Significance
The mammalian intestine provides a key interface with several essential environmental factors, including nutrients, toxins, resident microbiota, and pathogens. Consequently, the intestine undergoes major developmental transitions that correspond to dramatic changes in the environment: one at birth and the other at weaning. These transitions reflect both developmental and environmentally induced changes in intestinal gene expression. Here, we performed a systematic analysis of global gene expression that is associated with developmental timing versus the changes that are due to the innate immune signaling pathways mediated by toll-like receptor (TLR) and IL-1 receptor families. The results reveal distinct roles of these pathways in intestinal adaptation throughout postnatal development.
Keywords: intestine, innate immunity, microbiota, postnatal development, ontogeny
Abstract
Unlike mammalian embryogenesis, which takes place in the relatively predictable and stable environment of the uterus, postnatal development can be affected by a multitude of highly variable environmental factors, including diet, exposure to noxious substances, and microorganisms. Microbial colonization of the intestine is thought to play a particularly important role in postnatal development of the gastrointestinal, metabolic, and immune systems. Major changes in environmental exposure occur right after birth, upon weaning, and during pubertal maturation into adulthood. These transitions include dramatic changes in intestinal contents and require appropriate adaptations to meet changes in functional demands. Here, we attempt to both characterize and provide mechanistic insights into postnatal intestinal ontogeny. We investigated changes in global intestinal gene expression through postnatal developmental transitions. We report profound alterations in small and large intestinal transcriptional programs that accompany both weaning and puberty in WT mice. Using myeloid differentiation factor 88 (MyD88)/TIR-domain-containing adapter-inducing interferon-β (TRIF) double knockout littermates, we define the role of toll-like receptors (TLRs) and interleukin (IL)-1 receptor family member signaling in postnatal gene expression programs and select ontogeny-specific phenotypes, such as vascular and smooth muscle development and neonatal epithelial and mast cell homeostasis. Metaanalysis of the effect of the microbiota on intestinal gene expression allowed for mechanistic classification of developmentally regulated genes by TLR/IL-1R (TIR) signaling and/or indigenous microbes. We find that practically every aspect of intestinal physiology is affected by postnatal transitions. Developmental timing, microbial colonization, and TIR signaling seem to play distinct and specific roles in regulation of gene-expression programs throughout postnatal development.
Metazoan development is a product of gene–environmental interactions. However, some characteristics (for example, basic body plan) are hard-wired and relatively insulated from the environment whereas others (for example, body weight) have broad reaction norms indicating a strong environmental impact (1, 2). Nutrient and water availability, ambient temperature, and humidity have long been recognized as essential environmental factors that impact many aspects of animal physiology and development. The mammalian intestine is a particularly interesting example of environmental impact on developmental programs and physiological functions: similar to the skin and the upper respiratory tract, the intestine is directly exposed to the environment, including food and nutrients, microorganisms, and a variety of bioactive molecules delivered with breast milk (3, 4). Consequently, intestinal development and physiology are highly adaptable to environmental factors, including nutrients and microbial colonization (5, 6). In addition, the gastrointestinal tract faces two major transitions in environmental exposure. First, immediately after birth, the intestine is exposed to mother’s milk and undergoes initial colonization with microorganisms. Second, after weaning, the intestinal tract becomes exposed to solid foods and is no longer exposed to mother’s milk components, the host immune system matures, and the microbiota shifts.
Although it is widely recognized that these transitions have a major impact on intestinal development and function, the full spectrum of these effects remains to be characterized. Such an understanding will require systematic analyses of developmental, physiological, and microbiological changes that accompany each postnatal phase and transition. These analyses are complicated by the fact that both microbial colonization and abiotic (including nutrition) and hormonal (such as neonatal and pubertal variation in steroids) changes accompany postnatal intestinal development. In addition, the impact of the microbiota can be dependent on the immune recognition of microorganisms, as well as on their metabolic and other activities.
To gain insight into postnatal intestinal development, here, we analyzed global changes in intestinal gene expression that accompany intestinal colonization by microbiota and the transition from milk suckling to solid foods that occurs during weaning. To better evaluate the changes that occur due to postweaning colonization and changes caused by developmental timing, we included the analyses of adult mice after the pubertal transition. To distinguish between changes caused by innate immune recognition by toll-like receptors (TLRs) and the IL-1 receptor family from changes caused by other factors, we compared gene expression in the small intestine and colon between WT and MyD88/TRIF double knockout (DKO) littermates at each postnatal stage. In addition, to correlate these changes in intestinal gene expression, we performed 16S ribosomal RNA gene sequencing of the bacterial microbiota by function of genotype and postnatal stage. Our results provide an in-depth analysis of the transcriptional regulation of the diverse biological processes that occur during postnatal intestinal development and dissect the contribution of innate immune signaling and the microbiota to ontogeny.
Results
Experimental Design and Data Analysis.
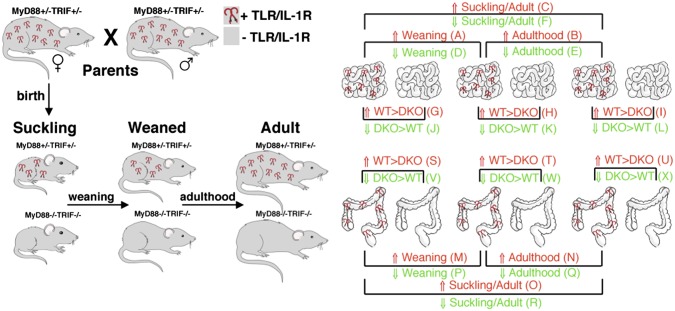
The goal of this study was to characterize postnatal intestinal development and the contribution of TLR/IL-1R (TIR) signaling while minimizing the contribution of multiple confounding factors (7, 8). For this purpose, we studied intestinal gene expression changes and microbiota composition in littermate mice that were either TIR signaling-sufficient (WT; MyD88+/− TRIF+/−) or -deficient (DKO; MyD88−/− TRIF−/−) (Fig. 1). Use of littermates controlled for potential contributions of maternal factors (nursing, microbiota), microbial exposure, and other environmental factors. Mothers were TIR signaling-sufficient (MyD88+/−TRIF+/−) to ensure gestation and nursing via a normative mother.
Fig. 1.
Experimental design and primary datasets. MyD88+/− TRIF+/− mice were bred to create WT and MyD88−/− TRIF−/− littermates. Whole genome intestinal gene expression was measured in the small and large intestine in suckling, weaned, and adult mice. Pairwise comparisons of gene expression over developmental stage or genotype (1° datasets) are listed in Dataset S1, Tables A–X.
The study was performed over a 2-y period, using over 30 litters. Both colon and small intestine (ileum) were harvested at the same time of day (midafternoon) and used for gene-expression analyses. The developmental time points used for analyses were as follows: postnatal day 16 (P16), postweaning day 26 (P26), and adult day 42 (P42). Weaning occurred on day 21. Additional ages were used for analysis of the microbiota by 16S rRNA gene analysis. There were no differences in mortality, life span, body weight, spleen size, small intestine, or colon length between WT and DKO mice across all time points.
RNA was isolated, and sex-matched samples were pooled from multiple litters with equal proportions of littermates in pooled samples. Illumina MouseWG-6 v2.0 Expression BeadChips for whole-genome expression profiling (covers >19,100 unique, curated genes in the National Center for Biotechnology Information RefSeq database) were used for whole-genome microarray expression profiling in duplicate using RNA samples pooled from different mice from different litters. Pairwise ratios of gene expression between samples by developmental stage, genotype, and organ were performed to create primary (1°) datasets (Fig. 1 and SI Appendix, Fig. S1). Probes included in 1° sets had to fit three inclusion criteria: (i) ≥1.5-fold difference between conditions, (ii) considered as an “expressed” gene product based on probe P value in both microarray analyses, and (iii) criteria i and ii in the duplicate microarrays (Fig. 1 and Dataset S1). For a complete description of the datasets and analytical methods, please see SI Appendix, SI Materials and Methods.
Major Trends and Patterns.
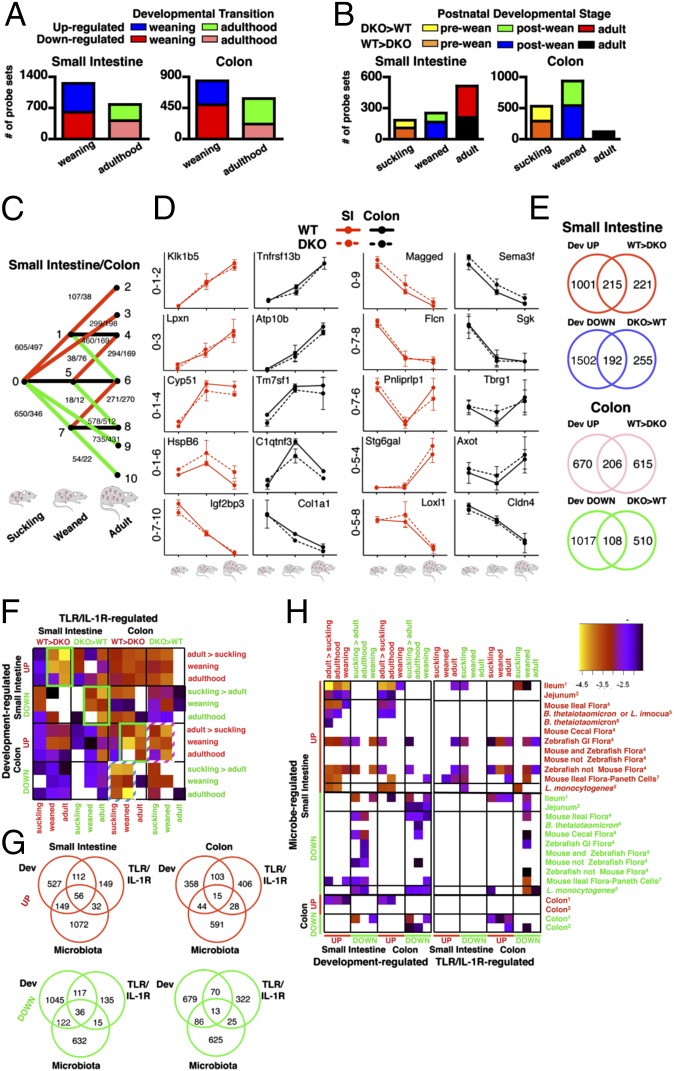
We first compared the number of probes regulated with normative (in TIR-sufficient mice) developmental transition in the small intestine and colon. In both organs, weaning more profoundly affected gene expression than did transition to adulthood although the latter also impacted transcriptional activity (Fig. 2A). Developmental changes in transcription were more abundant in the small intestine compared with the colon (Fig. 2A). Next, we determined the impact of MyD88/TRIF signaling on transcriptional regulation per postnatal developmental stage (Fig. 2B). In the small intestine, the number of MyD88/TRIF-regulated genes (WT > DKO and DKO > WT) increased at each postnatal stage, with the absence of TIR signaling most profoundly affecting gene expression in the adult compared with the preweaned (suckling) or postweaned (weanling) stages (Fig. 2B). In contrast, in the colon, TIR signaling most profoundly affected gene expression in suckling and weanling mice (Fig. 2B). These data reveal the differential roles of TIR signaling in the small intestine versus colon through the postnatal developmental stage. TIR signaling in the colon is more profound in neonatal states as opposed to in the small intestine where it affects gene regulation most at the adult states and suggests differential activation and/or permissiveness/dampening of TIR signaling (for example, by IL-10) (9) according to organ and developmental stage.
Fig. 2.
Trends in the developmental regulation of intestinal gene regulation. Number of probes up- and down-regulated by weaning and adulthood developmental transition (A) and by TLR/IL-R (TIR) signaling at each postnatal stage (B). (C) Number of probes demonstrating each of 10 different patterns of postnatal developmental kinetics, with specific examples in both the small intestine and colon (D). (E) Venn diagram of the number of probes present in secondary datasets for classification of genes with coordinate regulation by postnatal development and TLR/IL-R (TIR) signaling, with the inclusion of microbe-regulated genes compiled by metaanalysis (G) (SI Appendix, Fig. S1). (F) Heatmap of enrichment between developmentally and TLR/IL-R (TIR)-regulated 1° datasets, with the inclusion of microbe-regulated genes compiled by metaanalysis (H). Rows ending with numbers in superscript refer to microbe-regulated datasets from the study of the corresponding Supplemental Reference number. All heatmap colors are statistically significant enrichment by hypergeometric test (negative log P value < −1.5). Green and striped boxes are discussed in section Role of TIR Signaling on Postnatal Developmental Phenotypes. Error bars, SEM.
To determine the dynamics of transcription with developmental transitions, probes were classified into 10 subsets according to the patterns of expression (for example, probes induced with weaning were stratified into those that remained stable, or were induced or repressed with adulthood) (Fig. 2C), with representative examples (Fig. 2D). In the small intestine, the level of expression of 76.0% of weaning-inducible probes remained stable after the adulthood transitions, 17.7% were further induced, and 6.3% decreased in expression with adulthood (Fig. 2C). Of the weaning-repressible genes, 88.9% remained stably expressed through adulthood, 8.3% were further repressed, and 2.7% were up-regulated with adulthood (Fig. 2C). Similar trends were noted at the colon (Fig. 2C). This analysis demonstrates that, in both the small intestine and colon, the majority of genes undergo transcriptional stabilization after weaning.
We next sought to analyze the contribution of MyD88/TRIF signaling to postnatal developmentally regulated genes by creating secondary (2°) datasets of the 1° dataset (developmental- or TIR-regulated) (Fig. 1; SI Appendix, Fig. S1; and Dataset S1) intersections based on coordinate regulation (i.e., genes up-regulated with developmental transition and greater expression in WT compared with DKO mice and genes down-regulated with developmental transition and more highly expressed in DKO compared with WT mice (Fig. 2E and SI Appendix, Fig. S1 A and B) and statistical enrichment between 1° datasets (Fig. 2F). In the small intestine and colon, 17.7% and 23.5% of probes induced and 11.3% and 9.6% repressed, respectively, during development showed coordinate regulation by MyD88/TRIF (Fig. 2E). Of all probes demonstrated to be greater expressed in TIR-sufficient compared with TIR-deficient mice (WT > DKO), nearly half (49.3%) (in the small intestine) and one quarter (25.1%) (in the colon) were coordinately developmentally regulated (Fig. 2E). A similar role of TIR signaling (DKO > WT) in the repression of genes down-regulated during development were observed (Fig. 2E). These data demonstrate that, in both the small intestine and colon, a fair proportion (17.7% and 23.5%, respectively) of genes induced during development show coordinate regulation by TIR. This proportion of genes also applied to developmentally repressed probes, although to a lesser extent (11.3% and 9.6% in the small intestine and colon, respectively). Analysis for statistical enrichment among the developmental- and TIR-regulated 1° datasets revealed the highest enrichment between developmentally induced (UP) and TIR-inducible (WT > DKO) probes and developmentally repressed (DOWN) and TIR-repressed (DKO > WT) probes for the respective organ (Fig. 2F; green boxes).
This analysis revealed two significant enrichments among developmental and TIR 1° datasets (Fig. 2F; striped boxes) that did not reflect coordinate regulation. These enrichments were for genes repressed with development (DOWN) in the colon and with greater expression in WT compared with DKO mice (WT > DKO) (Fig. 2F; blue striped box) and genes induced with development (UP) in the colon and expressed at higher level in colons in the absence of TIR signaling (DKO > WT) (Fig. 2F; pink striped box). To gain biological insight, genes within these enrichments were subjected to bioinformatic analysis (see Role of TIR Signaling on Postnatal Developmental Phenotypes and see Fig. 4 C and D).
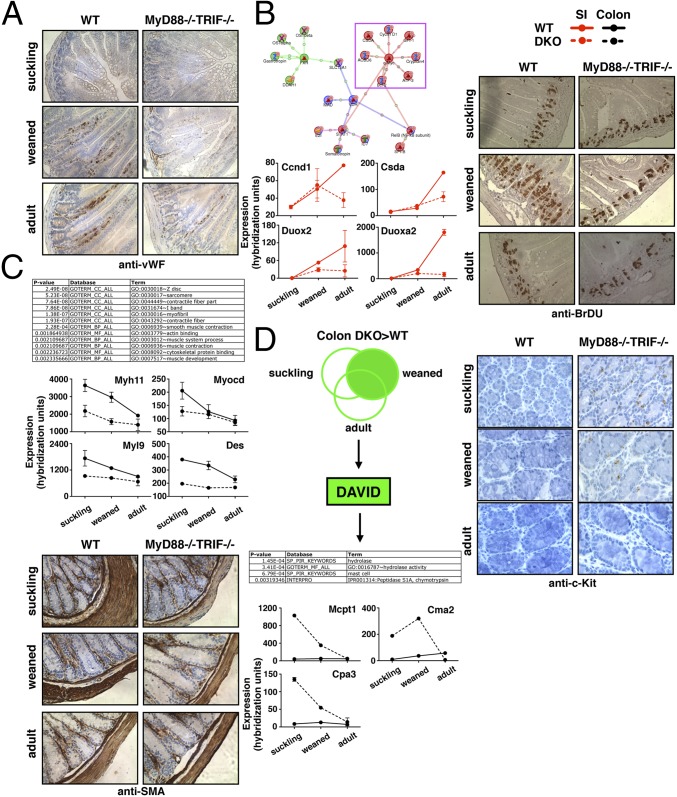
Fig. 4.
Role of TLR/IL-R signaling on postnatal developmental phenotypes. (A) Immunohistochemistry with αWF Ab to identify vascular endothelium in the small intestine. (B) GenoGo MetaRodent TF subnetwork enrichment for c-Myc among regulated developmentally induced and TIR coordinately regulated genes in the small intestine (Upper Left). Expression of selected genes in WT and DKO small intestines over postnatal developmental stage (Lower Left). Immunohistochemistry with αBrDU Ab at the small intestine of mice treated with BrDU pulse (Right). (C) DAVID bioinformatic analysis of developmentally repressed genes with increased expression in WT compared with DKO colons (Fig. 2F, blue striped box) (Top). Expression of selected genes in WT and DKO in the colon over postnatal developmental stage (Middle). Immunohistochemistry with αSMA Ab to identify smooth muscle cells present in the colon (Bottom). (D) DAVID bioinformatic analysis of genes overexpressed in DKO colons compared with WT at weaning (overlap with Fig. 2F, pink striped box) (Upper Left). Expression of selected genes in WT and DKO colons over postnatal development (Lower Left). Immunohistochemistry with αcKit Ab to identify mast cells present in the colon (Right). (Magnification: A–C, 200×; D, 400×.) Counterstains are hematoxylin. Error bars, SEM.
The microbiota has been shown to play a critical role in the normative development of the intestine (6, 10). In particular, studies of germ-free versus conventionally raised or germ-free mice either monoassociated with a single member of the microbiota or conventionalized with a consortium have demonstrated a profound effect of the microbiota on intestinal transcription in mice (see references in SI Appendix). In order to identify developmentally and/or TIR-regulated genes modifiable by the microbiota, we performed a cross-platform metaanalysis of studies identifying microbiota-regulated genes and referenced these genes to our 1° and 2° datasets (creating 3° datasets) (SI Appendix, Fig. S1 and Dataset S1, tabs AC–AF; see SI Appendix, SI Materials and Methods for details). In the small intestine, we identified 205 and 158 genes induced and repressed with postnatal development, respectively, coordinately regulated by the microbiota (Fig. 2F). This set of genes represented nearly one-quarter (24.3%) of developmentally induced genes and one-eighth (12.0%) of developmentally repressed genes. Of these developmentally- and microbiota-regulated genes, 56 (27.3%) and 36 (22.8%), respectively for those induced and repressed, showed coordinate dependence on TIR signaling. Together, these data demonstrate the extent that developmentally regulated and TIR-regulated genes are modified by the microbiota during intestinal ontogeny.
Hypergeometric-distribution analysis was performed to determine the enrichment of genes regulated by the microbiota (based on our metaanalysis) among both developmentally regulated and TIR-regulated genes. In general, we observed that there is good coordinate correlation of enrichment of developmentally regulated genes and microbiota-regulated genes (for example, developmentally induced genes are enriched among genes induced by the microbiota and developmentally repressed genes are enriched among genes repressed by the microbiota and for the most part, not vice versa) (Fig. 2H). The datasets produced by Rawls et al. (11) allowed for analysis of enrichment of genes regulated by the mouse versus zebrafish microbiota and those regulated developmentally and by TIR signaling at different postnatal stages. Interestingly, we found an enrichment of genes requiring TIR signaling for postnatal induction and those induced by the zebrafish, but not mouse, microbiota upon colonization of the germ-free mouse (Fig. 2H). Conversely, we found that genes repressed with development are enriched among those that are repressed by the mouse but not zebrafish microbiota (Fig. 2H). Although there are many interpretations of these correlations, they seem to reflect host-specific effects of microbiota (11, 12). An additional possibility is a differential abundance of microorganisms in different host species, which can in turn affect intestinal gene expression. Notably, in this regard, a major difference between the mouse and zebrafish is the relative scarcity of γ-proteobacteria in the mouse steady-state adult microbiota compared with that of the zebrafish (11).
Bioinformatic Analysis.
Having established the 1° and 2° sets of genes regulated during postnatal development and/or by MyD88/TRIF signaling, we performed a series of bioinformatic analyses to gain biological insights into small and large intestine postnatal ontogeny and mechanism of regulation (TIR signaling) (SI Appendix, Fig. S2). The Mouse Genome Informatics Mammalian Phenotype database compiles the phenotypes (including lethality) of gene deletion according to the pre- or postnatal age of phenotype penetrance (13). This analysis revealed that genes induced during weaning are significantly enriched for those important for survival during the peri- and postnatal time period (SI Appendix, Fig. S2). These data indicate a specialized function of these genes during the peri/postnatal stage and the critical role that developmental stage-specific expression of these genes plays in host survival. These genes may be particularly important for adaptation to the transition from intrauterine to extrauterine environments. The timing of expression of these genes may be an important factor in the pathogenesis of conditions associated with preterm delivery, including necrotizing enterocolitis (NEC) (14).
Having established profound transcriptional regulation during postnatal developmental transition and by TIR signaling, we parsed our datasets through GeneGo MetaRodent (MR) (portal.genego.com/) and the RIKEN database of transcription factors (TFs) (genome.gsc.riken.jp/TFdb/tf_list.html) to gain insight into the transcriptional regulation of TF themselves (RIKEN) and subnetworks centered on TF (transcriptional targets of TF; MR) by developmental transition, TIR, and the microbiota (SI Appendix, Fig. S3 and Dataset S2). We were able to determine developmental- and/or organ-specific (small intestine vs. colon) transcription factor subnetworks (SI Appendix, Fig. S3). For example, subnetworks surrounding members of the hepatocyte nuclear factor (HNF) family of transcription factors showed specific enrichment during different developmental transitions according to organ. HNF4α was found to be a subnetwork associated specifically with weaning (both induction and repression) in the small intestine and not with any other developmental transition in either the small intestine or colon (SI Appendix, Fig. S3), corroborating the role of HNF4 in small intestinal epithelial differentiation and biology (15). In contrast, HNF3α (FoxA1) was identified as a subnetwork specifically associated with weaning-repressed genes in both the small intestine and colon, and HNF3β (FoxA2) with developmental repression in the small intestine throughout weaning and adulthood transitions (SI Appendix, Fig. S3).
This analysis also enabled us to identify transcription-factor subnetworks enriched for developmentally regulated and TIR-regulated genes (SI Appendix, Fig. S3). For example, in the small intestine, c-myc, FXR, RelA, NF-κB, and VDR subnetworks were enriched among developmentally induced genes regulated by TIR whereas SRF, GATA-6, HNF3β, HIF1α, RARγ, and FKHR (FOXO1) subnetworks were enriched among developmentally repressed, TIR-repressed genes. We also identified scenarios in which the TF itself and its targets (subnetwork) were developmentally regulated, such as induction or repression of PPARγ and ATF3 and target subnetworks, respectively, during weaning and adulthood transition in the small intestine (SI Appendix, Figs. S2 and S3; and Dataset S2), suggesting that these TFs act as master regulators of postnatal intestinal developmental programs.
Datasets were parsed through Gene Ontology and Ingenuity Pathways Analysis (IPA) to identify biological processes regulated by postnatal developmental transition and/or TIR signaling. Genes involved in host defense and the immune system [such as antigen presentation, T-cell receptor (TCR) and B-cell receptor (BCR), glucocorticoid receptor, acute phase response, chemokine, IFN, and eicosanoid signaling pathways] were found to be induced with postnatal development, with certain processes showing greater enrichment during weaning versus adulthood (such as acute phase response and glucocorticoid, respectively) (SI Appendix, Figs. S2 and S4). A notable exception to this pattern was the postnatal regulation of genes involved in the coagulation system, which were repressed with weaning in both the small intestine and colon (SI Appendix, Fig. S4). The weaning transition at the small intestine showed the greatest degree of metabolic diversity (SI Appendix, Figs. S2 and S4). This transition was associated with a switch in regulation of genes encoding enzymes responsible for disaccharide utilization with repression of genes involved in starch, sucrose, and galactose metabolism and glycolysis/gluconeogenesis and the induction of those for fructose and mannose metabolism. There was developmental repression of genes encoding enzymes for the degradation of valine, leucine, isoleucine, and lysine and the metabolism of tryptophan, arginine, and proline, and developmental induction of those for phenylalanine, histidine, and glutamate metabolism. TIR signaling was identified as important for the coordinate developmental regulation of genes enriched in the IFN response, nuclear receptor and antigen presentation signaling, and xenobiotic, starch and sucrose, galactose, fatty acid, valine, leucine and isoleucine, and arachidonic acid metabolism (SI Appendix, Figs. S2 and S4).
Biological Process.
Using both bioinformatics approaches and manual curation of our datasets, we sought to provide deeper insight into the transcriptional regulation of postnatal intestinal ontogeny by analysis of two major biological processes affected by development: nutrition and metabolism and host defense and barrier function.
Nutrition and metabolism.
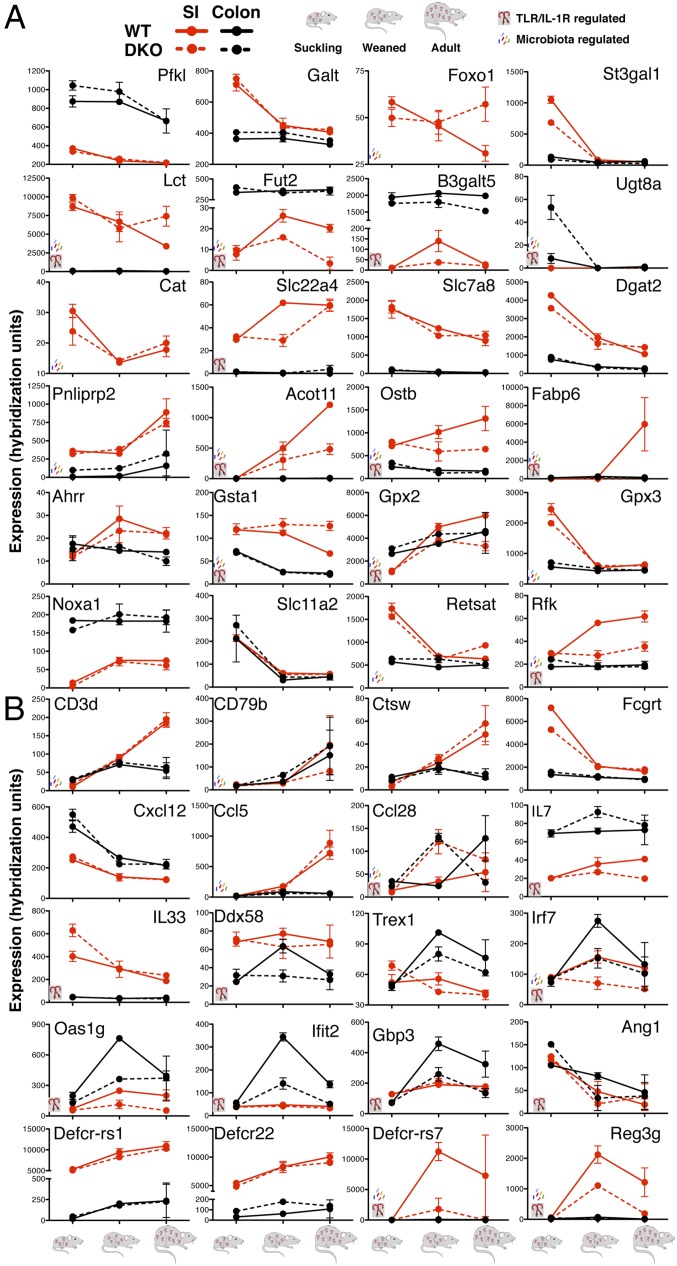
Our analysis allowed for the characterization of the transcriptional changes involved in carbohydrate, lipid, and amino acid metabolism during this transition, in addition to providing mechanistic insights of regulation by TIR signaling and the microbiota. Weaning was associated with a repression of genes involved in pyruvate metabolism and glycolysis/gluconeogenesis (Pklr, Pkm2, Pfkm, Pckl, and Fbp1 and -3) in the small intestine, which occurred independently of TIR signaling (Fig. 3A and Dataset S1). Interestingly, however, we identified a series of genes in this process (Foxo1/Fkhr, Pdh, and G6PC) as requiring TIR signaling for full postnatal repression (Fig. 3A and Dataset S1). Weaning was also associated with small intestinal switch genes encoding factors responsible for disaccharide utilization (SI Appendix, Figs. S2 and S4) and with the repression of genes involved in galactose (Galt, Pfkl, and Gaa) and starch/sucrose (Slc37a4) and the induction of genes involved in fructose/mannose utilization (Nudt5, Hk2, Gmds, Mpi, Sord, Glut5/Slc2a5) and sodium/glucose cotransport (SGLT3a and -b, Slc5a4a, and Slc5a4b) all occurring independently of TIR signaling and the microbiota (Fig. 3A and Dataset S1). Genes requiring intact TIR signaling for postnatal repression were enriched for starch and sucrose metabolism (SI Appendix, Fig. S4). Among these genes, we found Enpp2/autotaxin, Glut2/Slc2a2, and lactase (Lct), the latter a glycoside hydrolase of the β-galactosidase family of enzymes involved in the hydrolysis of the disaccharide lactose into constituent galactose and glucose, which we also found to be repressible by the microbiota in multiple germ-free/gnotobiotic studies (SI Appendix, Fig. S5 and Dataset S1). These data suggest that, in addition to changes in gene expression that are based on dietary substrate availability (for example the switch from lactose-rich mother’s milk to fructose-rich solid foods), the developmental switches in carbohydrate metabolism and absorption that occur postnatally may involve TIR- and/or microbiota-derived signals. This form of dual control by both nutrient availability and by microbial/TIR signaling has interesting implications for plasticity of the digestive system and its regulation by the microbiota.
Fig. 3.
Developmental, TLR/IL-R, and microbiota transcriptional regulation of metabolism and host defense. Gene expression by postnatal developmental stage in the small intestine and colon for metabolism (A) and host defense (B) related genes. TLR/IL-R (TIR) and/or microbe icons indicate regulation coordinate with the developmental trend. Error bars, SEM.
Postnatal intestinal development is manifest with many changes in the glycosylation states of mucus and transmembrane proteins (16). We find weaning associated with the up-regulation of Fut2 and B3galt5 and down-regulation of St3gal whereas adulthood is associated with up-regulation of St6gal (Fig. 3A and SI Appendix, Fig. S7). Fut2 has been shown to be up-regulated upon colonization of adult germ-free mice (16, 17). Interestingly, the developmental up-regulation of Fut2 and B3galt5 in the ileum is dependent on TIR signaling (Fig. 3A and SI Appendix, Fig. S7). It will be interesting to dissect the contributions of individual members of the microbiota such as segmented filamentous bacteria (18) and systemic signals acting through TLR (19) in TIR-dependent regulation of small intestinal fucosylation. Furthermore, we found that many UDP glucuronosyltransferases (Ugt) members show aberrant regulation in the colon in the absence of TIR signaling (Fig. 3A; SI Appendix, Fig. S7; and Dataset S2).
In addition to being used for polypeptide synthesis, amino acids also play a critical role as precursors for the synthesis of polyamines, glutathione, and nitric oxide, as well as signaling molecules and key regulators of various intestinal physiological functions, such as differentiation, barrier function, and inflammation (20–22). However, little is known about the regulation of amino acid metabolism and transport at different postnatal states. Bioinformatic analysis revealed postnatal developmental transitions, in particular weaning, to involve major shifts in the gene expression in many amino acid metabolic pathways (SI Appendix, Fig. S4). Most developmental regulation of genes involved in amino acid metabolism occurred independently of TIR signaling but interestingly showed coordinate regulation by the microbiota, including genes for tryptophan (Cat, Ehhadh, and Indo), selenoacid (Papss2), and arginine and proline (Oat, Ckb, and Aoc3) metabolism (Fig. 3C; SI Appendix, Fig. S5; and Dataset S2). We found amino acid/peptide and organic anion/cation transporters to undergo significant changes during postnatal developmental transitions (Fig. 3A and Dataset S2). Slc7a11 (cysteine/glutamate), Slc7a8 (aromatic/branched chain amino acid), Slc7a15 (aromatic amino acid), Slc15a1 and -2 (H+/peptide, also involved in innate immune recognition via transport of bacterial products) (23), Slc5a4b (neutral amino acids), Slco3a1 (anion), and Slco2a1 (anion/prostaglandins) to occur independently of TIR signaling, in addition to Slc3a1 (cystine/dibasic amino acids) and Slc1a1 (neuronal/epithelial high affinity glutamate transporter), which are coordinately modified by the microbiota (Fig. 3A and SI Appendix, Fig. S5). Of note, expression of Slc22a4 (Octn1), a transporter of organic cations and carnitine clusters at the IBD5 locus (24), is dependent on TIR signaling for full induction with weaning (Fig. 3A).
Consistent with the cessation of lipid-rich mother’s milk, weaning is enriched for genes involved in lipid metabolism, such as fatty acid and butanoate metabolism, bile acid biosynthesis, and the synthesis and degradation of ketone bodies (SI Appendix, Figs. S2 and S4). We found certain steps in lipid metabolism (from acyl and acetyl-CoA modification to fatty acid and lipoprotein synthesis to be TIR- and microbe-independent, including Dgat2, Acox2, Acaa2, Acot7 and -12, Srb1, ApoC1, Scd1 and -2, Fads 1 and 2, and Bdh 1 and 2 (Fig. 3A; SI Appendix, Fig. S7; and Dataset S1). Our cross-platform analysis demonstrated the microbiota as capable of modifying a number of developmentally regulated genes involved in lipid digestion, uptake, and transport, also independently of TIR signaling, such as Pnliprp2 and Clps, Slc27a2, Oxct1, Acsl1, Abcg5 and -8 and ApoA4, ApoB, and ApoC2 and ApoC3 (Fig. 3A and SI Appendix, Fig. S7). Interestingly, our analysis identified a number of postnatally regulated genes involved in lipid metabolism modules to be TIR-dependent, such as Npc1l1 (a putative brush border cholesterol transporter), Lpl (lipoprotein lipase), Acaa1b, Hmgcs2 (ketone body synthesis), and Acot11 (Fig. 3A and SI Appendix, Fig. S7).
We found enrichment for nuclear receptor transcriptional networks, such as RXR, LXR, and PPARα (SI Appendix, Fig. S3 and Dataset S2). Interestingly, we found an FXR-based transcription network enriched among developmentally induced and TIR up-regulated genes (SI Appendix, Fig. S3), including the apical bile acid transporter heterodimer subunits Osta and Ostb, Fgf15, and the intracellular bile acid transporter Fabp6/Gastropin, which were nearly completely dependent on TIR signaling for postnatal induction in the small intestine. This finding reveals a previously unidentified role of TIR in the regulation of bile acid homeostasis during ontogeny (Fig. 3A). These insights complement recent studies on the effect of the microbiota on bile acid biology (25).
In summary, genes involved in carbohydrate, amino acid, and lipid metabolism undergo complex changes during development and microbial colonization of the intestine. These changes in gene expression may fall into at least three main categories: genes that change as a direct result of nutrient shifts caused by transition from mother’s milk to solid foods, genes that change as a consequence of additional metabolites that become available due to microbial processing of metabolic precursors, and genes that change as a result of TIR-mediated sensing.
Bioinformatic analysis revealed that, in both the small intestine and colon, weaning was associated with a profound regulation (mostly repression) of genes involved in glutathione and xenobiotic metabolism (SI Appendix, Fig. S4). Many of these genes were regulated independently of TIR signaling yet showed modulation by the microbiota, such as Ahrr, Ltcs4, Mgst2, Nox1, Ephx2, Gpx3, and many members of alcohol and aldehyde dehydrogenase (Adh, Aldh), cytochrome P450 enzymes families (Fig. 3A and SI Appendix, Fig. S7). Interestingly, both developmentally repressed and TIR-repressed genes were enriched for genes involved in glutathione and xenobiotic metabolism (SI Appendix, Fig. S4). TIR signaling played a major role in the regulation of genes involved in these processes, such as Gpx2, dual oxidases, and numerous GST family members (Gst), the latter in the small intestine but not the colon (Fig. 3A and SI Appendix, Fig. S4). Furthermore, many of these genes showed coordinate regulation by the presence of the microbiota as determined by gnotobiotic studies (Fig. 3A and SI Appendix, Fig. S7). These data reveal the postnatal dynamics of the biology of redox reactions and xenobiotic metabolism and the role of TIR signaling in regulating oxidative stress.
Although there are known and important postnatal changes in the absorption and transport of vitamins, minerals, and various electrolytes, the details of the mechanisms of these processes, let alone their regulation, are still fragmentary (4, 22). We observed the postnatal regulation of subsets of genes involved in iron (Mfsd7, Slc40a1/ferroportin, Slc11a2/Nramp2, and Mfi2) and zinc (Slc39a14/Zip4, Slc30a7/Zip7, Slc30a1/Znt1, and metallothioneins Mt1 and -2) homeostasis, including regulatory roles of TIR in the neonatal colon (Prl1r, S100g, Slc40a1/ferroportin, and Mt1 and -2) (Fig. 3A and Dataset S1). We identified transcriptional changes in genes involved in metabolism of lipid (Vdr, Rbp1, -2, and -7, Bcmo1, Bcdo2, Retsat, and Rdh family members) and water soluble vitamins, such as folate (Slc29a4/Rfc, Folr1, and Slc46a1/Pcft), thiamine (Slc19a3), vitamin B2 (Rfk), B12 (Cubn, Amn), and vitamin C (Slc23a2) in addition to the role of TIR (for example, Rdh16) and the microbiota (for example, Rfk) in this regulation (Fig. 3A and Dataset S1). These data reveal previously unknown aspects of the ontogeny of vitamin and mineral homeostasis, unexpected aspects of regulation by TIR, and correlation with modulation by the microbiota.
Host defense and barrier function.
Bioinformatics analysis validated known changes in the postnatal development of the adaptive immune system (26) occurring locally at the intestine, such as T-cell and B-cell ontogeny (Fig. 3B). Analysis of WT and MyD88−/−TRIF−/− littermates revealed the postnatal accumulation of CD4+, CD8+, and IgA-producing cells to be TIR-independent (SI Appendix, Fig. S6). Postnatal developmental datasets in both the small intestine and colon revealed enrichment for genes involved in antigen processing and presentation and Ig transport, such as Ub, Psmb, Cts, Tap, and C2ta, major histocompatibility (MHC) genes Pigr and Fcrn/Fcgrt, and identification of those developmentally regulated and TIR- and microbiota-regulated (Fig. 3B and Dataset S1). At a finer level, our analysis revealed the developmental kinetics and role of TIR signaling in the expression of individual Ig genes in the small intestine and colon, revealing subsets regulated in both a TIR-dependent and -independent manner (SI Appendix, Fig. S6 and Dataset S2).
Our analysis allowed for the systematic regulatory analysis of the organ-specific developmental, TIR, and microbial regulation of chemokines and interleukins, mediators of innate and adaptive immune responses and their antagonists and receptors. Chemokine signaling pathways were significantly enriched among genes up-regulated in adulthood in the small intestine and colon (SI Appendix, Fig. S4). We identified postnatal developmentally down-regulated (Ccl21, 24, and Cx3cr1) and up-regulated (Ccl5 and -28, Il-15, and Il-18) chemokine and cytokine genes, occurring mostly independently of TIR signaling, although a handful of genes were modified by TIR signaling (Ccl8, Ccl28, Cxcl9, Il-7, Il-18bp, and Il-33) (Fig. 3B; SI Appendix, Fig. S7; and Dataset S1).
We found a number of trends with regard to the postnatal changes in expression of genes involved in antiviral processes. Weaning was associated with induction of genes involved in the recognition (Ddx58/RigI and Trex1), signaling (IRF and STAT family members), effector function (Gbp, Ifi, Ifit, and Oas) and regulation (Samdh1) of innate immune responses to viruses (Fig. 3B and SI Appendix, Fig. S7). For the majority of genes, this induction was more pronounced in the colon compared with the small intestine and was transient, with down-regulation occurring with transition to adulthood (Fig. 3B and SI Appendix, Fig. S7). Furthermore, in both the small and large intestine, a significant number of this developmentally regulated antiviral/IFN program was dependent on TIR signaling (Fig. 3B and SI Appendix, Fig. S7). These data may reflect changes in a TIR–microbiota–enteric virus axis through postnatal development (27–30).
Weaning was also associated with an extensive induction of antimicrobial genes, such as angiogenins (Ang), the regenerating gene family of bacteriocidal lectins (Reg), defensins (Def), defensing-related sequence (Defcr) and defensing-related sequence cryptdin peptides (Defcr-rs) (Fig. 3B and SI Appendix, Fig. S7). We found previously undescribed aspects of the postnatal dynamics and regulation of these factors, including repression with postnatal development (Ang1, Reg1, and Reg4). Notably, we found divergence in the mechanism of regulation of antimicrobial genes, even within the same family, because we identified those requiring TIR signaling for postnatal induction (Defa1, Defcr-rs1, -2, -4, -10, and -12, Pap, and Reg3g) and those that do not (Defcr8, -9, -13, -15, and -20, and Lyz) (Fig. 3B and SI Appendix, Fig. S7). Many of these factors show coordinate regulation by the microbiota (Defcr-rs10, Pap, Reg1, and Reg3g) (Fig. 3B and SI Appendix, Fig. S7) (27, 31–34). In addition, we found significant up-regulation of antimicrobial gene expression in the absence of TIR signaling (Reg4, Defa1, Defcr3, and Defcr-rs1) (Fig. 3B and SI Appendix, Fig. S7), many of these enriched among genes up-regulated in the colon and DKO > WT (Fig. 2F, pink striped box).
An intact intestinal barrier is critical for many functions, including host defense, and is involved in the pathophysiology of neonatal diseases such as NEC (35). The intestinal barrier undergoes significant transformations with postnatal development; however, our understanding of the mechanisms and regulation of this process is limited (35). Curation of our datasets allowed for a systematic approach to the molecular analysis of this process and revealed significant postnatal regulation of genes involved in the epithelial barrier (such as Cldn, Pcdh, Gja, Muc, Sprr, etc.) (SI Appendix, Fig. S7). Within these developmentally regulated genes, we identified those dependent on TIR signaling, such as Cldn2, Sprr2b, Adamts9, Mfap5, Mmp7, and Mep1a, and those coordinately regulated by the microbiota by cross-platform analysis, such as Mmp7, Sprr2b, Cldn10, Tff2, Adamdec1, Gja1/connexin43, and many collagen genes (SI Appendix, Fig. S7).
Analysis of the Intestinal Microbiota.
Our comparisons of mice sufficient and deficient in MyD88/TRIF revealed a striking role for TIR signaling in the regulation of a number of genes in the colon and small intestine. Given the role for TIR signaling in antimicrobial responses (27, 31–34) (Fig. 3B and SI Appendix, Fig. S7), we tested the hypothesis that the gene-expression patterns observed reflected changes in the configuration of the gut microbiota as a result of MyD88 and/or TRIF deficiency, rather than direct effects of TIR signals on host gene expression. To do so, we sequenced amplicons generated by PCR of variable region 2 (V2) region of bacterial 16S rRNA genes present in fecal samples collected from WT (MyD88+/−TRIF+/−), MyD88−/−TRIF+/−, MyD88+/−TRIF−/−, or MyD88−/−TRIF−/− mice at various time points during postnatal development and adulthood. We conducted pairwise comparisons of the microbial communities represented in all 101 samples collected using Unweighted UniFrac; this phylogenetic metric measures similarity based on the degree to which any two communities share branch length on a bacterial tree of life (36). Principal component analysis plots, based on UniFrac distances, revealed that TIR deficiency does not produce significant perturbations in community structure, in agreement with previous reports (8, 37). UniFrac measurements revealed differences in fecal microbiota configuration between suckling, weaning, and adult mice that were robust to genotype (SI Appendix, Fig. S9 A–E). Although no significant differences were observed in the proportional representation of family-level bacterial taxa in the microbiota of mice of each genotype within a given age bin (SI Appendix, Fig. S9F), Spearman correlations revealed significant reductions in Rikenellaceae and Porphyromonadaceae during the transition from the suckling to weaning phase and adulthood (SI Appendix, Fig. S9G). Collectively, the data indicate that, under cohousing conditions, neither MyD88 nor TRIF plays a major role in selection of the intestinal microbiota in SPF mice on a C57BL/6 background consuming the diet used for these experiments. It will be interesting to determine the role of MyD88/TRIF signaling on composition of the microbiota on different diets and during single housing in which exchange of the microbiota between genotypes is limited.
The lack of dramatic changes in microbiota composition in WT versus MyD88/TRIF DKO mice suggests that the changes in gene expression reported here are largely due to direct and indirect effects of MyD88-TRIF signaling, rather than a secondary effect of altered microbiota, although spatial aspects of microbial colonization may differ (37).
Role of TIR Signaling on Postnatal Developmental Phenotypes.
Angiogenesis.
The vasculature of the intestine is known to develop postnatally with the influence of the microbiota (38, 39). However, the developmental and microbiota-dependent regulation of this process remains poorly understood. We found an undeveloped vascular endothelium in both the small intestine and colon of suckling WT mice and the emergence of the vasculature after weaning (Fig. 4A). We found this postnatal angiogenesis to be nearly entirely TIR-dependent in the small intestine but not the colon (Fig. 4A). These data identify TIR as a postnatal regulator of small intestinal angiogenesis.
Epithelial cell proliferation.
Both weaning (40) and the microbiota (10) are associated with regulation of intestinal epithelial proliferation. Bioinformatics analysis using MetaRodent identified the transcriptional targets of c-myc to be highly enriched among genes that were both induced with postnatal development and TIR-dependent in the small intestine (Fig. 4B and SI Appendix, Fig. S3). This observation prompted us to investigate the role of TIR signaling in postnatal epithelial proliferation. We found small intestinal epithelial cells to undergo a burst in proliferation after weaning as demonstrated by the increase in the number of crypt cells, crypt height, and proliferating cells (BrDU+ 2-h pulse) after weaning and that this burst was almost completely dependent on TIR signaling (Fig. 4B). Candidate genes regulated by TIR during this process include cyclin D1 (also regulated by the microbiota), Csda (also known as Zonab) (41), and Duox2 and Duoxa2 (42), the latter of which have been implicated in activation of stem cells after injury (43, 44). These data include postnatal development within the spectrum of physiologic (tissue repair at the adult stage) (45, 46) and pathophysiologic (tumorigenesis) (47) processes regulated by TIR signaling.
Postnatal ontogeny of smooth muscle.
As noted above, we noticed a nonparsimonious enrichment between probes that were both down-regulated with development and expressed at lower levels in DKO colons (Fig. 2F, blue striped box). When this 2° dataset was analyzed through the DAVID Functional Annotation Clustering Algorithm (DAVID), we found enrichment of descriptors such as “Z disc” and “sarcomere” and “myofibril” and “muscle development,” including desmin (Des), myosin light chain kinase (Mylk), myosin heavy and light chain (Myl and Myh), and the smooth muscle differentiation factor myocardin (Myocd) (48) (Fig. 4C). We found that the neonatal colon is composed of thickened layers of smooth muscle that decreases with weaning and that, in the neonatal colon, this smooth muscle hyperplasia is TIR-regulated (Fig. 4C). These data suggest a previously unknown connection between innate immune recognition and the regulation of smooth muscle differentiation and contribute to an emerging role of the innate immune system and the microbiota in intestinal motility (49, 50).
Neonatal mast cell homeostasis.
Bioinformatics analysis of genes overexpressed in DKO colons during different developmental stages using DAVID identified the enrichment of Interpro and Swisspro descriptors such as “hydrolase activity” and “mast cell” in pre- and postweaned but not adult colons (Fig. 4D). This enrichment included genes such as mast cell protease (Mcpt) 1 and 5 and mast cell chymases (Cma) 2 and 3, and carboxypeptidases (Cpa) 3 and d (Fig. 4D). We found that, whereas at no developmental stage are mast cells detected in TIR-sufficient colons, TIR-deficient mice show a striking accumulation of mast cells both pre- and postweaned, but not at the adult steady state (Fig. 4D), demonstrating that TIR regulate mast cell homeostasis in a developmental stage-specific manner. This finding may have implications for the understanding of the relationship between early life environmental and microbial exposure and susceptibility to atopic disease, in addition to host defense to macroparasites (51–53).
Discussion
Early postnatal development is accompanied by exposure and adaptation to the external environment, particularly in tissues that directly interact with the outside world, such as skin, respiratory tract, and gastrointestinal tract. A unique and increasingly appreciated aspect of biology of these tissues is their colonization with a diverse and highly complex consortia of microorganisms, which can engage in both symbiotic and antagonistic interactions with their hosts. The lower intestinal tract (ileum and colon) is particularly densely colonized by bacterial, viral, fungal, and, in some cases, protozoan microorganisms (54, 55). The impact of the intestinal microbiota on mammalian physiology and pathology is being intensively studied and, whereas much remains to be learned, it is becoming increasingly clear that many physiological systems, including gastrointestinal and immune systems, do not functional properly in the absence of resident microbiota (6, 10, 56). In particular, little is known about the impact of microbial colonization on intestinal physiology, in part because of the complexity of the interactions between nutrients, microbiota, and digestive/absorptive and immune systems in the intestine.
Here, we provide a detailed analysis of the changes in gene expression in small intestine and colon during early postnatal development, focusing on the transition from suckling to weaning and to adulthood. Our study was designed to evaluate the impact of (i) the changes in nutrients (milk versus solid food), (ii) microbial colonization that naturally occurs during weaning, and (iii) MyD88 and TRIF signaling (which includes TLR-mediated sensing of microbiota as well as the inflammasome–IL-1R family pathways). In addition, we performed metaanalysis of the published data, examining changes in intestinal gene expression between germ-free and conventionally raised or monocolonized mice. We also performed analysis of the bacterial consortia and their changes during postnatal transitions in WT and MyD88/TRIF-deficient littermates. This analysis allowed us to partially deconvolute the complex effects of the nutrients, microbial colonization, and TLR/inflammasome-mediated sensing of microbiota, on intestinal gene expression throughout postnatal developmental transitions.
The data provided by this analysis provided a rich source of information about many aspects of intestinal development and function. We documented extensive changes in expression of genes involved in nutrient absorption and metabolism, epithelial proliferation, barrier function, intestinal immunity, and homeostasis. We dissected the changes that are caused by developmental timing, innate sensing of microbiota through TLR and inflammasome pathways, and the effect of microbial colonization that is independent of their ability to activate the TLR and inflammasome pathways. These data have important implications for understanding the disorders associated with premature birth, such as food intolerance and necrotizing enterocolitis, and provide necessary information on critical stages and windows of mammalian life.
Materials and Methods
MyD88−/− and Trif −/− mice were kindly provided by S. Akira, Laboratory of Host Defense, World Premier International Immunology Frontier Research Center, Osaka University, Osaka. Mice were bred to a C57BL/6 background and then crossed with each other to establish double heterozygote breeding lines as detailed in the main text. Animals were maintained under specific pathogen-free conditions at the animal facility of Yale University School of Medicine. Studies were approved by the Institutional Animal Care and Use Committee of Yale University.
See SI Appendix, SI Materials and Methods for additional sections.
Supplementary Material
Acknowledgments
We thank Aiping Lin (Keck Bioinformatics Resource), Charles Anicelli and Sophie Holley for animal care, and Sheila Umlauf (Keck Microarray Facility). This work was funded by NIH Grants AI046688, AI089771, and DK071754 (to R.M.) and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Raw 16S rRNA sequences have been deposited in the European Nucleotide Archive, www.ebi.ac.uk/ena (accession no. PRJEB8294).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424886112/-/DCSupplemental.
References
- 1.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford Univ Press; Oxford, UK: 2003. [Google Scholar]
- 2.Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci USA. 1998;95(15):8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandtzaeg P. The gut as communicator between environment and host: immunological consequences. Eur J Pharmacol. 2011;668(Suppl 1):S16–S32. doi: 10.1016/j.ejphar.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab. 2013;63(Suppl 2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 6.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 7.Goodrich JK, et al. Conducting a microbiome study. Cell. 2014;158(2):250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209(8):1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25(2):319–329. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 10.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127(2):423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw DR. Searching the Mouse Genome Informatics (MGI) resources for information on mouse biology from genotype to phenotype. Curr Protoc Bioinformatics. 2009;25(1.7):1.7.1–1.7.14. doi: 10.1002/0471250953.bi0107s25. [DOI] [PubMed] [Google Scholar]
- 14.Sharma R, Hudak ML. A clinical perspective of necrotizing enterocolitis: Past, present, and future. Clin Perinatol. 2013;40(1):27–51. doi: 10.1016/j.clp.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cattin A-L, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol. 2009;29(23):6294–6308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nanthakumar NN, et al. Regulation of intestinal ontogeny: Effect of glucocorticoids and luminal microbes on galactosyltransferase and trehalase induction in mice. Glycobiology. 2005;15(3):221–232. doi: 10.1093/glycob/cwi004. [DOI] [PubMed] [Google Scholar]
- 17.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273(5280):1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 18.Goto Y, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014;345(6202):1254009. doi: 10.1126/science.1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickard JM, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514(7524):638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciorba MA. Indoleamine 2,3 dioxygenase in intestinal disease. Curr Opin Gastroenterol. 2013;29(2):146–152. doi: 10.1097/MOG.0b013e32835c9cb3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blachier F, Davila AM, Benamouzig R, Tome D. Channelling of arginine in NO and polyamine pathways in colonocytes and consequences. Front Biosci (Landmark Ed) 2011;16:1331–1343. doi: 10.2741/3792. [DOI] [PubMed] [Google Scholar]
- 22.Pácha J. Development of intestinal transport function in mammals. Physiol Rev. 2000;80(4):1633–1667. doi: 10.1152/physrev.2000.80.4.1633. [DOI] [PubMed] [Google Scholar]
- 23.Ingersoll SA, et al. The role and pathophysiological relevance of membrane transporter PepT1 in intestinal inflammation and inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 2012;302(5):G484–G492. doi: 10.1152/ajpgi.00477.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peltekova VD, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36(5):471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 25.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2012;12(1):9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- 27.Larsson E, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2011 doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto M, et al. A microarray analysis of gnotobiotic mice indicating that microbial exposure during the neonatal period plays an essential role in immune system development. BMC Genomics. 2012;13:335. doi: 10.1186/1471-2164-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14(7):654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon C, Stappenbeck TS. Viral interactions with the host and microbiota in the intestine. Curr Opin Immunol. 2012;24(4):405–410. doi: 10.1016/j.coi.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105(52):20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204(8):1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frantz AL, et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5(5):501–512. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menendez A, et al. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J Innate Immun. 2013;5(1):39–49. doi: 10.1159/000341630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand RJ, Leaphart CL, Mollen KP, Hackam DJ. The role of the intestinal barrier in the pathogenesis of necrotizing enterocolitis. Shock. 2007;27(2):124–133. doi: 10.1097/01.shk.0000239774.02904.65. [DOI] [PubMed] [Google Scholar]
- 36.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99(24):15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinhardt C, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483(7391):627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummins AG, Steele TW, LaBrooy JT, Shearman DJ. Maturation of the rat small intestine at weaning: Changes in epithelial cell kinetics, bacterial flora, and mucosal immune activity. Gut. 1988;29(12):1672–1679. doi: 10.1136/gut.29.12.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sourisseau T, et al. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol. 2006;26(6):2387–2398. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer F, Bäckhed F. The gut microbiota engages different signaling pathways to induce Duox2 expression in the ileum and colon epithelium. Mucosal Immunol. 2014 doi: 10.1038/mi.2014.74. [DOI] [PubMed] [Google Scholar]
- 43.Ha E-M, et al. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat Immunol. 2009;10(9):949–957. doi: 10.1038/ni.1765. [DOI] [PubMed] [Google Scholar]
- 44.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459(7249):996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci USA. 2005;102(1):99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317(5834):124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 48.Wang D-Z, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev. 2004;14(5):558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology. 2012;143:1006–16.e4. doi: 10.1053/j.gastro.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller PA, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158(2):300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds LA, et al. MyD88 signaling inhibits protective immunity to the gastrointestinal helminth parasite Heligmosomoides polygyrus. J Immunol. 2014;193(6):2984–2993. doi: 10.4049/jimmunol.1401056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prioult G, Nagler-Anderson C. Mucosal immunity and allergic responses: Lack of regulation and/or lack of microbial stimulation? Immunol Rev. 2005;206:204–218. doi: 10.1111/j.0105-2896.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 53.Dubos R, Schaedler RW, Costello R. Lasting biological effects of early environmental influences. I. Conditioning of adult size by prenatal and postnatal nutrition. J Exp Med. 1968;127(4):783–799. doi: 10.1084/jem.127.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6(10):776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.