Significance
A layer of fat surrounds the heart in most mammals, including humans. The biology of this tissue has been speculated for centuries, but never subjected to experimental analysis because common experimental model species are thought to not have this tissue. In this study, we show that rodents have cardiac fat, albeit in a very specific location in the heart. We implicate the origin of this tissue from the epicardium (the outer epithelium of the heart) and the underlying mechanisms that account for its derivation. By comparing human and mouse epicardial cells, we provide an explanation for the prominent species differences in the presence and amount of cardiac adipose tissue.
Keywords: epicardial adipose tissue, EAT, PPARγ, epicardium to mesenchymal transformation
Abstract
The hearts of many mammalian species are surrounded by an extensive layer of fat called epicardial adipose tissue (EAT). The lineage origins and determinative mechanisms of EAT development are unclear, in part because mice and other experimentally tractable model organisms are thought to not have this tissue. In this study, we show that mouse hearts have EAT, localized to a specific region in the atrial–ventricular groove. Lineage analysis indicates that this adipose tissue originates from the epicardium, a multipotent epithelium that until now is only established to normally generate cardiac fibroblasts and coronary smooth muscle cells. We show that adoption of the adipocyte fate in vivo requires activation of the peroxisome proliferator activated receptor gamma (PPARγ) pathway, and that this fate can be ectopically induced in mouse ventricular epicardium, either in embryonic or adult stages, by expression and activation of PPARγ at times of epicardium–mesenchymal transformation. Human embryonic ventricular epicardial cells natively express PPARγ, which explains the abundant presence of fat seen in human hearts at birth and throughout life.
The human heart is surrounded by an extensive layer of fat, and for centuries, the biology of this tissue has been debated without experimental resolution (1–3). This tissue lies underneath the epicardium (the outer mesothelial layer of the heart) and is termed epicardial adipose tissue, or EAT, based on its anatomical location rather than from any understanding of its origins. EAT is present in fetal and newborn stages in humans (4, 5) and other species (6), which implies that its derivation is under developmental control. In adults, there is a tendency for more EAT with increasing obesity, which led to early speculation that EAT is pathological, and “fatty heart” was a common diagnostic explanation in the 17th and 18th centuries (7). With recognition that all humans have at least some EAT, attention turned in more recent times to the possibility that EAT might serve beneficial or detrimental functions in heart metabolism, insulation, response to injury, coronary artery disease, or many other speculated possibilities (1–3, 8). A major limitation in understanding the biology of EAT is the seeming absence of this tissue in virtually all commonly used experimental animal models. Thus, rodents, avians, and amphibians are generally considered to not have EAT, although all have noncardiac thoracic (paracardial) fat (which has sometimes been mischaracterized as EAT).
The epicardium is the outer noncontractile mesothelium of the heart. It migrates onto and spreads over the heart surface during early embryonic development, which in mice occurs during the embryonic day E9.5–10.5 period (9). Once formed, the epicardium serves as a source of secreted factors that influence mitogenic expansion of the ventricular myocardium and assembly of coronary blood vessels (10). In addition, the epicardium is a multipotent progenitor cell population (11). Starting from almost the time of its formation, the epicardium undergoes transformation to generate mesenchymal cells that first reside in the subepicardial space between the epicardium and myocardium. One subset of these cells assembles around nascent coronary endothelial tubes and constitutes the smooth muscle layer of the mature coronary vessels. A separate subpopulation migrates as single cells into the myocardium and becomes the predominant source of cardiac fibroblasts that secrete extracellular matrix needed for mature heart structure (12). These two fates are well established; additional speculated fates for the epicardium, including serving as a source of cardiomyocytes and of coronary endothelium, have not been confirmed, at least under normal conditions.
In this study, we show that mice have EAT, which is normally limited to a very specific location in the heart. Using genetic fate mapping approaches, we provide evidence that this tissue is derived by mesenchymal transformation of the epicardium. We explain differences in the presence and amount of EAT between species based on deployment of the peroxisome proliferator activated receptor gamma (PPARγ) molecular pathway, and we show in mice that we can eliminate normal EAT and induce the formation of ectopic ventricular EAT, based on these insights.
Results
The adult hearts of human and several other species show extensive adipose tissue over a significant portion of the entire surface of the ventricle (Fig. S1 A–I). In histological sections, this tissue contains adipocytes, fibroblasts or stromal cells, and blood vessels. In all cases, the adipose tissue lies underneath an outer mesothelium (i.e., the epicardium), an anatomical relationship that distinguishes EAT from thoracic or paracardial fat (3). In contrast, adult hearts from mice and rats show no ventricular EAT, and have direct contact of the epicardium to the myocardium (Fig. S1 J–O), even in locations where coronary vessels lie (Fig. S1P). We observed that adult mouse (Fig. 1A) and rat (Fig. S1Q) hearts have a depot of fat, limited to a very specific location between the atrial and ventricular chambers [the atrial–ventricular (AV) groove] on the dorsal to dorsolateral sides of the heart. Histologically, this tissue is subepicardial (Fig. 1 B and C). A second defining feature of EAT is its perfusion by coronary vasculature rather than from systemic vessels (3). By isolation of adult mouse hearts and retroaortic ink perfusion, we observed ink infiltration into small vessels of the AV groove tissue as well as into major and minor coronary vessels of the ventricle (Fig. S2 A–C). Human EAT is known to have brown adipose tissue character (1, 13), and several BAT-specific genes, as well as pan-adipose genes, were expressed in mouse AV groove fat (Fig. S2D). These three criteria imply that mouse AV groove fat tissue is EAT.
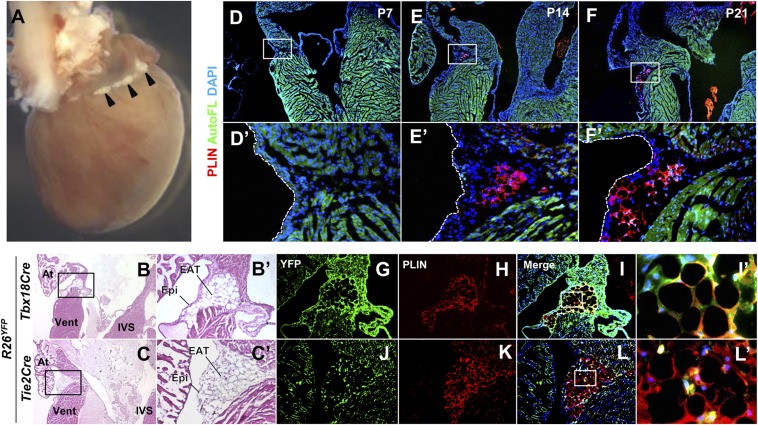
Fig. 1.
Tbx18Cre lineage-derived EAT is present in the AV groove of adult mice. (A) The dorsal surface of an adult (5 mo old) mouse heart. A small amount of fat (arrowheads) is visible in the AV groove. (B and C) Histological appearance of EAT. H&E-stained sections of adult mice show the presence of fat in the AV groove underneath the epicardium. The boxed regions are shown at higher magnification in B′ and C′. At, atrium; IVS, interventricular septum; Vent, ventricle. (D–F) Time course of appearance of AV groove fat. Staining with the adipocyte marker perilipin (PLIN) is absent until 2 wk after birth and increases with age. Autofluorescence (AutoFL) of the myocardium is included to delimit tissue borders. The boxed regions are shown at higher magnification in D′–F′. (G–L) Lineage origins of EAT adipocytes. The same sections are shown in the YFP (lineage marked) channel alone, the PLIN channel alone, or the merge of these and the DAPI channel. PLIN and YFP immunostaining colabel in Tbx18Cre lineage mapped tissue; small interstitial cells of unknown fate in the AV groove are labeled by Tie2Cre but these do not overlap PLIN+ adipocytes. The sections shown in G–L are adjacent to those used in B′ and C′. The boxed regions of I and L are shown at higher magnification in I′ and L′.
We examined the time course of appearance of AV groove adipocytes using the marker perilipin (PLIN), a protein present in lipid vacuoles of mature adipocytes (Fig. 1 D–F). Mesenchyme is particularly abundant in the AV groove at mid- and late- embryonic stages, but there were no PLIN+ cells in this region or elsewhere in the ventricle during embryonic stages or during early postnatal life. PLIN+ cells were first seen in the AV groove at 2 wk (Fig. 1E), intermixed with unlabeled mesenchymal cells. Positive cells at this time had multiple relatively small vacuoles rather than a single large vacuole with minimal cytoplasm that typifies mature adipocytes, suggesting that these cells are immature adipocytes or had only recently initiated terminal maturation. PLIN+ adipocytes increased in size and maturation progressively thereafter.
We used fate mapping to address the tissue origin of mouse EAT. In the heart, a Tbx18Cre driver line labels the entire epicardium from the time of its initial formation (14). In hearts from adult mice with Tbx18Cre and the conditional reporter allele R26YFP, highly efficient labeling of the epicardium, coronary vessel smooth muscle, and interstitial cells (likely fibroblasts) was evident (Fig. S3 A–C). Inspection of the AV groove revealed complete colocalization of PLIN+ cells with the lineage marker (Fig. 1 G–I). A parallel analysis with Tie2Cre, which labels endocardium and endothelium and their derived cells (Fig. S3 D–F), did not label AV groove EAT (Fig. 1 J–L). Thus, EAT adipocytes originate from a Tbx18Cre-expressing progenitor, which is likely to be the epicardium (see Discussion).
Primary epicardial cells can be grown from embryonic heart tissue explants (15). We compared mouse and human primary embryonic ventricular epicardial cells for their propensity to undergo adipogenesis in culture, either spontaneously or in response to an adipogenesis-inducing mixture containing dexamethasone, insulin, isobutyl methylxanthine (IBMX), rosiglitazone, and palmitate (DIIR-Pal). Adipocytes were visualized by Oil Red O staining, which accumulates in lipid vacuoles. In the absence of induction, human and mouse primary cell cultures showed no sign of adipocyte differentiation. Human cells treated with DIIR-Pal for 2 wk showed prominent adipocyte differentiation, whereas mouse cells under the same conditions did not at all (Fig. 2 A–D). This outcome corresponds with the presence and absence of EAT on the ventricular surface of human vs. mouse hearts and implies that primary epicardial cell culture could be a faithful means of studying epicardial cell differentiation in vivo. Adipocytes were also not evident when mouse epicardial cells from the embryonic AV groove were induced to differentiate (Fig. S4 A and B), showing that both subpopulations of mouse embryonic epicardial cells lack adipogenic potential. Therefore, the presence of EAT in the mouse AV groove starting around 2 wk after birth (Fig. 1E) represents a unique postnatal aspect of epicardial differentiation.
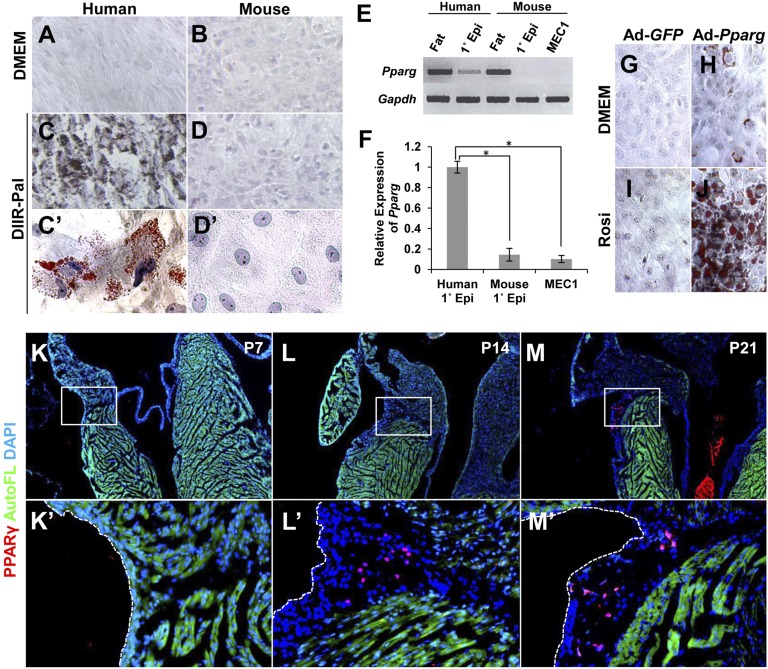
Fig. 2.
PPARγ controls epicardial adipogenesis. (A–D) In vitro adipogenic potential. Primary epicardial cells isolated from human and mouse embryonic ventricular tissue were cultured for 14 d in medium containing adipogenic inducers (dexamethasone, IBMX, insulin, rosiglitazone, palmitate) and stained with Oil Red O. (E) Pparg expression in epicardial cell cultures, analyzed by RT-PCR. Pparg was basally expressed in human cells, but was absent in mouse cells. Omental fat from adult human and adult mouse was used as positive controls. (F) Quantitation of normalized Pparg expression in human (n = 4 independent samples) and mouse (n = 3) primary epicardial cell cultures and MEC1 cell cultures (n = 3). The average human expression was set to 1.0. Statistically significant from human expression at *P = 0.0037 (mouse primary) and *P = 0.0012 (MEC1). (G–J) PPARγ transduction. PPARγ or GFP (used as a control) was transduced by adenovirus infection into primary mouse epicardial cells, and cells were then treated with rosiglitazone (Rosi). Lipid accumulation was visualized by Oil Red O staining. (K–M) PPARγ expression in vivo. PPARγ was detected in nuclei in the AV groove EAT of mouse heart 14 d after birth by immunostaining (red). Sections near those used for PLIN staining (Fig. 1 D–F) were used here for PPARγ immunostaining. The boxed regions are shown at higher magnification in K′–M′.
The ligand-dependent nuclear receptor PPARγ is a master regulator of adipocyte differentiation and lipid metabolism in several cell types (16). We compared cultured human and mouse primary embryonic ventricular epicardial cells and found that Pparg was expressed prominently in human cells, but at a 10-fold lower level in mouse cells (Fig. 2 E and F). Pparg was also minimally expressed in MEC1 immortalized mouse embryonic ventricular epicardial cells (Fig. 2 E and F), which also do not undergo adipogenic differentiation in response to the inducing mixture (Fig. S4 C and D). We virally expressed Pparg in primary mouse epicardial cells and in MEC1 cells; this resulted in a low degree of spontaneous adipocyte differentiation in the absence of treatment and a prominent level of differentiation in the presence of the PPARγ ligand rosiglitazone (Fig. 2 G–J and Fig. S4 E–H). PPARγ has a moderate level of ligand-independent transcriptional activity (17), which may explain the sporadic induction of differentiation when overexpressed in the absence of added ligand (Fig. 2H). Thus, the differential ability of human vs. mouse epicardial cells to initiate adipogenic differentiation can be explained by the level of endogenous PPARγ expression, and mouse epicardial cells are able to undergo adipogenic differentiation when they express PPARγ. Consistent with these observations, PPARγ was not present in the embryonic mouse heart and only became detectable in AV groove mesenchyme starting around 2 wk after birth (Fig. 2 K–M), coincident with the appearance of EAT (Fig. 1E).
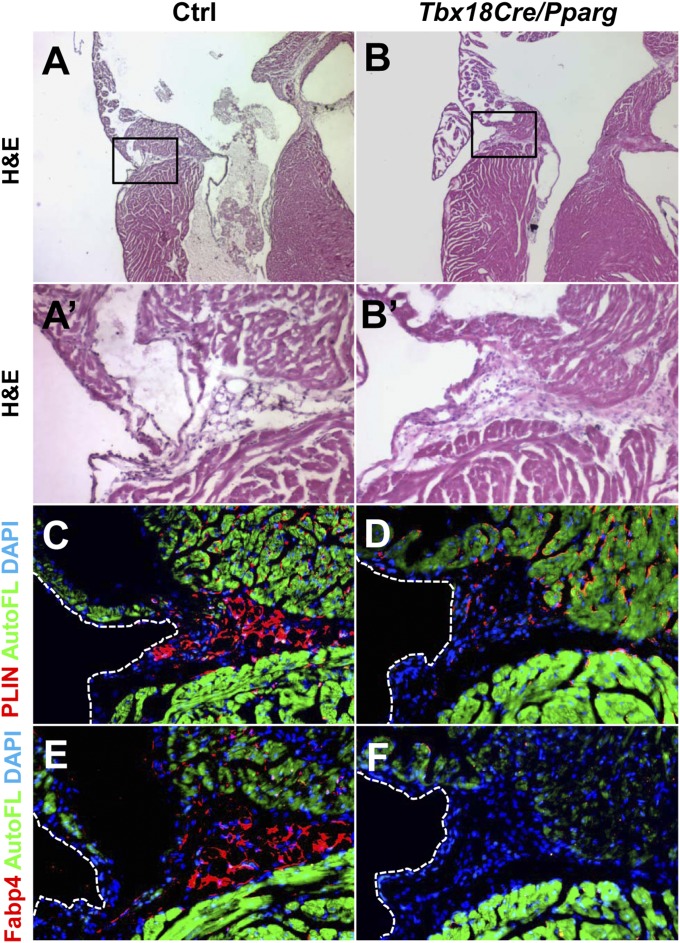
To determine if PPARγ activity is required for mouse EAT formation in vivo, we crossed a Pparg conditional loss-of-function allele with Tbx18Cre. Mutant mice survived into adulthood and were seemingly normal and contained typical amounts of fat in noncardiac locations, including the kidney fat pad, intestinal omentum, and female reproductive tract (Fig. S5). In the AV groove of Tbx18Cre/Pparg conditional mutant mice, mesenchyme was still present (Fig. 3B), but mature PLIN+ adipocytes were eliminated (Fig. 3D). A small number of interstitial PLIN+ cells of uncertain identity were observed, but the absence of adipocytes was confirmed by immunostaining for fatty acid binding protein 4 (FABP4) as a second adipocyte marker (Fig. 3F). Thus, the PPARγ pathway is required for formation of mouse AV groove adipocytes. We cannot yet say if mesenchymal cells in the AV groove in these conditional mutants are of an alternative lineage (e.g., fibroblast) that is also present in normal hearts, or if these are immature preadipocytes that have failed to mature to the point of expressing PLIN or FABP4. In either case, these observations imply that PPARγ activity does not influence the process of epicardial mesenchymal transformation, but rather the ability of mesenchyme to adopt an adipogenic fate.
Fig. 3.
PPARγ is required for development of EAT. Sections through the AV groove of 6-wk-old mice are shown, stained either by H&E (A and B) or nearby sections stained for PLIN (C and D) or FABP4 (E and F). The locations of the high magnification views of the AV groove shown in A′, B′, and C–F are indicated by the boxed region in A and B. The scattered PLIN+ cells seen in Tbx18Cre/Pparg hearts are of unknown fate but are not adipocytes, as evidenced by their morphology and lack of staining by FABP4. Green is autofluorescence of the myocardium. Control mice (n = 4) were littermates of the Tbx18Cre/Pparg mice (n = 4).
Conversely, we also used Tbx18Cre to force PPARγ expression in a gain-of-function manipulation. Because an allele that conditionally expresses wild-type PPARγ is not available, for this purpose we used a conditional allele that expresses a Pax8–PPARγ fusion protein (PPFP) after recombination; the fusion protein was previously shown to behave like normal PPARγ, including the induction of adipogenic differentiation, in the presence of an appropriate PPARγ ligand (18). We first tested this genetic manipulation in primary cell culture. Primary ventricular epicardial cells derived from Tbx18Cre/PPFP embryos displayed typical epicardial morphology and showed no basal adipogenic differentiation, but underwent active adipogenic differentiation in the presence of rosiglitazone (Fig. 4 A–D). This mirrors the behavior of genetically normal primary mouse epicardial cells in which PPARγ expression is forced by viral infection (Fig. 2 G–J).
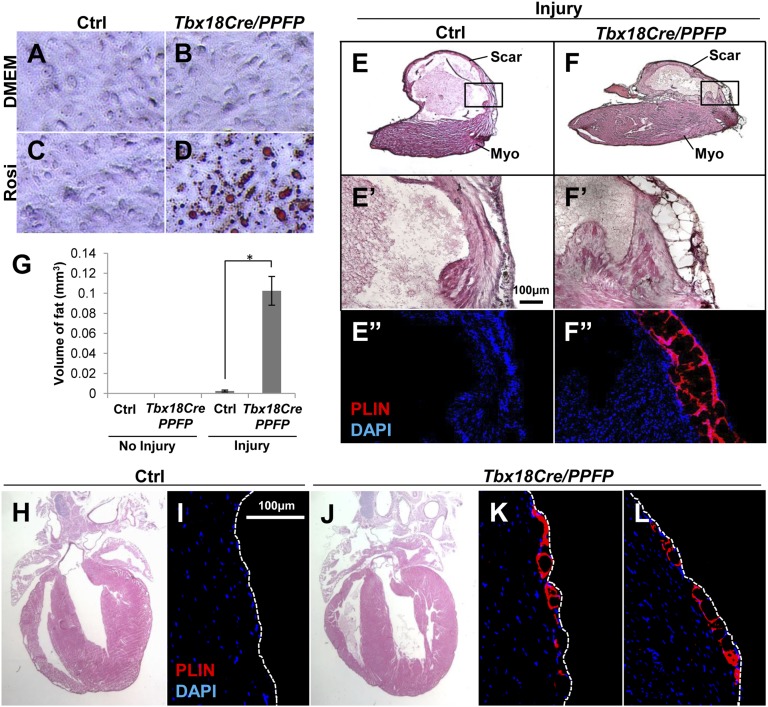
Fig. 4.
PPARγ activity and mesenchymal transformation induce adipogenesis in vivo. (A–D) In vitro induction of adipogenesis by PPFP. Primary embryonic ventricular epicardial cells were derived from Tbx18Cre/PPFP embryos and control littermates, treated with rosiglitazone, and lipid accumulation visualized by Oil Red O staining. (E and F) In vivo adult adipogenesis. Cryoinjury was performed on Tbx18Cre/PPFP adult mice and controls; a high fat diet and rosiglitazone were provided for 3 mo after surgery. In transverse sections near the apex, ventricular adipocytes were detected by PLIN immunostaining and DAPI counterstaining. Images of uninjured hearts are in Fig. S6 C and D. The boxed areas at the transition between normal myocardium (myo) and scar tissue are shown in E′, E′′, F′, and F′′; E′ and E″, and F′ and F″, are adjacent or nearby sections. (G) Quantitation of ventricular fat. The volume of ventricular fat per heart was derived from PLIN- and Oil Red O-stained serial sections. There was no fat in uninjured control (n = 4) or uninjured Tbx18Cre/PPFP (n = 5) hearts, a small amount in injured controls (n = 4), and substantially more (*P = 0.0004) in injured Tbx18Cre/PPFP (n = 5) hearts. Fat was only present in injury-adjacent areas. (H–L) Induction of embryonic adipogenesis. Pregnant females were fed a high fat diet and provided with rosiglitazone starting at E10.5 and continuing during lactation, and weaned pups were kept on the same regimen until 3 mo of age. The H&E images (H and J) show normal morphology of the treated adult hearts; immunostaining for PLIN (I, K, and L) shows the presence of adipocytes in two different locations of the ventricle in Tbx18Cre/PPFP mice but not in controls. These observations were repeated in six Tbx18Cre/PPFP mice and four control littermates (bearing only Tbx18Cre, or only PPFP, or neither allele).
We raised control and Tbx18Cre/PPFP mice to adulthood. The amount of AV groove EAT was comparable in both (Fig. S6 A and B), and ventricular chamber EAT was absent in both even when mice were provided a high fat diet supplemented with rosiglitazone starting in adulthood (Fig. S6 C and D). Thus, forced expression and ligand activation of PPARγ (specifically PPFP) in the Tbx18Cre lineage in the adult mouse does not override the signals that support or prevent adipogenic differentiation in vivo.
The observation that Tbx18Cre/PPFP epicardial cells in culture efficiently initiate adipogenesis in response to rosiglitazone (Fig. 4 A–D) suggested that epicardial–mesenchymal transformation (EMT) might be an important step in adipogenic differentiation, as primary epicardial cell cultures are at least partially mesenchymal during early growth (e.g., as manifested by expression of vimentin and smooth muscle actin). To test this model, we cryoinjured the heart surface of adult mice, because injury is known to activate EMT in the adult heart (19). Transdiaphragmatic cryoinjury may be a preferable injury model for these studies, as this procedure avoids thoracic cavity surgery, which is associated with formation of pericardial adhesions to the myocardial surface, and such adhesions can include paracardial fat (from the thoracic wall), which is distinct from but which might appear to be EAT. In control mice provided with a high fat diet supplemented with rosiglitazone, the injured surface of the heart was covered with an extensive fibrotic matrix. This scar tissue lacked adipocytes, although a small localized cluster of adipocytes was typically observed in the injury-adjacent area (Fig. 4 E and G and Fig. S6 E and F). The observation that this occurs at all implies that injury is accompanied by some degree of relaxation in the negative control of adipogenesis, and presumably also in expression of PPARγ. In contrast, in injured Tbx18Cre/PPFP mice, prominent accumulations of adipocytes were present in the injury-adjacent region (Fig. 4F), although fibrotic scar tissue was still extensive. Quantitation indicated a more than 50-fold increase in the amount of postinjury EAT in Tbx18Cre/PPFP hearts compared with littermate controls (Fig. 4G). A layer of epicardium identified by the marker podoplanin overlaid the induced ventricular EAT (Fig. S6 G and H), which was immunolabeled with both PLIN and FABP4 (Fig. S6 H and I). Mesenchyme induced by adult heart cryoinjury therefore almost completely adopts a fibrotic fate in control mice, whereas the expression of PPARγ (PPFP) promotes or allows significantly more adipogenic differentiation.
Mesenchymal transformation of the epicardium occurs only sporadically if at all during normal postnatal life, but occurs actively during normal embryonic development. We therefore treated mice with rosiglitazone in utero during the period when this process is actively underway. When examined in adulthood, rosiglitazone exposure of control embryos had no effect (Fig. 4 H and I), whereas in Tbx18Cre/PPFP embryos, this treatment resulted in broad domains of ventricular fat in the subepicardial space (Fig. 4 J–L and Fig. S6 J and K). The morphology of the adult heart (Fig. 4J) and the viability of these mice into adulthood was not compromised, which implies an adequate level of differentiation by the embryonic epicardium to coronary vascular smooth muscle and fibroblast fates, even though a subpopulation of the epicardium-derived mesenchyme underwent adipogenic differentiation.
Discussion
Our results demonstrate that rodents normally have EAT in the atrial–ventricular groove and support the conclusion that this tissue is derived from the epicardium. Very recently, two reports reached related conclusions. Using Wt1Cre as a lineage marker, Chau et al. (20) indicated that all visceral fat, including epicardial fat, is derived from the mesothelium; as noted above, the epicardium is the mesothelium of the heart. Liu et al. (21) described mouse AV groove EAT as we observed, and using MslnCre and Wt1Cre, concluded derivation from the epicardium. Our analysis with Tbx18Cre, bolstered with our demonstration that epicardial cells can become adipocytes in vitro, complements these recent reports. Although any single Cre driver can have experimental caveats related to efficiency and specificity, the aggregate data show that adipogenic differentiation is a third and new fate of the multipotent epicardium. In both of the recent reports, the experimental approaches resulted in only moderately efficient epicardial recombination, so the possibility of a mixed derivation could not be addressed. Tbx18Cre is highly efficient in the epicardium, and we observed virtually complete labeling of AV groove EAT with this lineage marker (Fig. 1 G–I), indicating that the epicardium is likely to be the sole source of EAT adipocytes. However, it should be cautioned that additional nonepicardial sites of Tbx18Cre expression (22, 23) could in principle contribute cells to this tissue.
Collectively, our cell culture and in vivo results indicate that mouse epicardium-derived cells can adopt an adipocyte fate after mesenchymal transformation and if they express PPARγ. The requirement for PPARγ is not unexpected, as adipogenic differentiation requires PPARγ activity (24). Our conclusions related to the requirement for EMT rest on several observations: adult heart injury induces EMT and induces fat when PPARγ is expressed (Fig. 4F); an adipocyte fate is induced in embryonic heart during the period when normal EMT is underway (Fig. 4 J–L); epicardial cells in culture are partially mesenchymal and this unleashed adipogenic potential when they express PPARγ (Figs. 2 G–J and 4 A–D); and finally, adipocytes are a mesenchymal cell type and if derived from the epicardium, it is almost inescapable that EMT must occur before adipocyte differentiation can occur. Formally, however, definitive confirmation of this model would require experimental manipulation to prevent EMT and then determine the consequence to adipogenesis. Liu et al. (21) also noted induction of ventricular fat after adult heart injury, although interestingly, their observations were conducted in mice that were genetically unmanipulated for PPARγ. In our study, this fate was minimal (Fig. 4 E and G and Fig. S6 E and F), unless PPARγ expression was forced in the Tbx18Cre domain (Fig. 4 F and G and Fig. S6 H and I). This discrepancy might be the result of biological or technical differences in the injury models (coronary ligation vs. transdiaphragmatic cryoinjury).
AV groove EAT in mouse appears between the first and second postnatal weeks (Fig. 1 D–F), coincident with the appearance of PPARγ+ nuclei (Fig. 2 K–M) and in a PPARγ-dependent manner (Fig. 3). As a working model, we propose that embryonic epicardial transformation in mice is dedicated to fibroblast and smooth muscle fates, in a manner that prevents expression of PPARγ. Forced expression of PPARγ and provision of ligand reprogram a subset of these cells in the embryonic ventricle to an adipogenic fate (Fig. 4 J–L). Conditions in the AV groove of postnatal mice might support continued mesenchymal transformation and in a manner that allows PPARγ expression and activation, which could account for the postnatal formation of EAT in this location. Alternatively, preexisting undifferentiated mesenchyme derived from embryonic EMT might be induced to initiate PPARγ expression and adipogenic differentiation by conditions in the postnatal AV groove; the available evidence using Tbx18Cre (this study) or Wt1Cre (21) do not distinguish these alternatives. Regardless, these fate controls are seemingly less restrictive in the human heart, in that human embryonic ventricular epicardial cells express PPARγ and readily adopt an adipocyte fate. This behavior was seen explicitly in culture (Fig. 2 A–D, E, and F) and we infer also occurs in vivo, in a manner that results in the presence of EAT over the entire ventricle in newborn human infants and thereafter throughout adult life.
In the normal human heart, surface fat (EAT) infiltrates into the myocardium to some degree (25). At the extreme, however, arrhythmogenic right ventricular cardiomyopathy (ARVC), a leading cause of sudden adult death, involves transmural replacement of cardiomyocytes by fat and fibrous tissue (26). This pathology is initiated by desmosomal dysfunction, which is thought to then modulate signals that induce fibroblast and adipogenic differentiation (27). In principle, this pathology may represent excess activation of the same signals used in normal differentiation of EAT.
Materials and Methods
Additional methods are provided in SI Materials and Methods.
Mice.
All mouse lines in this study have been previously described: Tbx18Cre (14), Tie2Cre (28), conditional R26YFP (29), conditional PPFP (18), and conditional Pparg (30). All mice were maintained in accordance with Institutional Animal Care and Use Committee guidance.
Cell Culture.
MEC1 is a stably immortalized mouse embryonic ventricular epicardial cell line (31). Derivation and isolation of primary mouse epicardial cells were described previously (15, 31); briefly, mouse embryonic day E13.5 ventricular heart tissue was minced coarsely into pieces and allowed to settle on gelatin-coated dishes, from which epicardial cells migrate and expand. Primary human embryonic ventricular epicardial cells were grown in an identical manner from fetal tissue obtained from Novogenix Laboratories following informed consent and elective termination. All primary epicardial cell cultures were used without passage.
Adenovirus Infection.
Adenovirus expressing full-length PPARγ1 (32) was generously provided by Hidekazu Tsukamoto, University of Southern California, Los Angeles. Cells were infected for 8 h, washed with PBS, and then provided with fresh medium containing vehicle/DMEM or 10 μM rosiglitazone.
Cardiac Cryoinjury.
Transdiaphragmatic cardiac injury was performed on 8-wk-old mice under isoflurane anesthesia, using a procedure adapted from a previously described protocol (33). Briefly, a 2-cm vertical incision was made on the ventral surface through skin and peritoneum along the midline, 1.5 cm below the diaphragm. A plastic sleeve was inserted and placed against the diaphragm such that most of the right dorsal ventricular wall and a part of the dorsal left ventricular wall were in direct contact with the diaphragm. A 0.5-mm metal blunt probe precooled in liquid nitrogen for 1 min was inserted through the sleeve and pushed against the heart and diaphragm for 10 s. Sham-operated mice received the same procedure but with a room temperature probe.
High Fat Diet and Rosiglitazone Administration.
A single daily dose of rosiglitazone (10 mg/kg body weight) was administered intraperitoneally immediately after surgery and for the next 3 d. 1 μM rosiglitazone was also coadministered in the drinking water. The high fat diet was purchased from Harlan Laboratories (TD06414). The water and diet were provided for 3 mo from the day of surgery until tissue harvest. To administer rosiglitazone in utero, a single daily dose of 10 mg/kg body weight was administered intraperitoneally to pregnant females from E10.5 to E13.5. The females were provided with a high fat diet and with drinking water containing 1 μM rosiglitazone from E10.5 until pups were weaned. Pups continued to receive the water and high fat diet until tissue harvest at 3 mo of age. Control mice were littermates of the experimental mice and therefore were exposed to the same conditions.
Quantification of Fat Volume.
Serial sections of samples were generated and stained with Oil Red O. Sections were then photographed, and Oil Red O positive surface area was quantified using ImageJ. Volume was calculated, and Student’s t test was performed for statistical significance.
Supplementary Material
Acknowledgments
The authors acknowledge the technical assistance of Mr. Danny Lee in these studies. This work was supported by NIH Grant HL070123 and by a pilot award from the University of Southern California Diabetes & Obesity Research Institute (to H.M.S.). Y.Y. was supported by a predoctoral fellowship from the American Heart Association and H.S. was supported by a postdoctoral fellowship from the California Institute for Regenerative Medicine.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417232112/-/DCSupplemental.
References
- 1.Iacobellis G, Bianco AC. Epicardial adipose tissue: Emerging physiological, pathophysiological and clinical features. Trends Endocrinol Metab. 2011;22(11):450–457. doi: 10.1016/j.tem.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabkin SW. Epicardial fat: Properties, function and relationship to obesity. Obes Rev. 2007;8(3):253–261. doi: 10.1111/j.1467-789X.2006.00293.x. [DOI] [PubMed] [Google Scholar]
- 3.Sacks HS, Fain JN. Human epicardial adipose tissue: A review. Am Heart J. 2007;153(6):907–917. doi: 10.1016/j.ahj.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Fletcher BD, Jacobstein MD, Abramowsky CR, Anderson RH. Right atrioventricular valve atresia: Anatomic evaluation with MR imaging. AJR Am J Roentgenol. 1987;148(4):671–674. doi: 10.2214/ajr.148.4.671. [DOI] [PubMed] [Google Scholar]
- 5.Jornet A, Reig J, Ruiz C, Uson M, Petit M. The intervenous tubercle (of lower): Morphological characteristics and changes in relation to age. Arch Anat Histol Embryol. 1990;73:21–32. [PubMed] [Google Scholar]
- 6.Marchington JM, Mattacks CA, Pond CM. Adipose tissue in the mammalian heart and pericardium: Structure, foetal development and biochemical properties. Comp Biochem Physiol B. 1989;94(2):225–232. doi: 10.1016/0305-0491(89)90337-4. [DOI] [PubMed] [Google Scholar]
- 7.Bedford E. The story of fatty heart. A disease of Victorian times. Br Heart J. 1972;34(1):23–28. doi: 10.1136/hrt.34.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayes-Genis A, et al. Human progenitor cells derived from cardiac adipose tissue ameliorate myocardial infarction in rodents. J Mol Cell Cardiol. 2010;49(5):771–780. doi: 10.1016/j.yjmcc.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Virágh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201(1):157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- 10.Sucov HM, Gu Y, Thomas S, Li P, Pashmforoush M. Epicardial control of myocardial proliferation and morphogenesis. Pediatr Cardiol. 2009;30(5):617–625. doi: 10.1007/s00246-009-9391-8. [DOI] [PubMed] [Google Scholar]
- 11.Riley PR. An epicardial floor plan for building and rebuilding the mammalian heart. Curr Top Dev Biol. 2012;100:233–251. doi: 10.1016/B978-0-12-387786-4.00007-5. [DOI] [PubMed] [Google Scholar]
- 12.Acharya A, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139(12):2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sacks HS, et al. Uncoupling protein-1 and related messenger ribonucleic acids in human epicardial and other adipose tissues: Epicardial fat functioning as brown fat. J Clin Endocrinol Metab. 2009;94(9):3611–3615. doi: 10.1210/jc.2009-0571. [DOI] [PubMed] [Google Scholar]
- 14.Cai CL, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454(7200):104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, et al. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol. 2002;250(1):198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- 16.Spiegelman BM, Hu E, Kim JB, Brun R. PPAR gamma and the control of adipogenesis. Biochimie. 1997;79(2-3):111–112. doi: 10.1016/s0300-9084(97)81500-3. [DOI] [PubMed] [Google Scholar]
- 17.Molnár F, Matilainen M, Carlberg C. Structural determinants of the agonist-independent association of human peroxisome proliferator-activated receptors with coactivators. J Biol Chem. 2005;280(28):26543–26556. doi: 10.1074/jbc.M502463200. [DOI] [PubMed] [Google Scholar]
- 18.Dobson ME, et al. Pioglitazone induces a proadipogenic antitumor response in mice with PAX8-PPARgamma fusion protein thyroid carcinoma. Endocrinology. 2011;152(11):4455–4465. doi: 10.1210/en.2011-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B, Pu WT. Epicardial epithelial-to-mesenchymal transition in injured heart. J Cell Mol Med. 2011;15(12):2781–2783. doi: 10.1111/j.1582-4934.2011.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chau YY, et al. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat Cell Biol. 2014;16(4):367–375. doi: 10.1038/ncb2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, et al. Epicardium-to-fat transition in injured heart. Cell Res. 2014;24(11):1367–1369. doi: 10.1038/cr.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christoffels VM, et al. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458(7240) doi: 10.1038/nature07916. E8–E9; discussion E9–E10. [DOI] [PubMed] [Google Scholar]
- 23.Christoffels VM, et al. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006;98(12):1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 24.Anghel SI, Wahli W. Fat poetry: A kingdom for PPAR gamma. Cell Res. 2007;17(6):486–511. doi: 10.1038/cr.2007.48. [DOI] [PubMed] [Google Scholar]
- 25.Tansey DK, Aly Z, Sheppard MN. Fat in the right ventricle of the normal heart. Histopathology. 2005;46(1):98–104. doi: 10.1111/j.1365-2559.2005.02054.x. [DOI] [PubMed] [Google Scholar]
- 26.Iyer VR, Chin AJ. Arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D) Am J Med Genet C Semin Med Genet. 2013;163C(3):185–197. doi: 10.1002/ajmg.c.31368. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Gras E, et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116(7):2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisanuki YY, et al. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 29.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 30.He W, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P, et al. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138(9):1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazra S, et al. Peroxisome proliferator-activated receptor gamma induces a phenotypic switch from activated to quiescent hepatic stellate cells. J Biol Chem. 2004;279(12):11392–11401. doi: 10.1074/jbc.M310284200. [DOI] [PubMed] [Google Scholar]
- 33.Leferovich JM, et al. Heart regeneration in adult MRL mice. Proc Natl Acad Sci USA. 2001;98(17):9830–9835. doi: 10.1073/pnas.181329398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.