Significance
We introduce a novel method for controlling orientation of proteins during immobilization on solid-state surfaces using electric fields. Atomic force microscopy measurements and fluorescence-based readout are used to confirm this phenomenon. This method of immobilization has broad implications for improvement in performance of both affinity-based protein assays and probe-free detection assays.
Keywords: protein immobilization, electrokinetic, microfluidics, sample preparation, protein assay
Abstract
The controlled immobilization of proteins on solid-state surfaces can play an important role in enhancing the sensitivity of both affinity-based biosensors and probe-free sensing platforms. Typical methods of controlling the orientation of probe proteins on a sensor surface involve surface chemistry-based techniques. Here, we present a method of tunably controlling the immobilization of proteins on a solid-state surface using electric field. We study the ability to orient molecules by immobilizing IgG molecules in microchannels while applying lateral fields. We use atomic force microscopy to both qualitatively and quantitatively study the orientation of antibodies on glass surfaces. We apply this ability for controlled orientation to enhance the performance of affinity-based assays. As a proof of concept, we use fluorescence detection to indirectly verify the modulation of the orientation of proteins bound to the surface. We studied the interaction of fluorescently tagged anti-IgG with surface immobilized IgG controlled by electric field. Our study demonstrates that the use of electric field can result in more than 100% enhancement in signal-to-noise ratio compared with normal physical adsorption.
Immobilization and attachment of proteins on solid-state surfaces has wide application in various types of optical (1, 2), electronic (3, 4), and magnetic (5–8) biosensing platforms. Both affinity-based sensing platforms (9) and probe-free (10)–based platforms can benefit from controlled and uniform immobilization of proteins on sensor surfaces. In the case of affinity-based sensors, probe molecules such as antibodies are used to capture the target protein of interest. Given that the probe protein has specific sites or epitopes where binding occurs, the orientation of the protein as it is immobilized onto the surface can affect whether the target protein of interest will be able to bind properly or not. Thus, depending on the morphology of the probe antibody and the assay configuration, an affinity-based immunosensor with uniformly oriented immobilized probe antibodies with binding sites all accessible to the incoming target proteins can have higher capture efficiency of target molecules compared with a sensor where the probe antibodies are randomly oriented with a percentage of the binding sites being inaccessible to the incoming target antigen. For affinity-based immunosensors, this controlled immobilization of probe antibodies is performed by using surface functionalization techniques (11) or protein engineering techniques (12) allowing only the constant fragment (Fc) region of the antibody to bind to the sensor surface.
Probe-free sensors refer to sensors that do not require the use of probe molecules such as antibodies to attain specificity in detection. The specificity in general is achieved through some sort of spectroscopic technique where physical and chemical signatures of the molecule under investigation are identified. One example of a probe-free biosensing technique is surface enhanced Raman scattering (13, 14). Another emerging probe-free technique is biosensing based on quantum electronic tunneling (15, 16). For both surface Raman and tunneling-based sensing, the binding molecule of interest will generate a spectroscopic response such that a unique signature of the molecule will be obtained. In both cases, the “hot spot” or the sensing region extends on the order of 3 nm or so above the sensing surface (17). As a result, consistency of measurements will be affected by the random orientation of target proteins becoming randomly oriented on the surface. Also, uniform surface immobilization of target proteins that have a high degree of asymmetry will result in an amplified signal due to consistency of the motif attached to the surface, thus enhancing sensitivity as well.
In general, controlled and consistent immobilization of protein is achieved using surface functionalization techniques where specific motifs on the surface of the protein attach to the functional groups on the sensor surface (11, 12). This can become difficult and tedious in the case where large scale multiplexing is desirable where a unique chemistry will be required for each individual element of the sensor array. A tunable technique like the use of electric field would be much more amenable to use on a large scale in certain scenarios depending on the binding region and charge distribution of the proteins.
Previously, our group developed a simulation procedure to predict the orientation of a protein upon its immobilization on a solid-state support in presence of the electric field (18). To this end, exploiting the charge distribution and polarizability properties of various proteins may be possible.
In this study, we sought to experimentally demonstrate a proof of concept of the idea of using electric fields for tunable control of the orientation of proteins during immobilization. Without a suitable sensor at hand ready for direct measurement of the orientation of a protein, we set out to develop an experimental test bed using atomic force microscopy (AFM) to directly measure the orientation of immobilized proteins. In this manuscript, we describe the test bed we developed using AFM to image the IgG molecules immobilized onto the surface and also the application of controlled immobilization to the improvement in performance of a fluorescence protein assay involving enhanced binding of fluorescent anti-IgG molecules to IgG molecules, which are immobilized with controlled orientation.
In all cases, we make use of a lateral electric field across a microchannel as a proof of concept. The reason we applied a lateral field is because when applying dc signals, faradaic electrodes (like Ag/AgCl) are necessary to generate electric fields across the bulk solution; otherwise if the electrodes are inert, the electric field will get screened entirely by the double layer at the surface. Achievement of a vertical field would be possible by patterning electrodes on the surface; however, because it was more complicated to fabricate Ag/AgCl electrodes on the surface (19), as a proof of concept, here, we demonstrate the idea using lateral fields.
Theoretical Analysis
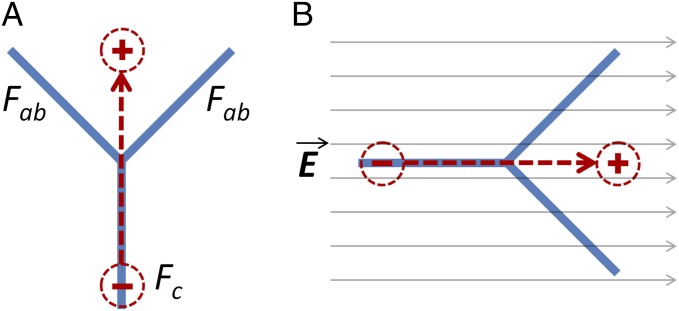
As can be seen from Fig. 1A, the structure of the antibody can be illustrated as a joint composition of two identical Fab (fragment antigen-binding) arms and one Fc that altogether form a “Y”-shaped molecule. The Fab fragments contain the variable regions that have the ability to specifically bind to antigens. The isoelectric point (IEP) of the (Fab)2 fragment is often larger than that of the Fc fragment and the whole antibody (20, 21). Hence, in an intermediary buffer pH level condition [i.e., pH level between IEP of the (Fab)2 and IEP of the Fc], the (Fab)2 fragment is positively charged and the Fc fragment is negatively charged (18, 19). As a result, in this condition, the antibody molecule as a whole can be effectively modeled as an electric dipole, where the dipole moment vector is pointing from the negatively charged Fc to the positively charged (Fab)2. Therefore, it is possible to use an external electric field to control and manipulate the orientation of antibody molecules (Fig. 1B).
Fig. 1.
(A) Simplified structure of the antibody as a joint composition of two Fab fragments and one of Fc. At the IEP of the whole antibody, the antibody molecule can be modeled as an electric dipole, with dipole moment vector pointing from Fc to Fab. (B) The dipole-like IgG molecule gets aligned in the presence of electric field.
Previously, the effect of electric field on the orientation of the antibody upon its adsorption on charged surfaces was simulated (20) and demonstrated (21). In such cases the electric field (produced as a result of the charged surface) was perpendicular to the surface and aligned with antibody’s “end-on” orientation on positively charged surfaces and ‘‘head-on’’ orientation on negatively charged surfaces. Here, we are exploiting the antibody dipole property to investigate the effect and benefit of using external lateral electric field during the antibody surface adsorption process.
As a proof of concept, we used the reasonably well-modeled IgG antibody in our experiments. In a series of experiments, during the adsorption step of IgG molecules, we applied lateral electric fields of varying strengths. In our approach, the relatively uniform external electric field produces a net torque on the dipole-like IgG molecules, which tends to align them with the field. This deterministic alignment is counterbalanced by the entropic forces of diffusion, resulting in a stochastic orientation distribution about the direction of the applied field. An analytical model for describing the orientation distribution of particles in presence of deterministic and entropic forces has been previously developed and experimentally verified in the context of alignment of striped metallic microrods (22, 23). However, to analytically model the effect of electric field on proteins in nanoscale, more detailed study combined with molecular dynamics simulation is required.
As mentioned earlier, in our study of the alignment of adsorbed IgG molecules, we used AFM to directly measure the orientation distribution of the molecules. Furthermore, in the context of a protein assay, we used fluorescence detection to indirectly verify the modulation of the orientation of proteins bound to the surface. In this case, using an external lateral electric field during the adsorption process, we may enable antibodies to achieve an orientation state that would favorably enhance the analyte detection capability of the immunoassay as a whole. The enhancement in the capture of analyte (i.e., response of the immunoassay) can in turn be used as a measure of effectiveness of the applied field in controlling the orientation of the immobilized antibody. In this context, the preference basis for the orientation state of the antibody is determined by the interaction and surface chemistry under study. In the stated examples from the literature for the charged surfaces, the Fab fragments on the immobilized antibody were specific and meant to target the analyte of interest in the sample. On the other hand, here, the Fab fragments of the analyte (i.e., the subsequently introduced anti-IgG) are specific to and are meant to target the Fc fragment of the immobilized IgG. Thus, by using a lateral electric field we seek to immobilize the IgG molecule on its side such that the Fc region of the molecule is exposed to the bulk electrolyte, and hence, the incoming fluorescent anti-IgG molecule's Fab region will be able to bind successfully to the immobilized IgG molecule's Fc region.
Results and Discussion
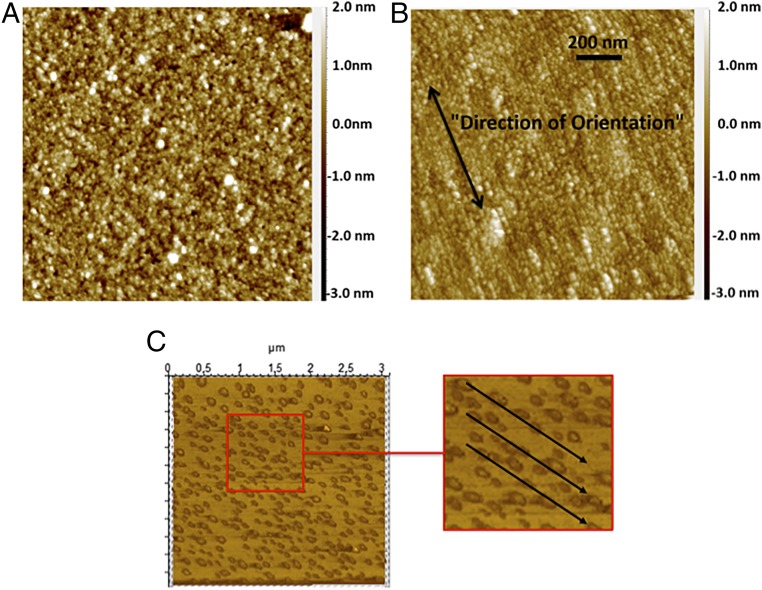
Our AFM images illustrated clear differences between the surfaces with and without electric field. Surfaces without electric fields (Fig. 2A) show images of amorphous films with random orientation. The application of electric field results in AFM images that have a crystalline structure, where structures qualitatively appear to be oriented in straight lines, indicating uniform orientation and lining up of the antibodies during the immobilization step (Fig. 2B). We diluted the IgG sample even further (1,000 times) to be able to image single antibodies. Again, qualitatively the results clearly showed that IgG molecules were uniformly oriented in a single direction (Fig. 2C) as predicted by our hypothesis.
Fig. 2.
Comparison of AFM image of antibody coated surface when the field is (A) off to when the field is (B) on during IgG immobilization step. (C) AFM image of antibody coated surface when a 1,000 times diluted IgG sample was used, indicating alignment at a molecular level. The results indicate that the surface-bound antibodies were oriented in a uniform direction when the field was applied during the immobilization step.
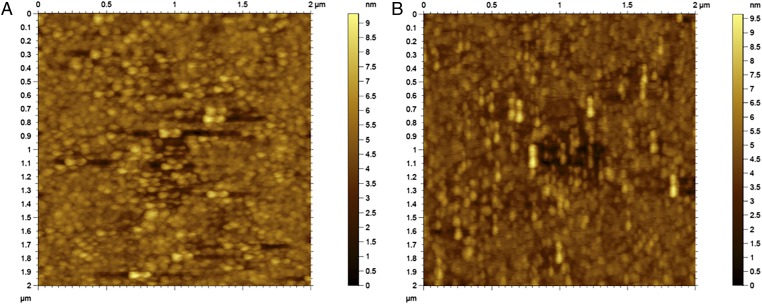
To verify that the orientation effect we observed, was not an artifact of AFM stamping of a single molecule, as control experiments we rotated our images by 90° and reimaged with an AFM. The 90° rotation of the substrate resulted in a 90° rotation of the features on the AFM image, as expected (Fig. 3 A and B). Had the orientation simply been an artifact resulting from an IgG molecule getting attached to the AFM tip, regardless of the orientation of the substrate, we would have observed no change in the orientation of the features on the image. Furthermore, by rescanning the molecule coated surface we validated that the molecules’ arrangement and their orientation do not get disturbed by the AFM tip.
Fig. 3.
Comparison of AFM image of antibody coated surface where the AFM imaging was done at (A) 0° and (B) 90° with respect to the direction of the originally applied 8 V/cm electric field during the IgG immobilization step. This illustrates that the observed orientation of molecules is not due to AFM stamping artifact.
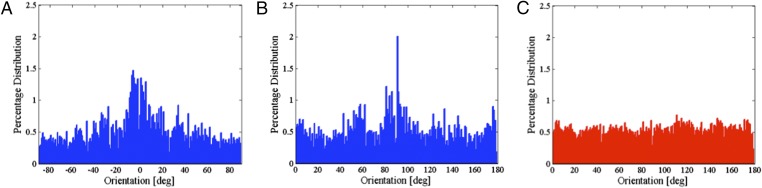
We also quantitatively analyzed the distribution of the orientation of the surface features on the AFM images using commercially available software (Agilent Pico Image software). As can be seen from Fig. 4A, in the case where the field is applied and AFM imaging is done in the direction of the field, a significant portion of the orientation distribution is concentrated at about 0°. Similarly, when the imaging is done at a 90° angle with respect to the applied field, the peak of the distribution takes place at 90° (Fig. 4B). On the other hand, as illustrated in Fig. 4C, in the case with no applied field during IgG immobilization, the distribution of the surface texture orientation is similar to that of a uniform distribution with no distinguished peak.
Fig. 4.
Representative orientation distribution of the texture of the IgG coated surface where the AFM imaging was done at (A) 0° and (B) 90° with respect to the direction of the originally applied 8 V/cm electric field during the IgG immobilization step. (C) Representative orientation distribution of the texture of the IgG coated surface where no field was applied during the IgG immobilization step (control experiment). The results indicate that the orientation of immobilized IgG molecules is similar to that of a uniform distribution, whereas in the case where the field is applied during the immobilization step, the significant portion of the orientation distribution is concentrated about the axis of the applied field.
Our AFM analysis of five samples shows that in the case with field, on average 4.9% of the overall distribution is concentrated within 5° of the peak, which is 75% more than the distribution in a 5° interval for the case of a uniform distribution. Furthermore, our results indicate that in the case with field, on average 50% of the distribution about the peak takes place within a 76° interval, which is a narrower interval than the 50% distribution interval of a uniform distribution (i.e., 90°).
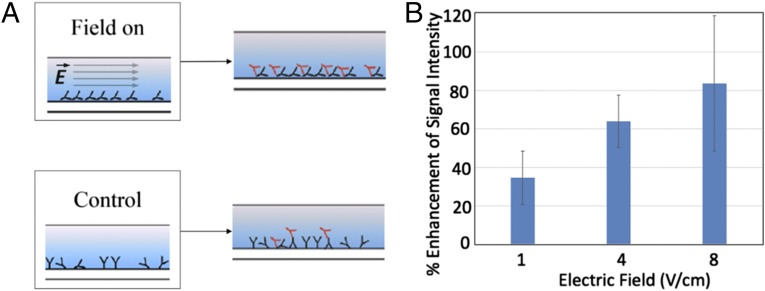
Moreover, we sought to apply the effect of immobilization with controlled orientation to improve the performance of fluorescent affinity-based protein assays. To be able to quantify the effect of the applied field on the response of the immunoassay, we used fluorescently labeled anti-IgG. More specifically, we used fluorescein isothiocyanate (FITC)-labeled anti-IgG to perform a fluorescence assay (Fig. 5A). As opposed to the AFM assay, where the channel was removed before AFM analysis, this assay was performed entirely in a polydimethylsiloxane (PDMS) microchannel.
Fig. 5.
(A) Experimental setup for the fluorescence assay (including the control experiment). The electric field is applied during the IgG immobilization step. The immobilized IgG molecules then target the subsequently injected FITC-labeled anti-IgG molecules. (B) Improvement in signal intensity of the anti-IgG-IgG assay, with respect to the control experiment (no field applied), as a result of applying electric field during the IgG immobilization step. Error bars represent the SE in our measurements.
To compare the difference in the distribution of the assay for the two cases of electric field on and off, we used fluorescence imaging. Here, the case of no electric field served as the control experiment, which was performed with no electrodes inserted in the channel during the immobilization of IgG. The channel for the control experiment was prepared in parallel and adjacent to the channels that were used for the actual experiments. To quantify the effect of the electric field, we used ImageJ software to measure the mean gray value as the signal intensity representing the anti-IgG and IgG bindings in the channel. The experiment corresponding to each electric field strength was repeated at least three times and normalized with respect to the measured signal from the corresponding control channel (SI Appendix). The collective results for the fluorescence assay are presented in Fig. 5B, which shows the improvement in signal intensity (with respect to the control experiment) as we increased the applied electric field.
It is worth noting that before performing the experiments, with the aid of an impedance spectroscope (Zurich Instruments; HF2IS) we measured the low-frequency (dc) resistance of the channel as 400 kΩ, resulting in the current range of 2.5 µA to 20 µA for the applied voltage range of 1 V to 8 V. Given such relatively low voltage values, the effect of electroosmotic flow in transportation of the IgG molecules was negligible. We validated this hypothesis by performing a separate experiment where we injected 2.8 µm beads in our channel during the application of lateral electric field. We observed no flow or migration of the beads, implying that electroosmotic flow was negligible. Furthermore, our calculations based on the IgG transport properties (24) showed that the electrophoretic force at most (8 V/cm case) would contribute to effective IgG transport length of 1.5 mm only. In fact, in our experiments, we did not observe a distribution gradient along the channel to vindicate the significance of the electrophoretic transport phenomenon.
Conclusions
Overall, our results demonstrated the successful use of lateral electric field to control the orientation of the antibodies. We further illustrated the application of the method to enhance the performance of an immunoassay. Our results indicate that we can achieve more than 100% enhancement in signal-to-noise ratio with applied electric field. Although we demonstrated our proof of concept testing the control of orientation of IgG molecules on their side, we emphasize that the findings here can be used for a wide range of proteins and can apply to a wide range of both affinity-based and probe-free sensing platforms. We envision incorporating various different electrode configurations, including patterning faradaic electrodes on the base of the microchannel, to control the field to orient the proteins in various directions, including vertically. Vertical orientation of antibodies during immobilization would be useful for antigen capture assays where the anitbody Fab region should be exposed to the solution to maximize antigen capture rate. Future work will also involve the use of the electronic quantum tunneling sensor (14), undergoing development in our group, to verify the orientation control, based on direct electronic detection of the motif of the protein that gets attached to the sensor surface.
Materials and Methods
Fabrication.
We carried out our experiments using rectangular microchannels to ensure uniform electric field across the channel. The fabricated PDMS-based channels used to perform the experiments were 200 μm wide, 50 μm high, and 1 cm long. The master mold for the channel was patterned onto a silicon substrate using SU-8 photoresist. PDMS (10:1 prepolymer/curing agent) was poured onto the master mold and allowed to cure at 80 °C overnight. The PDMS channel was removed from the mold once it was formed. Then, to create the channel’s inlet and outlet ports for injecting the sample into the channel and inserting electrodes (to apply external electric field), two holes of 3 mm diameter were punched, one at each end. Finally, the PDMS channel was bonded to a micro slide (Corning) after oxygen plasma treatment.
Sample Preparation and Surface Chemistry.
To prepare the channel surface, goat IgG (originally 0.02 mg/mL) was injected into the channel and allowed to incubate for 20 min so that the IgG molecules physically adsorbed onto the channel surface. This surface chemistry was used both for AFM imaging and fluorescence assay response experiments.
For the fluorescence assay, to eliminate nonspecific binding, 1 mg/mL BSA introduced in the channel. This was followed by flushing the channel and injecting the sample FITC-labeled mouse anti-goat IgG (originally 1 mg/mL, diluted 30 more times by PBS). To test for binding specificity, on a separate chip, the above steps were performed minus the immobilization of IgG, which as expected minimized binding of anti-IgG due to nonspecific binding.
Experiment Setup.
To establish lateral electric field inside the channel, we inserted Ag-AgCl pellet electrodes (In Vivo Metric) at the inlet and outlet ports of the microchannel. Using a signal generator (Agilent; 33220A) we excited the electrodes with dc voltages up to 8 V. To characterize the channel resistance we used an impedance spectroscope (Zurich Instruments; HF2IS) and a transimpedance amplifier (Zurich Instruments; HF2TA).
Supplementary Material
Acknowledgments
The authors thank Yang Liu, Klint A. Rose, and Juan G. Santiago for sharing knowledge and expertise with regard to the reported findings. Fabrication of the devices was performed in the Stanford Nanofabrication Facility. This work was supported by the Defense Advanced Research Projects Agency Grant DARPA-ICLF10-56 and National Institutes of Health Grant P01 HG000205.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424592112/-/DCSupplemental.
References
- 1.Kim Y-G, Moon S, Kuritzkes DR, Demirci U. Quantum dot-based HIV capture and imaging in a microfluidic channel. Biosens Bioelectron. 2009;25(1):253–258. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol. 2003;7(1):55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 3.Javanmard M, et al. Electrical detection of protein biomarkers using bioactivated microfluidic channels. Lab Chip. 2009;9(10):1429–1434. doi: 10.1039/b818872f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok J, Mindrinos MN, Davis RW, Javanmard M. Digital microfluidic assay for protein detection. Proc Natl Acad Sci USA. 2014;111(6):2110–2115. doi: 10.1073/pnas.1323998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, et al. Nanomagnetic competition assay for low-abundance protein biomarker quantification in unprocessed human sera. J Am Chem Soc. 2010;132(12):4388–4392. doi: 10.1021/ja910406a. [DOI] [PubMed] [Google Scholar]
- 6.Mulvaney SP, Myers KM, Sheehan PE, Whitman LJ. Attomolar protein detection in complex sample matrices with semi-homogeneous fluidic force discrimination assays. Biosens Bioelectron. 2009;24(5):1109–1115. doi: 10.1016/j.bios.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Osterfeld SJ, et al. Multiplex protein assays based on real-time magnetic nanotag sensing. Proc Natl Acad Sci USA. 2008;105(52):20637–20640. doi: 10.1073/pnas.0810822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Chen Y, Hassibi A, Scherer A, Hajimiri A. 2009. A Frequency-Shift CMOS Magnetic Biosensor Array with Single-Bead Sensitivity and No External Magnet, IEEE International Solid-State Circuits Conference (ISSCC, San Francisco) Dig Tech Papers, pp 438–439.
- 9.Squires TM, Messinger RJ, Manalis SR. Making it stick: Convection, reaction and diffusion in surface-based biosensors. Nat Biotechnol. 2008;26(4):417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 10.Xu Z, Chen Y, Gartia MR, Jiang J, Liu GL. Surface plasmon enhanced broadband spectrophotometry on black silver substrates. Appl Phys Lett. 2011;98(24):241904–241904-3. [Google Scholar]
- 11.Peluso P, et al. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal Biochem. 2003;312(2):113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 12.Kanno S, Yanagida Y, Haruyama T, Kobatake E, Aizawa M. Assembling of engineered IgG-binding protein on gold surface for highly oriented antibody immobilization. J Biotechnol. 2000;76(2-3):207–214. doi: 10.1016/s0168-1656(99)00186-8. [DOI] [PubMed] [Google Scholar]
- 13.Jeanmaire DL, Van Duyne RP. Surface Raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J Electroanal Chem Interfacial Electrochem. 1977;84(1):1–20. [Google Scholar]
- 14.Lu Y, Liu GL, Lee LP. High-density silver nanoparticle film with temperature-controllable interparticle spacing for a tunable surface enhanced Raman scattering substrate. Nano Lett. 2005;5(1):5–9. doi: 10.1021/nl048965u. [DOI] [PubMed] [Google Scholar]
- 15. Gupta C, et al. (2012) Electrochemical Quantum Tunneling for Electronic Detection and Characterization of Biological Toxins, Proceedings of SPIE Defense, Security and Sensing (Baltimore), p 837303.
- 16.Chang S, et al. Electronic signatures of all four DNA nucleosides in a tunneling gap. Nano Lett. 2010;10(3):1070–1075. doi: 10.1021/nl1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Liu GL, Kim J, Mejia YX, Lee LP. Nanophotonic crescent moon structures with sharp edge for ultrasensitive biomolecular detection by local electromagnetic field enhancement effect. Nano Lett. 2005;5(1):119–124. doi: 10.1021/nl048232+. [DOI] [PubMed] [Google Scholar]
- 18.Talasaz AH, et al. Prediction of protein orientation upon immobilization on biological and nonbiological surfaces. Proc Natl Acad Sci USA. 2006;103(40):14773–14778. doi: 10.1073/pnas.0605841103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polk BJ, Stelzenmuller A, Mijares G, MacCrehan W, Gaitan M. Ag/AgCl microelectrodes with improved stability for microfluidics. Sens Actuators B Chem. 2006;114(1):239–247. [Google Scholar]
- 20.Zhou J, Tsao H-K, Sheng Y-J, Jiang S. Monte Carlo simulations of antibody adsorption and orientation on charged surfaces. J Chem Phys. 2004;121(2):1050–1057. doi: 10.1063/1.1757434. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Liu L, Zhou J, Jiang S. Controlling antibody orientation on charged self-assembled monolayers. Langmuir. 2003;19(7):2859–2864. [Google Scholar]
- 22.Rose KA, Meier JA, Dougherty GM, Santiago JG. Rotational electrophoresis of striped metallic microrods. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;75(1 Pt 1):011503. doi: 10.1103/PhysRevE.75.011503. [DOI] [PubMed] [Google Scholar]
- 23.Rose KA, Hoffman B, Saintillan D, Shaqfeh ESG, Santiago JG. Hydrodynamic interactions in metal rodlike-particle suspensions due to induced charge electroosmosis. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79(1 Pt 1):011402. doi: 10.1103/PhysRevE.79.011402. [DOI] [PubMed] [Google Scholar]
- 24.Berk DA, Yuan F, Leunig M, Jain RK. Fluorescence photobleaching with spatial Fourier analysis: Measurement of diffusion in light-scattering media. Biophys J. 1993;65(6):2428–2436. doi: 10.1016/S0006-3495(93)81326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.