity of the triple mutant (AL-Ala) with that of purified WT kinase domain protein in an in vitro assay. Although WT protein autophosphorylated robustly, the AL-Ala mutant protein was significantly less active (although not completely inactive like the KD mutant) (Fig. 4B). Thus, phosphorylation of the AL is critical for in vitro Plk4 kinase activity.
To test the effects of mutating the AL of Plk4 expressed in cells, we generated an inducible AL-Ala mutant within full-length Plk4-EGFP. Similar to KD-Plk4, the protein level of AL-Ala-Plk4 was stabilized dramatically compared with WT-Plk4 and migrated on SDS/PAGE mostly as a sharp, high-mobility band (as would be expected for a non- or low-phosphorylated protein) (Fig. 4C, lanes 1–3 and Fig. 4D, lane 4). Moreover, when coexpressed with 3xFLAG-Ubi, AL-Ala is clearly less ubiquitinated than and binds less Slimb than WT (Fig. 4D, lanes 1 and 4), further demonstrating its inability to autophosphorylate and recruit Slimb. AL-Ala-Plk4 localized to centrioles (Fig. 5C) and induced significant centriole loss (P < 0.0001) (Fig. 5D). Taken together, these results suggest that phosphorylation of the Plk4 AL is crucial for its kinase activity in cells, but some activity (approximately fivefold less) is present if the AL is not phosphorylated.
Fig. 5.
L1 autoinhibits Plk4 kinase activity in vitro, and expression of Plk4 phospho-mutants influences centriole numbers. (A and B) In vitro autophosphorylation of purified Plk4-deletion constructs reveals an autoinhibitory mechanism. (A) Schematic of Plk4 constructs used in this in this assay. All constructs were fused to GST to induce stable dimer formation. (B, Upper Top) Coomassie-stained SDS/PAGE gel; (Upper Bottom) corresponding autoradiograph. (Lower) Quantitation of autophosphorylation activity (normalized by protein load and graphed relative to the normalized activity of GST-602). Asterisks indicate significant differences from GST-602. Error bars indicate SEM. (C) S2 cells coexpressing the indicated Plk4-EGFP construct (green) and Nlp-EGFP (green nuclei) were immunostained for PLP (red) to mark centrioles. DNA is shown in blue. (D) Transfected S2 cells were induced to express Plk4-EGFP constructs for 3 d, then were immunostained for PLP, and their centrioles were counted. Nlp-EGFP–transfected cells were used as control. Average percentages of cells containing the indicated number of centrioles are shown (n = 300 cells in each of three experiments). Asterisks mark significant differences (relative to control) for comparisons mentioned in the text. Error bars indicate SEM.
Our findings reveal a potential autoinhibitory mechanism within Plk4. Previous studies have shown that a region of Plk1 autoinhibits the kinase by preventing phosphorylation of its AL. In this case, the interdomain linker (IDL), a region immediately downstream of the kinase domain, masks the activation loop from Aurora A phosphorylation (16, 27, 28). During mitotic entry, the protein Bora relieves this autoinhibition by binding Plk1 and exposing the AL (27, 28), allowing its phosphorylation and preventing further inhibition by the IDL. We hypothesized that a similar mechanism might operate within Plk4, specifically that L1 prevents phosphorylation of the AL and as a result reduces Plk4 activity significantly. Furthermore, based on our finding that PB3 stimulates Plk4 activity, we suggest that PB3 functions to relieve L1-mediated autoinhibition of the kinase domain, thereby allowing autophosphorylation of the AL. Finally, because L1 itself is phosphorylated (Fig. 4A), autophosphorylation of L1 may prevent further autoinhibition.
MS analysis of Plk4 identified two phosphorylated residues within L1 (S374/S378) (Fig. 4A and Fig. S4A). If L1 is responsible for autoinhibition, then constructs containing L1 (but lacking PB3) should display reduced catalytic activity. To test this hypothesis, we first generated a series of Plk4-deletion constructs lacking the C-terminal regions, L2 to PB3, and examined their activities by measuring their autophosphorylation in vitro (Fig. 5A). Purified Plk4 kinase domain (amino acids 1–317) can trans-autophosphorylate and was stimulated by fusing it to GST (317-GST) to induce dimerization artificially (Fig. 5B), as in human Plk4 (41). Strikingly, GST-602 autophosphorylates significantly less than 317-GST (Fig. 5B), supporting the hypothesis that the presence of L1 (without PB3) results in partial autoinhibition. [Interestingly, removal of GST from the 1–602 protein decreases kinase activity (Fig. S4F), demonstrating that the N-terminal fusion of GST partially activates this otherwise autoinhibited protein.]
If autophosphorylation of L1 blocks autoinhibition, then substituting L1 residues Ser374 and Ser378 with nonphosphorylatable Ala should impair kinase activity severely. Indeed, the double-Ala mutation S374A/S378A within L1 (GST-L1-Ala-602) (Fig. 5A) decreased catalytic activity further, but not as completely as observed with a KD protein (GST-KD-602) (Fig. 5B). PB1–PB2 is unlikely to contribute to autoinhibition in the GST-602 constructs because (i) PB1–PB2 does not bind the kinase domain when both are coexpressed in cells (Fig. S1B), and (ii) purified PB1–PB2 does not inhibit kinase activity when mixed with 1–317-His6 protein (18). These results suggest that L1 impairs autophosphorylation in vitro and that, when not phosphorylated, suppresses kinase activity.
To test the effects of mutating L1 on Plk4 activity in cells, we generated expression constructs of double-Ala and phosphomimetic (PM) (S374D/S378D) mutants within full-length Plk4-EGFP. Both L1-Ala and L1-PM mutants matched WT-Plk4 in all respects. Protein levels of both mutants were similar to those of WT-Plk4, associated with Slimb, and were ubiquitinated efficiently (Fig. 4 C and E). Likewise, L1-Ala localized to centrioles (Fig. 5C), and expression of either mutant amplified centrioles (Fig. 5D). Taken together, these findings suggest that mutations within L1 have no obvious effects on Plk4 regulation or activity within the context of full-length Plk4 where PB3 is present, unlike the truncated proteins we analyzed in vitro which lack PB3 to relieve autoinhibition.
Relief of Autoinhibition.
We propose that an important function of PB3 is to contribute to autoinhibition relief. So far, our results indicate that PB3 is required for full kinase activity and that nonphosphorylated L1 autoinhibits Plk4 (Fig. 5B). We hypothesize that PB3 specifically prevents L1-mediated inhibition, thus predicting that L1 is responsible for inhibiting kinase activity in the PB3-deletion mutant. To test this possibility, we generated a new inducible Plk4-EGFP construct containing the L1-PM mutation but lacking PB3 (L1-PM–ΔPB3). If our hypothesis is correct, then the inhibition caused by the ΔPB3 mutation should be rescued by the second L1-PM mutation. Strikingly, expression of the combined L1-PM and ΔPB3 mutant in cells resulted in a more slowly migrating (presumably more phosphorylated) species with a dramatically decreased protein level compared with ΔPB3 (Fig. 4F). Moreover, expression of L1-PM–ΔPB3 rescued the loss of centrioles observed in cells expressing ΔPB3 (Figs. 3E and 5D) and significantly decreased the percentage cells containing fewer than two centrioles compared with ΔPB3 (ΔPB3 vs. L1-PM–ΔPB3, fewer than two centrioles, P = 0.0008). Therefore, our data support the remarkable conclusion that Plk4 itself contains domains that are responsible for both the suppression (L1) and rescue (PB3) of its activity. Moreover, PB3 is not necessary for Plk4-induced centriole assembly because expressed L1-PM–ΔPB3 supports centriole duplication.
We attempted to reconstitute PB3-mediated relief of autoinhibition in vitro using bacterially expressed protein, but were unable to express long Plk4 constructs containing L2 and PB3. Using purified proteins, we found that PB3 mixed with GST-602 failed to rescue its full kinase activity in vitro. PB3-EGFP also failed to coimmunoprecipitate with either the kinase domain or a Plk4 fragment containing both the kinase domain and L1 (amino acids 1–381) (Figs. S1B and S4G). Taken together, these results suggest that PB3 does not bind L1 and release autoinhibition on its own but possibly binds and positions an unidentified protein to fulfill this function.
Plk4 Kinase Activity Induces Dimer Separation Mediated by L2 Autophosphorylation.
Plk4 has been described as a suicide kinase that is active as synthesized and then robustly autodestructs. Our results suggest that newly translated Plk4 is briefly inactivated by L1-mediated autoinhibition, thus forestalling its degradation. Subsequently, Plk4 homodimerizes and gains activity as autoinhibition is relieved. Possibly dimerization is a regulated event controlled by autoinhibition.
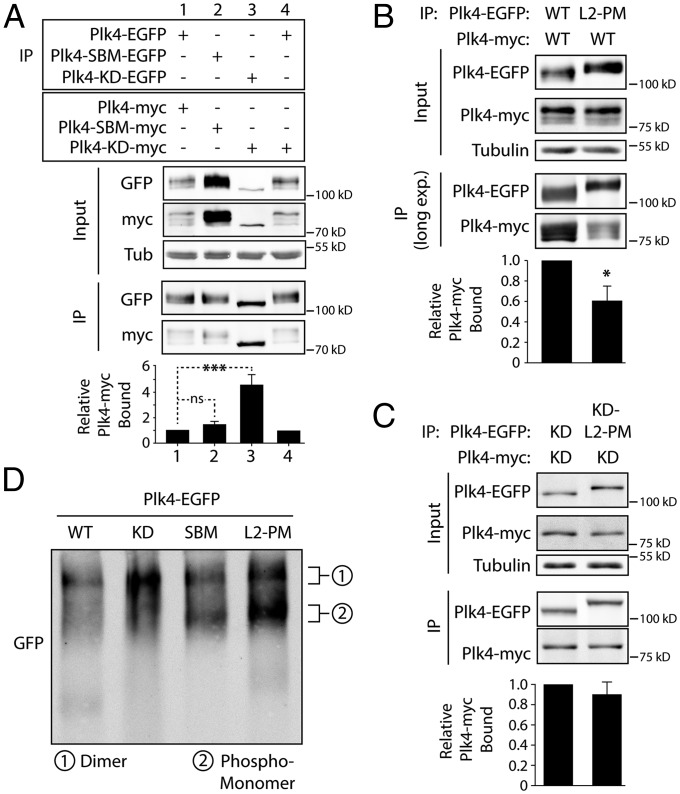
Therefore, we examined whether kinase activity influenced dimerization in cells coexpressing different EGFP- and myc-tagged Plk4 constructs. Following immunoprecipitation of EGFP-tagged constructs from cell lysates, quantitative immunoblots were used to measure the amounts of copurifying Plk4-myc. Although WT-myc binds WT-EGFP, a surprisingly small amount of WT-myc was recovered in the immunoprecipitate relative to WT-EGFP (Fig. 6A, lane 1). This difference was not caused by the rapid turnover of the proteins, because similar results were obtained using nondegradable SBMs in both proteins (Fig. 6A, lane 2). Identical results also were observed by first depleting Slimb using RNAi to stabilize the WT-Plk4 proteins (Fig. S4H). Strikingly, we observed a significant, approximately fourfold, increase in the amount of KD-Plk4-myc that associated with KD-Plk4-EGFP compared with the kinase-active forms used in this assay (Fig. 6A, lane 3). However, dimerization was decreased when one subunit in the heterodimer was kinase-active (Fig. 6A, lane 4); in this case, we have shown that WT-Plk4 efficiently trans-phosphorylates KD-Plk4 and promotes its degradation (41). These results support the surprising conclusion that Plk4 kinase activity promotes dimer separation and that one active kinase domain in a heterodimer is sufficient to induce separation.
Fig. 6.
Plk4 autophosphorylation of L2 promotes homodimer separation. (A–C) Anti-GFP immunoprecipitations were prepared from lysates of S2 cells transiently co-overexpressing the indicated inducible EGFP- and myc-tagged Plk4 constructs. (Upper) Blots of the input lysates and IPs were probed for α-tubulin, GFP, and myc. (Lower) Average amounts of Plk4-myc bound to Plk4-EGFP. For each treatment, a ratio of the measured amount of coprecipitated Plk4-myc to the measured amount of precipitated Plk4-EGFP was calculated. The graphed values are the calculated ratios relative to the Plk4-EGFP/Plk4-myc ratio in lane 1 of the respective blots. Error bars indicate SEM. ns, not significant. (A) Plk4 proteins lacking kinase activity (KD/KD, lane 3) are bound to a significantly greater extent than kinase-active proteins (WT/WT, lane 1) (P = 0.0002). (B) The presence of the L2-PM mutation significantly reduced the binding between Plk4 proteins with WT kinase domains (P = 0.03). (C) In contrast, the L2-PM mutation did not significantly alter binding between KD proteins. (D) Lysates of S2 cells expressing the indicated inducible Plk4-EGFP construct were resolved by native PAGE and then were immunoblotted with anti-GFP. Depending on the Plk4 transgene, up to two populations of Plk4 are apparent and are predicted to correspond to dimeric (1) and phospho-monomeric (2) Plk4 species.
L2 is a target of extensive autophosphorylation (Fig. 4A), and the function of these modifications is unknown. Because L2 is adjacent to the PB2 dimerization domain, we hypothesized that kinase activity promotes homodimer separation by autophosphorylating L2. To test the effects of L2 phospho-mutants on Plk4 activity and oligomerization in cells, we generated expression constructs harboring 11 Ala (L2-Ala) or PM (Asp/Glu) (L2-PM) substitution mutations within full-length Plk4-EGFP. Similar to WT, the L2-Ala protein was capable of autodestruction (Fig. 4C, lane 6), associated with Slimb, and incorporated 3xFLAG-Ubi (Fig. 4D, lane 2), indicating that L2-Ala is able to dimerize and trans-autophosphorylate. Moreover, L2-Ala localized to centrioles (Fig. S3A) and induced significant centriole amplification (more than two centrioles, P < 0.0001) (Fig. 5D), demonstrating normal functionality. Thus, L2-Ala mutations do not compromise Plk4 regulation or activity.
In contrast, L2-PM mutations dramatically increased Plk4 protein levels (Fig. 4C, lane 7), and measurements of protein turnover showed enhanced stability compared with WT-Plk4 (Fig. S2 E and F). Consistent with its increased stability, Slimb association with L2-PM was reduced, as was its ubiquitination, compared with WT-Plk4 (Fig. 4D). However, L2-PM localized to centrioles and amplified centrioles when overexpressed (Fig. 5 C and D), suggesting it retains normal activity.
One explanation for the increased stability of L2-PM is that this mutant does not form stable homodimers and therefore fails to generate its Slimb-binding phosphodegron efficiently and recruit normal amounts of Slimb. We examined whether L2-PM influenced dimerization by immunoprecipitating EGFP from lysates of cells coexpressing different EGFP- and myc-tagged Plk4 constructs and then used quantitative immunoblots to measure the amounts of bound Plk4-myc. Heterodimers of L2-PM and WT are less stable than WT/WT homodimers: L2-PM binding was significantly less than the WT control (Fig. 6B). (Long exposures of the immunoblots were necessary to visualize and measure WT-Plk4-myc in this assay.) In contrast, dimerization was unaffected when KD-L2-PM was coexpressed with KD-Plk4 (Fig. 6C). Taken together, these findings suggest that phosphorylation of L2 influences dimer stability. Because the L2-PM mutation fails to affect dimerization of two KD subunits, this mutation is unlikely to inhibit dimer formation, and autophosphorylation of additional residues (besides those mutated in L2-PM) probably is necessary for efficient dimer separation.
Thus far, our results suggest that Plk4 exists as three different oligomeric species in cells: (i) a newly translated monomeric and autoinhibited form, (ii) an active dimer, and (iii) a phosphorylated monomeric species (formerly dimeric). To identify these different populations, we examined WT and Plk4 mutants expressed in S2 cells using immunoblots of whole-cell lysates resolved by native PAGE. Our analysis revealed two different Plk4 species which, based on their mutation and relative size, predict a slowly migrating dimer form and a faster migrating phosphorylated monomeric species. WT and KD-Plk4 were present only in the larger dimeric species (Fig. 6D, lanes 1 and 2). In contrast, SBM-Plk4 was clearly present as two distinguishable populations, i.e., as dimer and phospho-monomer bands on the native gel (Fig. 6D, lane 3). In the case of SBM, this kinase-active but nondegradable protein would persist after dimer separation, allowing detection of the phospho-monomeric species. In contrast, KD-Plk4 accumulates as a dimer because separation is impaired, and, in the case of WT-Plk4, phospho-monomers are rapidly degraded. Similar to SBM, L2-PM also is detected in both dimer and monomer pools (Fig. 6D). This result is consistent with our finding that L2-PM is capable of forming dimers that likely are unstable. Thus, the presence of two mutant populations of Plk4 in cell lysates supports the hypothesis that Plk4 kinase activity promotes dimer separation and that phospho-monomeric Plk4 normally is degraded promptly. We predict that newly synthesized monomeric, autoinhibited Plk4 dimerizes rapidly and is too transient to detect.
Discussion
Multiple mechanisms constrain the activities of the Plk family to short durations within the cell cycle, and for good reason, because Plk overexpression is observed in a variety of cancers (5–9). Therefore, Plk members are attractive drug targets for developing anticancer therapies and are the focus of several small-molecule screens (48–50). Unlike monomeric Plk1 which autoinhibits and requires multiple external inputs for its activity, Plk4 was not known to autoinhibit but instead was thought to rely on degradation (stimulated by trans-autophosphorylation) as its sole means of regulation. Thus, inhibition of degradation activates Plk4 by allowing its protein levels to rise. During mitosis, protein phosphatase 2ATwins fulfills this role by counteracting Plk4 autophosphorylation of its Slimb-binding domain (47).
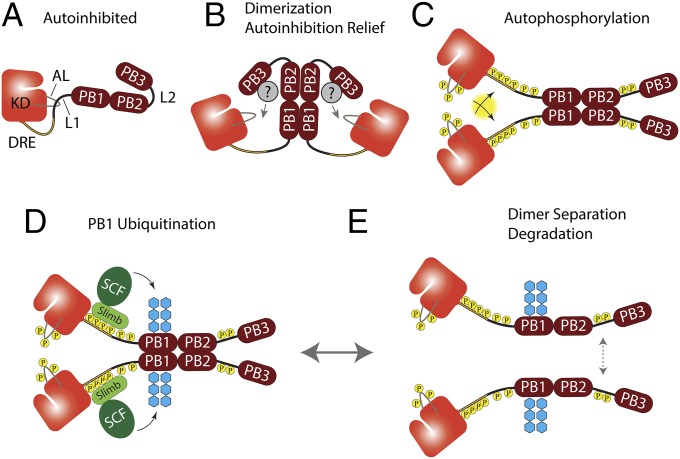
Our findings demonstrate that Plk4 does autoinhibit in a process that is as complex as that of Plk1, and suggest a pathway for its regulation and activation (Fig. 7). According to our model, newly synthesized Plk4 (like Plk1) is autoinhibited by L1 (Fig. 7A). Dimerization of Plk4 monomers is mediated by PB1–PB2, without the participation of PB3. However, we find that PB3 has an important regulatory role because it is needed to relieve autoinhibition of the Plk4 dimer (a process that may require the assistance of an unidentified binding partner) (Fig. 7B). Increased kinase activity results in autophosphorylation (Fig. 7C) of the ALs of the kinase domains (fully activating Plk4), of the DRE regions [generating the phosphodegron recognized by Slimb (Fig. 7D)] and of additional downstream elements which promote dimer separation (Fig. 7E). It is interesting and perhaps physiologically significant that, because of autoinhibition, the most probable initial act of the de-inhibited dimer is to transautophosphorylate its subunits. The phosphorylated product would be a fully active dimer, primed for SCFSlimb-mediated ubiquitination and prone to disassemble. On SDS/PAGE, Plk4 migrates as a series of phosphorylated polypeptides, appearing as a diffuse band or even a short ladder (e.g., Fig. 3B), potentially corresponding to several distinct phosphorylation steps. Although the order in which the multiple phosphorylations occur is unknown, we propose that autophosphorylation of the AL likely occurs first. Notably, a mutant Plk4 kinase containing Ala substitutions in its AL is not kinase dead but retains limited activity, which may be an important feature because our results suggest that Plk4 relies on autophosphorylation of these key residues for activation.
Fig. 7.
Speculative multistep model for Plk4 activation and regulation. (A) Newly synthesized Plk4 is initially autoinhibited, allowing homodimer formation via PB1–PB2 interaction. (B) PB3 relieves autoinhibition by positioning an unidentified binding partner to move L1 away from the AL. (C) Plk4 autophosphorylates several domains with assorted consequences; our data do not mandate an order of phosphorylation. (1) AL phosphorylation fully activates the kinase; (2) trans-autophosphorylation of the DRE generates the Slimb-binding phosphodegron; (3) L1 phosphorylation further relieves autoinhibition; (4) L2 phosphorylation promotes dimer separation. (D) SCFSlimb is recruited and ubiquitinates PB1 (blue hexagons). (E) Kinase activity promotes dimer separation through L2 phosphorylation and additional unidentified residues. We propose that dimer separation causes the disassembly of higher-order Plk4 complexes.
We find that the linker regions of Plk4 also are important for regulation. L1 is a cis-acting inhibitor of Plk4 kinase activity whose effect can be undone by autophosphorylation of residues within L1. Drawing on the seemingly analogous situation with human Plk1 (16), we propose that L1 of Plk4 interacts with the AL to inhibit the kinase, but future studies that resolve the atomic structure of the relevant regions of Plk4 are needed. On the other hand, L2 affects the stability of the Plk4 dimer. L2 is phosphorylated extensively, and mutation of L2 residues with PM substitutions decreases the binding of Plk4 monomers. Therefore, our results suggest that autophosphorylation of L2 (and probably additional sites) stimulates dimer separation, but the mechanistic explanation for this effect is unknown.
What is the function of Plk4-induced dimer separation? One possibility is that dimer separation causes disassembly of higher-order Plk4 complexes. During mitosis, Plk4 forms an aggregate on the surface of parent centrioles which is thought to serve as a platform for a newly emerging daughter centriole (11, 31, 33). Likely, Plk4 forms a higher-order structure within this aggregate which may require disassembly before degradation of the individual subunits. In this regard, induced dimer separation could regulate disassembly of such a structure.
A key feature of this pathway is that kinase activity facilitates homodimer separation. Without a mechanism of temporary autoinhibition, kinase activity could prevent dimer formation, which is required to generate the phosphodegron via trans-autophosphorylation within the homodimer. Consequently, Plk4 levels would accumulate and induce centriole amplification. Thus, our findings suggest that autoinhibition controls Plk4 oligomerization and degradation. Under physiological conditions, we propose that autoinhibition delays autodestruction, thus allowing the accumulation of a minor amount of Plk4 that is insufficient to initiate harmful centriole amplification but serves as a seed for a centriole-assembly complex when needed.
Although this study has focused on identifying the regulatory properties of different structural domains within Plk4, it is appropriate to acknowledge the importance of other centriolar proteins in licensing centriole duplication. Furthermore, the regulation and proper function of Plk4 undoubtedly requires other proteins. Notably, the N terminus of Asl binds PB1–PB2 (43–45), and this region in Cep152 (the human homolog of Asl) also is a Plk4 substrate (45). Although Asl targets Plk4 to centrioles (44), future studies of the Asl–Plk4 interaction will be necessary to determine if the phosphorylation state of Asl regulates Plk4 dimerization.
The Plk4 autophosphorylation pattern is complex and may dictate either down-regulation or activation, depending on which residues are modified. During mitosis, when the Plk4 protein level peaks, its kinase activity somehow stimulates centriole assembly. A major future challenge will be to understand how Plk4’s phosphorylation pattern and those of its centriolar targets are regulated spatially and temporally as cells navigate through mitosis, so that Plk4 can induce centriole duplication without being inactivated.
Materials and Methods
Cell Culture and dsRNAi.
Drosophila S2 cell culture, in vitro dsRNA synthesis, and RNAi treatments were performed as previously described (51). Briefly, cells were cultured in SF-900 II serum-free medium (Life Technologies). RNAi was performed in six-well plates. Cells (50–90% confluency) were treated with 5 μg of dsRNA in 1 mL of medium and were replenished with fresh medium/dsRNA every day for 4–7 d. Control dsRNA was synthesized from a control DNA template amplified from a non-GFP sequence of the pEGFP-N1 vector (Clontech) using the primers 5′-CGCTTTTCTGGATTCATCGAC and 5′-TGAGTAACCTGAGGCTATGG (all primers used for dsRNA synthesis begin with the T7 promoter sequence 5′-TAATACGACTCACTATAGGG). Slimb dsRNA was synthesized from cDNA using the primers 5′-GGCCGCCACATGCTGCG and 5′-CGGTCTTGTTCTCATTGGG.
Constructs and Transient Transfections.
Full-length cDNA of Drosophila Plk4 was subcloned into a pMT vector containing an in-frame coding sequence for EGFP or myc and the inducible metallothionein promoter. Phusion polymerase (Thermo Fisher) was used according to the manufacturer’s instructions to generate the series of Plk4 deletion and point mutants. Transient transfections of S2 cells were performed as described (52). Briefly, ∼2–5 × 106 cells were gently pelleted by centrifugation, resuspended in 100 µL of nucleofection solution (5 mM KCl, 15 mM MgCl2, 60 mM Na2PO4, 60 mM NaH2PO4, 50 mM d-mannitol, pH 7.2) containing 1–2 µg of purified plasmid, transferred to an electroporation cuvette (2-mm gap size), and then electroporated using a Nucleofector 2b (Lonza), program G-030. Transfected cells were diluted immediately with 1 mL of SF-900 II medium and plated in a six-well cell-culture plate. Typically, cells were allowed 24 h to recover before further manipulation. Expression of all Plk4 constructs (and GFP control) was induced by the addition of 50 μM–2 mM copper sulfate to the culture medium. Transfection frequencies range between 70–80% on average. CHX (Sigma) was used at final concentration of 100 μM. The image in Fig. 2B was generated with Chimera software (University of California, San Francisco) using coordinates from Protein Data Bank ID code 4G7N.
Immunofluorescence Microscopy.
For immunostaining, S2 cells were fixed and processed as previously described (51) by spreading S2 cells on Con A-coated, glass-bottomed dishes and fixing with 10% (wt/vol) formaldehyde. Primary antibodies were diluted to concentrations ranging from 1–20 μg/mL and included mouse anti-GFP (JL-8; Clontech), rabbit anti-PLP, and guinea pig anti-Asl. Secondary antibodies [conjugated with Cy2, Rhodamine red-X, or Cy5 (Jackson ImmunoResearch Laboratories)] were used at the manufacturer-recommended dilutions. Hoechst 33342 (Life Technologies) was used at a final dilution of 3.2 μM. Cells were mounted in PBS, 0.1 M n-propyl galate, 90% (vol/vol) glycerol. Specimens were imaged using a DeltaVision Core system (Applied Precision) equipped with an Olympus IX71 microscope, a 100× objective (NA 1.4), and a cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics). Images were acquired with softWoRx v1.2 software (Applied Science).
Immunoblotting.
S2 cell extracts were produced by lysing cells in cold PBS and 0.1% Triton X-100. Laemmli sample buffer was then added and boiled for 5 min. Samples of equal total protein were resolved by SDS/PAGE, blotted, probed with primary and secondary antibodies, and scanned on an Odyssey imager (Li-Cor Biosciences). Care was taken to avoid saturating the scans of blots. Transfected ninein-like protein (Nlp)-EGFP (a constitutively expressed nuclear protein) (11) was used as loading control and transfection marker. Antibodies used for Western blotting include anti-Slimb (47), anti-Asl (41), anti-GFP (JL-8; Clontech), anti-myc (9B11; Cell Signaling Technologies), anti–α-tubulin (DM1A; Sigma), and anti-FLAG (M2; Sigma) at 1:1,000 dilutions. IRDye 800CW secondary antibodies (Li-Cor Biosciences) were prepared according to the manufacturer’s instructions and were used at 1:1,500 dilutions.
Native PAGE.
S2 cells were transfected with plasmid encoding a Plk4-GFP construct and then were induced overnight to express protein. Cells were harvested and lysed in cold NativePAGE sample buffer (Invitrogen) without additional detergents using brief, gentle trituration and then were clarified by centrifugation (20,000 × g, 30 min, 4 °C). Lysates were resolved by normal native discontinuous PAGE (7.5% T, 2.7% C, Tris-buffered resolving gel), Tris/glycine running buffer, and using low current (5–10 mA) to minimize heating. Blots of gels were probed with anti-GFP to visualize Plk4-EGFP bands as described above.
GFP Immunoprecipitation Assays.
GFP-binding protein (GBP) (53) was fused to the Fc domain of human IgG, tagged with His6 in pET28a (EMD Biosciences), expressed in Escherichia coli, and purified on HisPur resin (Thermo Fisher) according to the manufacturer’s instructions (54). Purified GBP was bound to Protein A-coupled Sepharose, then was cross-linked to the resin by incubation with 20 mM dimethyl pimelimidate dihydrochloride in PBS, pH 8.3, for 2 h at 22 °C; the coupling reaction was quenched by incubation with 0.2 M ethanolamine, pH 8.3, for 1 h at 22 °C. Antibody-coated beads were washed three times with 1.5 mL of cell lysis buffer (CLB) [50 mM Tris (pH 7.2), 125 mM NaCl, 2 mM DTT, 0.1% Triton X-100, and 0.1 mM PMSF]. Transfected cells expressing recombinant proteins were lysed in CLB, and the lysates were clarified by centrifugation. GBP-coated beads were rocked with lysate for 1 h at 4 °C, washed two times with 1 mL CLB, and then boiled in Laemmli sample buffer. In vivo ubiquitination assays were performed by coexpressing Plk4-EGFP constructs with triple FLAG-tagged Drosophila ubiquitin (CG32744) (also under the metallothionein promoter and Cu-induced) (54) and then probing the immunoblot of the cell lysate with anti-FLAG antibody (Sigma).
In Vitro Autophosphorylation Assays.
Drosophila Plk4 kinase domain + DRE (amino acids 1–317) C-terminally tagged with FLAG-His6 (also called “317-His”) was cloned into the pET28a vector, expressed in BL21(DE3) bacteria, and purified on HisPur resin (Thermo Fisher) according to the manufacturer’s instructions. Purified Plk4 was autophosphorylated by incubation with 50 µM total ATP for 1–2 h at 25 °C in reaction buffer [40 mM Na Hepes (pH 7.3), 150 mM NaCl, 5 mM MgCl2, 0.5 mM MnCl2, 1 mM DTT, 10% (vol/vol) glycerol]. (To identify phosphorylated residues that are not generated by autophosphorylation, a sample of the same purified Plk4 was left untreated.) Samples were resolved on SDS/PAGE, Coomassie stained, and then processed for MS. To identify autophosphorylated regions downstream of the kinase domain and DRE, various Plk4 domains were cloned into pGEX-6p2, pGEX-JDK, or pET28a vectors, expressed in BL21(DE3) bacteria, purified, and mixed with 317-His + ATP. These domains include PB1–PB2-His6, PB3-His6, GST-L1–PB3 (amino acids 318–741), and GST-PB1–PB3 (amino acids 382–741). Single GST-tagged constructs that contained the kinase domain plus downstream regions (up to amino acid 602) were also examined for autophosphorylation (Fig. 5A).
MS.
MS/MS was performed at the National Heart, Lung, and Blood Institute Proteomics Core Facility (National Institutes of Health). Following resolution of protein samples by SDS/PAGE, selected Coomassie-stained bands were cut from the gel and were destained, reduced, alkylated, and trypsin digested; then the peptides were extracted. Peptide samples were loaded onto a Zorbax C18 trap column (Agilent Technologies) to desalt the peptide mixture using an on-line nano-LC ultra HPLC system (Eksigent). The peptides then were separated on a 10-cm PicoFrit BioBasic C18 analytical column (New Objective). Peptides were eluted over a 90-min linear gradient of 5–35% acetonitrile/water containing 0.1% formic acid at a flow rate of 250 nL/min, ionized by electrospray ionization in positive mode, and analyzed on a LTQ Orbitrap Velos (Thermo Electron) mass spectrometer. All LC MS analyses were carried out in the data-dependent mode in which the top six most intense precursor ions detected in the MS1 precursor scan (m/z 300–2,000) were selected for fragmentation via collision-induced dissociation. Precursor ions were measured in the Orbitrap at a resolution of 60,000 (m/z 400), and all fragment ions were measured in the ion trap.
LC MS/MS data acquired from tryptic digests were searched independently using the MASCOT algorithm. All data were searched against the Drosophila National Center for Biotechnology Information nonredundant protein database for peptide and protein identifications. Trypsin or chymotrypsin was specified as the digestion enzyme, allowing up to two missed cleavage sites. Carbamidomethylation (C) was set as a static modification, and Oxidation (M) and Phosphorylation (S, T, Y) were selected as variable modifications. Precursor and fragment ion mass tolerances were set to 20 ppm and ±0.8 Da, respectively. After MASCOT searches, database search results were combined to obtain a comprehensive map of all peptides identified from Plk4.
Statistical Analysis.
Means of measurements were analyzed for significant differences by two-tailed t test or one-way ANOVA (followed by Dunnett’s posttest to evaluate pairwise differences between treatments and control) using Prism 6 (GraphPad) software. (Tukey’s posttest was used to evaluate all possible pairwise comparisons in Fig. 4B.) Means are assumed to be significantly different if P < 0.05. P values shown for the Dunnett’s posttests are adjusted for multiplicity. In figures, “*” indicates 0.05 > P ≥ 0.01, “**” indicates 0.01 > P ≥ 0.001, “***” indicates 0.001 > P ≥ 0.0001, and “****” indicates 0.0001 > P for the indicated pairwise comparison. Error bars in all figures indicate SEM.
Supplementary Material
Acknowledgments
We thank P. Krieg and S. Rogers for comments on the manuscript, D. Morgan for use of his facilities, and S. Aguirre for technical assistance. G.C.R. was supported by National Cancer Institute (NCI) Grant P30 CA23074, NCI/NIH Gastrointestinal Specialized Programs of Research Excellence Grant P50 CA95060, National Science Foundation Grant 1158151, the Arizona Biomedical Research Commission Grant 1210, and the Phoenix Friends. N.M.R. is supported by the Division of Intramural Research at the NIH/National Heart, Lung, and Blood Institute Project 1ZIAHL006104.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417967112/-/DCSupplemental.
References
- 1.Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5(6):429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 2.Petronczki M, Lénárt P, Peters JM. Polo on the rise: From mitotic entry to cytokinesis with Plk1. Dev Cell. 2008;14(5):646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Archambault V, Glover DM. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10(4):265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 4.Lowery DM, Lim D, Yaffe MB. Structure and function of Polo-like kinases. Oncogene. 2005;24(2):248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- 5.Holtrich U, et al. Induction and down-regulation of PLK, a human serine/threonine kinase expressed in proliferating cells and tumors. Proc Natl Acad Sci USA. 1994;91(5):1736–1740. doi: 10.1073/pnas.91.5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi T, et al. Polo-like kinase 1 (PLK1) is overexpressed in primary colorectal cancers. Cancer Sci. 2003;94(2):148–152. doi: 10.1111/j.1349-7006.2003.tb01411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24(2):267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 8.Pellegrino R, et al. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51(3):857–868. doi: 10.1002/hep.23467. [DOI] [PubMed] [Google Scholar]
- 9.Marina M, Saavedra HI. Nek2 and Plk4: Prognostic markers, drivers of breast tumorigenesis and drug resistance. Front Biosci (Landmark Ed) 2014;19:352–365. doi: 10.2741/4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunha-Ferreira I, et al. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19(1):43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 11.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184(2):225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem Sci. 2012;37(12):534–542. doi: 10.1016/j.tibs.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Avidor-Reiss T, Gopalakrishnan J. Building a centriole. Curr Opin Cell Biol. 2013;25(1):72–77. doi: 10.1016/j.ceb.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivers DM, Moreno S, Abraham M, Ahringer J. PAR proteins direct asymmetry of the cell cycle regulators Polo-like kinase and Cdc25. J Cell Biol. 2008;180(5):877–885. doi: 10.1083/jcb.200710018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonner AM, et al. Binding of Drosophila Polo kinase to its regulator Matrimony is noncanonical and involves two separate functional domains. Proc Natl Acad Sci USA. 2013;110(13):E1222–E1231. doi: 10.1073/pnas.1301690110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Shen C, Wang T, Quan J. Structural basis for the inhibition of Polo-like kinase 1. Nat Struct Mol Biol. 2013;20(9):1047–1053. doi: 10.1038/nsmb.2623. [DOI] [PubMed] [Google Scholar]
- 17.Jana SC, Bazan JF, Bettencourt-Dias M. Polo boxes come out of the crypt: A new view of PLK function and evolution. Structure. 2012;20(11):1801–1804. doi: 10.1016/j.str.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Slevin LK, et al. The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure. 2012;20(11):1905–1917. doi: 10.1016/j.str.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 2003;22(21):5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang YJ, Lin CY, Ma S, Erikson RL. Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc Natl Acad Sci USA. 2002a;99(4):1984–1989. doi: 10.1073/pnas.042689299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mundt KE, Golsteyn RM, Lane HA, Nigg EA. On the regulation and function of human polo-like kinase 1 (PLK1): Effects of overexpression on cell cycle progression. Biochem Biophys Res Commun. 1997;239(2):377–385. doi: 10.1006/bbrc.1997.7378. [DOI] [PubMed] [Google Scholar]
- 22.Qian YW, Erikson E, Maller JL. Mitotic effects of a constitutively active mutant of the Xenopus polo-like kinase Plx1. Mol Cell Biol. 1999;19(12):8625–8632. doi: 10.1128/mcb.19.12.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem. 2002b;277(46):44115–44120. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- 24.Elia AE, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115(1):83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 25.van de Weerdt BC, et al. Uncoupling anaphase-promoting complex/cyclosome activity from spindle assembly checkpoint control by deregulating polo-like kinase 1. Mol Cell Biol. 2005;25(5):2031–2044. doi: 10.1128/MCB.25.5.2031-2044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson TM, Antrobus R, Johnson LN. Plk1 activation by Ste20-like kinase (Slk) phosphorylation and polo-box phosphopeptide binding assayed with the substrate translationally controlled tumor protein (TCTP) Biochemistry. 2008;47(12):3688–3696. doi: 10.1021/bi702134c. [DOI] [PubMed] [Google Scholar]
- 27.Macůrek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455(7209):119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 28.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320(5883):1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15(24):2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 30.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7(11):1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 31.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17(10):834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316(5827):1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 34.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63(6):1398–1404. [PubMed] [Google Scholar]
- 36.Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci. 2010;123(Pt 13):2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- 37.Leung GC, et al. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol. 2002;9(10):719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- 38.Korzeniewski N, et al. Cullin 1 functions as a centrosomal suppressor of centriole multiplication by regulating polo-like kinase 4 protein levels. Cancer Res. 2009;69(16):6668–6675. doi: 10.1158/0008-5472.CAN-09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188(2):191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunha-Ferreira I, et al. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr Biol. 2013;23(22):2245–2254. doi: 10.1016/j.cub.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 41.Klebba JE, et al. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr Biol. 2013;23(22):2255–2261. doi: 10.1016/j.cub.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sylvestersen KB, Young C, Nielsen ML. Advances in characterizing ubiquitylation sites by mass spectrometry. Curr Opin Chem Biol. 2013;17(1):49–58. doi: 10.1016/j.cbpa.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Cizmecioglu O, et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191(4):731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467(7316):714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 45.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191(4):721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mennella V, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14(11):1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brownlee CW, Klebba JE, Buster DW, Rogers GC. The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J Cell Biol. 2011;195(2):231–243. doi: 10.1083/jcb.201107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degenhardt Y, Lampkin T. Targeting Polo-like kinase in cancer therapy. Clin Cancer Res. 2010;16(2):384–389. doi: 10.1158/1078-0432.CCR-09-1380. [DOI] [PubMed] [Google Scholar]
- 49.Christoph DC, Schuler M. Polo-like kinase 1 inhibitors in mono- and combination therapies: A new strategy for treating malignancies. Expert Rev Anticancer Ther. 2011;11(7):1115–1130. doi: 10.1586/era.11.61. [DOI] [PubMed] [Google Scholar]
- 50.Marzo I, Naval J. Antimitotic drugs in cancer chemotherapy: Promises and pitfalls. Biochem Pharmacol. 2013;86(6):703–710. doi: 10.1016/j.bcp.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3(4):606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- 52.Nye J, Buster DW, Rogers GC. The use of cultured Drosophila cells for studying the microtubule cytoskeleton. Methods Mol Biol. 2014;1136:81–101. doi: 10.1007/978-1-4939-0329-0_6. [DOI] [PubMed] [Google Scholar]
- 53.Rothbauer U, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7(2):282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Buster DW, et al. SCFSlimb ubiquitin ligase suppresses condensin II-mediated nuclear reorganization by degrading Cap-H2. J Cell Biol. 2013;201(1):49–63. doi: 10.1083/jcb.201207183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.