Significance
In cyanobacteria, genome-wide transcriptional oscillations and cell division are under the control of the global response regulator RpaA. Here we show that regulator of phycobilisome-associated B (RpaB), a closely related response regulator that is essential in the model cyanobacterium Synechococcus elongatus PCC 7942, is involved in complex regulatory interactions with the RpaA signaling system. RpaB influences circadian timing and cell dimensions by modulating the RpaA∼P levels and by controlling a subset of the RpaA target genes. Our results suggest that regulatory interactions between RpaA and RpaB are required for transmission of updated time information to the level of gene expression, paving the way to uncover regulatory mechanisms involved in integration of temporal and environmental information.
Keywords: signaling network, transcription regulation, chronobiology
Abstract
The response regulator RpaB (regulator of phycobilisome associated B), part of an essential two-component system conserved in cyanobacteria that responds to multiple environmental signals, has recently been implicated in the control of cell dimensions and of circadian rhythms of gene expression in the model cyanobacterium Synechococcus elongatus PCC 7942. However, little is known of the molecular mechanisms that underlie RpaB functions. In this study we show that the regulation of phenotypes by RpaB is intimately connected with the activity of RpaA (regulator of phycobilisome associated A), the master regulator of circadian transcription patterns. RpaB affects RpaA activity both through control of gene expression, a function requiring an intact effector domain, and via altering RpaA phosphorylation, a function mediated through the N-terminal receiver domain of RpaB. Thus, both phosphorylation cross-talk and coregulation of target genes play a role in the genetic interactions between the RpaA and RpaB pathways. In addition, RpaB∼P levels appear critical for survival under light:dark cycles, conditions in which RpaB phosphorylation is environmentally driven independent of the circadian clock. We propose that the complex regulatory interactions between the essential and environmentally sensitive NblS-RpaB system and the SasA-RpaA clock output system integrate relevant extra- and intracellular signals to the circadian clock.
The genomes of cyanobacteria, phototrophic organisms that perform oxygenic photosynthesis, contain more genes that encode signal-transducing two-component systems (TCS) per unit genome size than other bacteria (1). TCSs are widely used in signal transduction and adaptation to environmental changes in bacteria, archaea, lower eukaryotes, and higher plants (2). In the prototypical system, the sensor histidine (His) protein kinase autophosphorylates on a conserved His residue and transfers this phosphoryl group to an aspartate (Asp) of a cognate response regulator. The Asp is a conserved feature of an N-terminal phosphoacceptor receiver (sensor) domain, and its phosphorylation transmits conformational changes to a C-terminal effector (output, often DNA-binding) domain, modifying its affinity for target genes. Finally, the response regulator is dephosphorylated by its own phosphatase activity or by the cognate His kinase (3). Multidomain signaling components, phosphoacceptor intermediaries, “branched” pathways where one His kinase recognizes several response regulators, and “orphan” components with unknown partners or ancillary regulatory proteins increase the complexity of this basic scenario by allowing integration of multiple signals and generating sophisticated signaling networks similar to those found in eukaryotes (4, 5).
Consistent with the transient nature of most signals that regulate them, the great majority of TCSs are not required for survival under laboratory conditions. However, the most conserved TCS of cyanobacteria, the NblS (nonbleaching sensor)-RpaB (regulator of phycobilisome-associated B) pair and its specific phosphorylatable His or Asp residues, are essential for viability (6) in Synechococcus elongatus PCC 7942 (hereafter S. elongatus), the model for cyanobacterial circadian rhythm studies. RpaB binds to high light regulatory 1 (HLR1) sequences (7) to control a large number of genes in response to a variety of signals, including changes in light intensity (8, 9). The abundance of the OmpR/PhoB-like response regulator RpaB remains constant under all tested conditions (10–13), but the ratio of phosphorylated (RpaB∼P) to nonphosphorylated RpaB is transiently modified by changes in light intensity (12). These and other findings (6) argue against RpaB acting by a simplistic on/off model in which the protein binds to target promoters only when it is phosphorylated, as proposed for OmpR/PhoB-like response regulators that respond to discrete signals. Recently, RpaB has also been implicated in the control of cell dimensions, for which nonphosphorylated RpaB appears to be the main player (13), and in the transcriptional oscillation of clock-regulated genes (10).
The cyanobacterial circadian clock generates genome-wide transcriptional oscillations and regulates the timing of cell division (14–16) using the TCS SasA-RpaA (17). Phosphorylated RpaA regulates the expression of clock genes, generating feedback on the core oscillator and on additional circadian effectors involved in orchestration of genome-wide transcriptional rhythms (18). Despite the known importance of environmental signals in resetting and adjusting the clock, little is known of the pathways that integrate circadian and environmental signals to modulate gene expression, or how the abundant and essential regulator RpaB affects clock output signaling. Here, we demonstrate regulatory interactions between RpaA and RpaB that affect clock gene expression, as well as cell dimensions and viability, showing that: (i) the paradigmatic RpaA-regulated genes kaiBC and purF are under negative control by RpaB, (ii) RpaB affects gene expression and the timing of cell division by altering RpaA phosphorylation as well as by RpaA-independent mechanisms, (iii) the RpaB∼P/RpaB ratio undergoes clock-independent oscillations during light:dark (LD) cycles, and (iv) the effect of RpaB and RpaB variants on viability of rpaA-null mutants differs between constant light (LL) and LD conditions.
Results and Discussion
RpaB Is Involved in Circadian Expression of kaiBC and purF Genes.
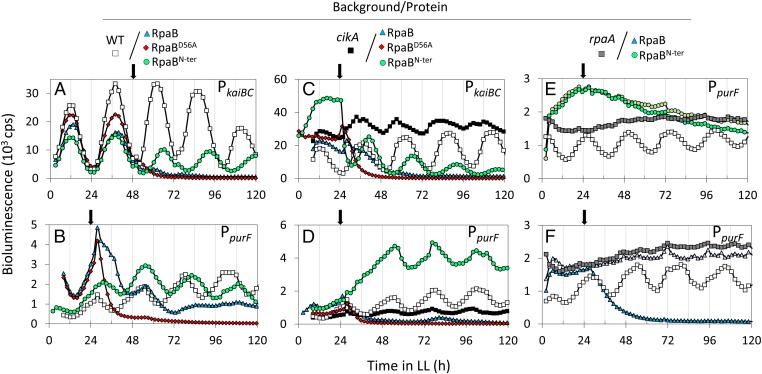
Coregulation of RpaA target genes by RpaB has already been proposed. In particular, RpaB was shown to bind to the class 1 kaiBC promoter in a circadian fashion (10), presumably at the HLR1 element that overlaps with the recently described RpaA-binding site (18) (Fig. S1A). We used PkaiBC::luc or PpurF::luc reporter fusions to examine the circadian effects of overexpressing RpaB from an isopropyl β-d-1 thiogalactopyranoside (IPTG)-inducible promoter on the paradigmatic kaiBC (class 1, dusk-peaking) and purF (class 2, dawn-peaking) promoters (Fig. 1 A and B and Fig. S1). RpaB overexpression rapidly depleted the bioluminescence signal from both PkaiBC::luc and PpurF::luc (Fig. 1 A and B), suggesting that RpaB represses both promoters, at least when present at excess. Overexpression of a derivative in which the receiver domain cannot be phosphorylated, RpaBD56A, had the same effect as WT RpaB, consistent with expected direct interaction of the DNA-binding domain at the kaiBC and purF promoters (Fig. 1 A and B). Fluorescence staining using LIVE/DEAD BacLight showed that cells remain viable for at least 4 d following RpaB induction (Fig. S2), suggesting that the signal loss reflects repression of the promoter rather than cell death. These data led us to investigate the mechanisms by which RpaA and RpaB may coregulate genes in the wider regulons in which each has been implicated (6, 12, 13, 18), and to understand how RpaB domains contribute to the observed phenotypes.
Fig. 1.
Effect of overexpressing RpaB, RpaBD56A, or RpaBN-ter on the rhythmic expression of kaiBC and purF. Bioluminescence activity of PkaiBC::luc or PpurF::luc reporters was assayed in the WT (A and B), cikA (C and D), and rpaA (E and F) genetic backgrounds. The arrows indicate the time of IPTG addition. For blue and green colors, dark and light correspond, respectively, to the activity assayed with and without IPTG. Averaged bioluminescence traces are shown. cps, counts per second.
Overexpression of the RpaB Receiver Domain Decreases Cell Size, but Not the RpaB∼P/RpaB Ratio.
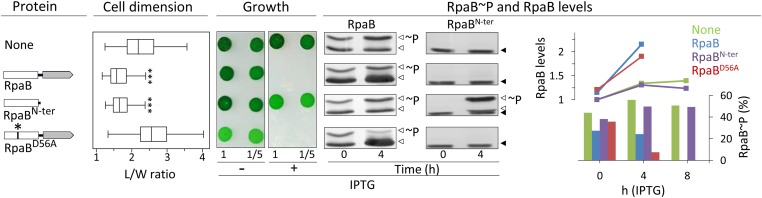
The phenotypes of decreased length/width (L/W) and arrested cell division that occur when RpaB is overexpressed (13) provided a simple assay for the independent contributions of the RpaB receiver and effector domains to RpaB function. We characterized strains that express RpaB, RpaBD56A, or a variant composed of only the receiver domain (RpaBN-ter) (Table 1). Strikingly, overproduction of RpaBN-ter, but not of RpaBD56A, altered cell dimensions in the same way as full-length RpaB (Fig. 2 and Fig. S3), indicating that a functional receiver is all that is required to generate this phenotype, independently of the DNA-binding domain. On the other hand, overproduction of RpaBD56A, which has an intact DNA-binding domain but a nonfunctional receiver, dramatically suppressed growth, whereas RpaBN-ter had no effect. Thus, culture arrest requires the effector (DNA-binding) domain, and is independent of the RpaB effect on the receiver-dependent L/W ratio.
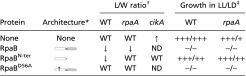
Table 1.
Overexpression phenotypes of RpaB variants in WT and clock mutants
 |
Schematic representation of the RpaB derivatives. Point mutation D56A (asterisk), receiver (white rectangle) and output (gray rectangle) domains are shown.
WT, WT L/W ratio; ↑ and ↓ indicate increase and decrease, respectively. ND, not determined.
The plus and minus signs indicate relative growth based on visual analysis from no growth (−) to WT growth (+++).
Fig. 2.
Increased levels of RpaB affect cell dimensions, growth, and RpaB phosphorylation ratio. Schematic representation of the RpaB derivatives overexpressed in the experiments shown to the right. Receiver and output domains are shown as rectangles and arrowheads, respectively, and the D56A point mutation is indicated with an asterisk (*). Results from control and overexpressing strains are always shown in the same order. Cell dimension: Boxplot representation of cell L/W ratio. Statistically significant differences to the (none) control in a Mann–Whitney–Wilcoxon Test are indicated (***P < 10−15). Growth: Strains were plated on BG11 at the dilutions indicated below and cultured in LL. RpaB and RpaB∼P levels: Representative immunodetections of RpaB and RpaBN-ter and their phosphorylated forms (∼P) from cells collected before (time 0) or 4 h (time 4) after adding IPTG are indicated with empty arrowheads. The position of a nonspecific band is indicated with solid arrowheads. Values of the quantified bands, alongside values for the indicated 8-h time points, were plotted. Curves (Upper) and bars (Lower) correspond, respectively, to total RpaB (RpaB + RpaB∼P) and RpaB∼P. RpaB levels are plotted relative to that in the control before inducer addition (set arbitrarily to 1).
We previously found a simple correlation between the levels of nonphosphorylated (and total) RpaB levels and cell dimensions, with high RpaB levels correlating with low L/W ratios (13). As shown in Fig. 2, overexpression of RpaBN-ter, which had no effect on the RpaB∼P/RpaB ratio, affected cell dimensions, whereas overexpression of nonphosphorylated RpaBD56A, which decreased the RpaB∼P levels, did not affect cell L/W. Therefore, the results indicate that a specific function of intact receiver domain is the factor that affects dimensions.
The finding that RpaB, and to a much greater extension RpaBD56A, decreased the RpaB∼P/RpaB ratio was in line with previous work showing that relatively small increases in the amount of RpaB had a negative impact on the RpaB∼P levels and suggests a complex regulation of the relative levels of the phosphorylated and nonphosphorylated forms (13). In addition, the drastic decrease of RpaB∼P levels after RpaBD56A overexpression suggests the ability of mutant polypeptides to associate with RpaB and prevent phosphorylation of heterodimers.
The lack of effect of RpaBN-ter overexpression on native RpaB∼P/RpaB challenged the assumption that overexpressed receiver domains alter phenotypes by competing with full-length response regulators for phosphorylation by the cognate His kinase. Our results suggest that overexpressed RpaB receiver affects phenotypes such as altered cell dimensions, or the previously described induction of RpaB-repressed targets, such as hliA and rpoD3 (19), by a mechanism other than by decreasing the RpaB∼P/RpaB ratio or overall RpaB levels. Such alternative mechanisms include that the overexpressed RpaBN-ter decreases the levels of functional (presumably homodimeric) RpaB, perhaps by formation of heterodimers between the overexpressed receiver and the full-length protein, or has an indirect effect on RpaB activity.
Overexpressed RpaB Receiver Domain Inhibits RpaA Phosphorylation.
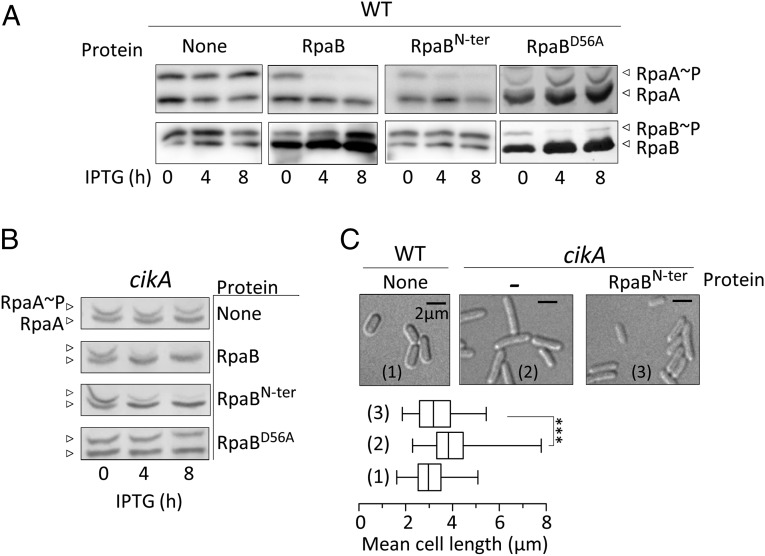
Because high levels of RpaA∼P or of a mutant RpaA derivative that mimics RpaA∼P (17) produces elongated cells (14, 18), an opposite phenotype to that caused by overexpression of RpaB or RpaBN-ter, we assessed whether the cell-size phenotypes observed in Fig. 2 also reflected abnormal intracellular levels of RpaA∼P. As shown in Fig. 3A, whereas overexpression of RpaBD56A had no impact, both RpaB and RpaBN-ter dramatically decreased RpaA∼P levels, which were almost undetectable 4 h after IPTG induction. Therefore, the receiver domain of RpaB can affect the RpaA phosphorylation pathway in S. elongatus.
Fig. 3.
Overexpression of RpaBN-ter interferes with RpaA phosphorylation and suppresses a cikA phenotype. RpaB (A) and RpaA (A and B) Phos-tag analysis of the indicated strains, expressed RpaB variant, and time points. Cells were entrained to two 12:12 LD cycles and IPTG was added at ZT 4 (A) or ZT 12 (B). (C) Mean cell length from at least 200 measurements for each strain, micrographs showing cell appearance. Error bars show SEM. ***P < 10−3 (one-way ANOVA).
Because the clock-related kinase CikA is an RpaA∼P phosphatase (20), a potential mechanism for cross-talk might be through interaction of the RpaB receiver with CikA. Thus, we determined the effect of overproducing RpaB, RpaBN-ter, or RpaBD56A in cikA-null mutants. As shown in Fig. 3B, overexpression of RpaBN-ter or RpaB, but not of RpaBD56A, produced a significant decrease in the RpaA∼P/RpaA ratio in the cikA background, indicating that the ability of RpaB to decrease RpaA∼P is not mediated by CikA.
A high RpaA∼P level in cikA-null cells blocks cell division and results in an elongated phenotype that can be suppressed by rpaA disruption (14) or by prevention of RpaA phosphorylation (18). To test whether the cell-size phenotypes of RpaB- or RpaBN-ter-overexpressing cells are the consequence of their impact on the RpaA∼P/RpaA ratio, we tested the ability of RpaBN-ter to suppress the cikA elongated-cell phenotype. As shown in Fig. 3C and Table 1, the overexpression of RpaBN-ter caused a modest, but significant, decrease in the mean length of cikA-null cells. Taken together, the results suggest that both CikA and RpaB affect cell dimensions by altering RpaA∼P levels. RpaB (and RpaBN-ter, but not RpaBD56A) may impair the RpaA∼P/RpaA ratio by interfering with the SasA-dependent phosphorylation of RpaA, an idea supported by in vitro phosphotransfer data (20, 21).
In line with the ability of the RpaB receiver to counteract RpaA∼P levels and functions, RpaBN-ter overexpression affected the amplitude and period lengths of the PkaiBC::luc expression rhythms, with no significant change in that of PpurF::luc (Fig. 1 A and B). However, in a cikA-null background (Fig. 1 C and D), RpaBN-ter overexpression resulted in significant respective decrease and increase of PkaiBC::luc and PpurF::luc expression, and in partial recovery of the amplitude of the rhythms that are characteristically damped when CikA is absent.
In an rpaA-null background, where PpurF::luc bioluminescence is constitutive at levels corresponding to circadian peaks, RpaBN-ter overexpression did not alter the PpurF::luc bioluminescence levels or pattern. However, overexpression of RpaB brought the signal to almost zero (Fig. 1 E and F). These findings confirm that, whereas the phenotypic effects of RpaBN-ter require functional RpaA, the full-length protein performs RpaA-independent functions, presumably via binding promoters through the output domain.
The RpaB Effector Domain Antagonizes RpaA Regulation of Cell Division and Growth.
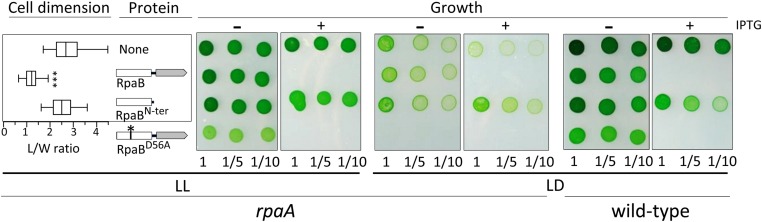
We inactivated rpaA in strains that overexpress RpaB or its variants and measured cell dimensions from the corresponding cultures. Consistent with the hypothesis that the receiver domain of RpaB affects cell phenotype by decreasing the RpaA∼P/RpaA ratio, overexpression of RpaBN-ter did not affect L/W in the rpaA-null background (Fig. 4, Table 1,and Fig. S3). However, overexpression of full-length RpaB, but not of RpaBD56A, dramatically decreased the L/W ratio, suggesting that intact phospho-accepting receiver and the effector domain are together necessary for RpaA-independent regulatory mechanisms.
Fig. 4.
Overexpression of RpaBN-ter restores growth and normal cell dimensions to rpaA-null strains. Panels show the phenotypic impact of ectopic overexpression of RpaB and derivatives in rpaA mutants under LL and LD conditions. Additional controls and details as in Fig. 2.
The known LD growth defect of rpaA-null mutants led us to examine the role of RpaB in this phenotype as well. We compared the effects of ectopic expression of RpaB derivatives on the growth of S. elongatus cultures in both LL and LD cycles. As observed for the WT, overexpression of RpaB or RpaBD56A in the rpaA background prevented growth in the presence of the inducer in both LL and LD (Figs. 2 and 4, and Table 1). However, overexpression of RpaBN-ter partially rescued the LD growth defects of the rpaA-null mutants (Fig. 4). This result is consistent with the hypothesis that RpaBN-ter forms nonfunctional heterodimers with the endogenous RpaB, lowering RpaB levels and compensating for the absence of its antagonist, RpaA, under LD. Even without inducer, the rpaA strain with basal ectopic expression of RpaBD56A was more impaired in growth than the rpaA-null parent, indicating that low levels of RpaBD56A suffice to increase the severity of the rpaA phenotype. This unexpected result, combined with the very low RpaB∼P/RpaB ratio detected in rpaA-mutants overexpressing RpaBD56A (Fig. S4), suggested that RpaB∼P or a relatively high RpaB∼P/RpaB ratio might be particularly important under LD cycles, raising the question of whether RpaB phosphorylation is also subject to LD oscillations.
RpaB Phosphorylation Oscillates in LD Cycles in a Clock-Independent Manner.
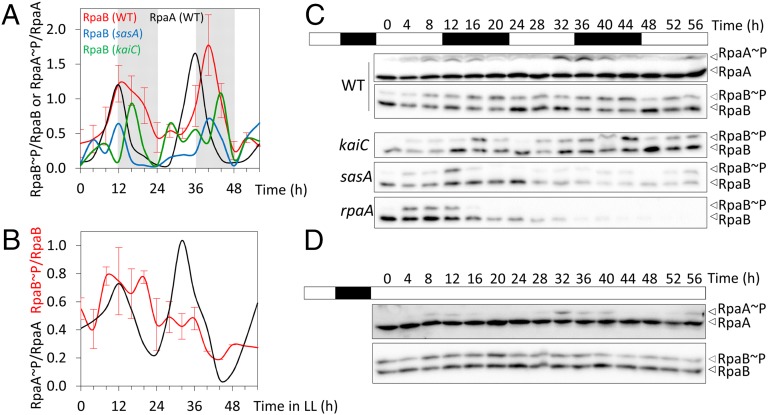
To determine whether the RpaB∼P/RpaB ratio changes during LD cycles, we compared cell extracts from synchronized cultures released in LL with those of cultures maintained in LD cycles. A reproducible cyclic pattern of RpaB phosphorylation was observed in LD cycles (Fig. 5 A and C) peaking at 12–14 h after lights on (zeitgeber time, ZT 12–14) with a trough at ZT 24, but not under LL (Fig. 5 B and D), indicating that the oscillations on the RpaB∼P/RpaB ratio are dependent on environmental changes and not on the clock. Control samples assayed in parallel showed RpaA∼P cycles in LD, as well as the previously reported cyclic pattern of phosphorylation in LL (18, 20) with RpaA∼P peaks at subjective dusk (8–12 h after lights on) and troughs at subjective dawn. The oscillations in the ratio of RpaB phosphorylation were less pronounced than for RpaA in LD, and RpaB∼P peaked several hours later than RpaA∼P.
Fig. 5.
RpaB phosphorylation is clock-independent and oscillates in LD. RpaA∼P/RpaA and RpaB∼P/RpaB phosphorylation profiles from synchronized cultures of indicated backgrounds released in LD (A) and LL (B). Mean values with SE are shown for RpaB∼P quantification in WT (n = 4 and 2 for LD and LL, respectively). Representative blots are shown in C and D.
To address potential circadian influence on the cyclic pattern of RpaB phosphorylation in LD we analyzed the RpaB∼P/RpaB ratio in strains in which the clock oscillator or output pathway is interrupted by inactivation of kaiC, sasA, or rpaA (Fig. 5 A and C). Because all three mutants retained cyclic oscillations of the RpaB∼P/RpaB ratio in LD, we concluded that these cycles are driven by the LD cycle. However, the phase relationship of RpaB∼P peaks to LD cues appears to be altered in rpaA mutants. Although the absolute levels of RpaB and RpaA are constant in WT S. elongatus (10–12, 20), RpaB levels dropped dramatically in LD in the absence of a functional SasA-RpaA pathway, suggesting that the clock output pathway is required to maintain the physiological levels of RpaB in LD cycles.
The clock-independent oscillations of the RpaB∼P levels during LD cycles indicated that the potential for RpaB to inhibit RpaA∼P levels varies depending on environmental conditions, and supports the physiological relevance of cross-talk between the environmentally sensitive NblS-RpaB TCS and the clock output system as a means of integrating external and intracellular information.
A Model for RpaB and RpaA Interactions in Connection with the Cyanobacterial Circadian Program.
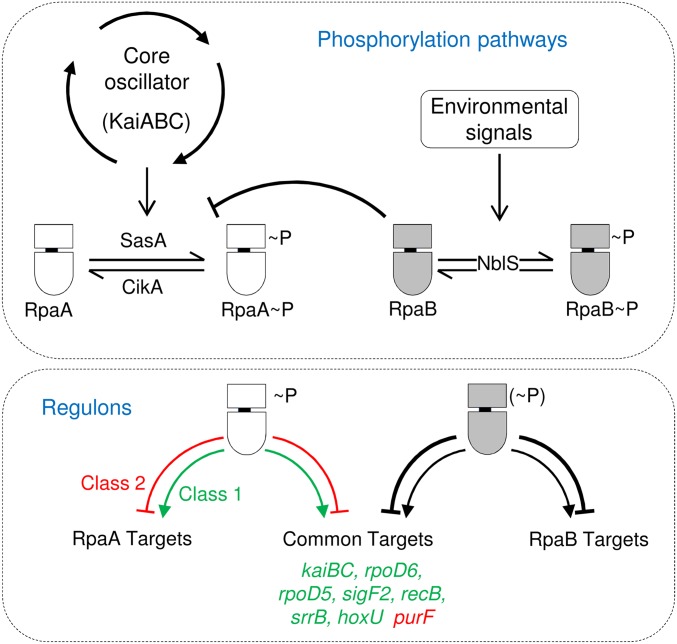
This work establishes RpaB as a modulator of the master clock regulator RpaA. RpaB influences RpaA at different regulatory levels. We propose that transient RpaB dephosphorylation in response to environmental cues would be expected to change the RpaA∼P/RpaA ratio, which in turn would affect RpaA∼P-mediated regulation of cell division and gene expression. Additionally, RpaB (and RpaB∼P presumably to a greater extent) would also bind to a subset of promoters in the RpaA regulon, including kaiBC and purF, as well as to target genes important for functions that affect viability and cell dimensions. The coincidence of RpaA and RpaB binding sites at the paradigmatic class 1 and 2 promoters (Fig. S1) suggests how RpaB directly contributes to the diverse array of dynamic transcriptional responses in which RpaA∼P is the main player (18): directly, by modulating the RpaA∼P levels, and indirectly, by binding to the same promoters (Fig. 6).
Fig. 6.
Model for RpaB contribution to circadian transcriptional responses. Kai oscillator transduces time information into RpaA phosphorylation via SasA and CikA, producing oscillations in RpaA∼P (20). In response to environmental cues, NblS controls phosphorylation of RpaB. In turn, RpaB would affect RpaA activity by decreasing the RpaA∼P levels (by competition for SasA kinase, a function mapping to the RpaB receiver) and by binding to common gene targets (a function requiring the RpaB effector domain). Only class 1 (green) and 2 (red) genes for which there is experimental evidence of being controlled by both regulators are listed. RpaA∼P and RpaB(∼P) represent the different requirement of phosphorylation for response regulator binding to promoters [RpaA (18), and RpaB (10)]. The different thickness of traces from RpaB(∼P) indicates that RpaB appears to function mainly as a repressor.
Because circadian binding of RpaB to kaiBC and other target genes (10) has been observed in LL conditions in which RpaB∼P levels do not show circadian oscillations (Fig. 5), it is tempting to propose the involvement of the circadian oscillations of RpaA∼P in that phenomenon. The strict dependence on phosphorylation of RpaA, but not of RpaB, for binding to target promoters (18) and the overlap between the binding sites of the two regulators at relevant targets support this additional scenario for regulatory interactions. In summary, it appears that the transmission of updated time information to the level of gene expression lies in multiple and complex regulatory interactions between the response regulators RpaA and RpaB, and that their cooperation serves to integrate temporal and environmental information.
Materials and Methods
Growth Conditions.
S. elongatus strains were routinely grown photoautotrophically at 30 °C under LL conditions (∼30 µE⋅m−2⋅s−1) or subjected to alternating 12-h LD cycles. BG11 medium (22) was used for all cyanobacterial culture (BG110 plus 17.5 mM NaNO3 and 10 mM Hepes/NaOH pH 7.8). Cell density was measured at OD750nm (Ultrospec 2100 pro; Amersham Biosciences). For growth on plates, 1% (wt/vol) agar was added. S. elongatus strains were transformed as previously described (23), and transformants were selected on antibiotic-containing BG11 plates (2 µg/mL gentamycin; 5 µg/mL streptomycin). When needed, BG11 media were supplemented with IPTG to a final concentration of 1 mM. To test growth on solid media, exponentially growing cultures were adjusted to 0.5 (OD750nm) before dropping 5 µL of the cell suspensions onto BG11 plates with or without IPTG and incubated in LL (5 d) or in LD (9 d).
Construction of Plasmids and Strains.
All cloning procedures were carried out in Escherichia coli DH5α using standard techniques, and all generated constructs were analyzed by automated dideoxy DNA sequencing. Strains and plasmids are listed in Table S1. Oligonucleotides are listed in Table S2. To obtain overexpression constructs of RpaBN-ter (residues 1–136) or RpaBD56A, PCR amplification products from pUAGC758 (with primers RpaB-PTRC-1F and RpaB-HCN-2R) or pUAGC773 (with primers RpaB-PTRC-1F and RpaB-HCN-1R), were cloned into EcoRI-BamHI sites of pUAGC280, yielding pUAGC283 and pUAGC284, respectively. Strains carrying Ptrc constructs were obtained following transformation with pUAGC280, pUAGC282, pUAGC283, and pUAGC284. Mutants null for rpaA, sasA, or cikA were obtained by transformation with pAM4523, pAM2176, and pAM2152, respectively.
Bioluminescence Measurements.
Bioluminescence assays were conducted as described previously (24–28). Cultures were synchronized in LD for 2 d, and then returned to LL for bioluminescence monitoring using a Packard TopCount luminometer (PerkinElmer Life Sciences). When appropriate, 1 mM of IPTG was added to each sample well after the daily first cycle. In the absence of inducer, Ptrc-strains show WT activity for both PkaiBC::luc or PpurF::luc (Fig. S5).
Data were analyzed using the BRASS (Biological Rhythms Analysis Software System; A. J. Millar laboratory, University of Edinburgh, Scotland, United Kingdom, millar.bio.ed.ac.uk/PEBrown/BRASS/BrassPage.htm) import and analysis program with Microsoft Excel. Data shown for each strain are the averages of at least 6 separate wells from a 96-well plate.
Protein Sample Preparation and Phos-Tag Acrylamide Gels.
Protein sample preparation, Phos-tag acrylamide gel preparation, and immunoblot analysis were carried out essentially as described previously (29), except 5% (wt/vol) nonfat dry milk/Tris-buffered saline was used for the blocking step and a tank buffer system was used to transfer proteins to PVDF membranes. Samples of 10 or 30 μg of protein extract, respectively, were used for RpaB or RpaA immunodetections. Primary antibody dilutions were 1:2,500 (RpaA) and 1:20,000 (RpaB), and goat anti-rabbit-HRP was used as secondary antibody. Immunoreactive bands were detected using either SuperSignal West Femto Maximum Sensitivity Substrate and detection in an Alpha Innotech FluorChem HD2 Imaging System (Alpha Innotech) (chemiluminescence), or Pierce ECL2 Western blotting substrate and detection in a Typhoon 9410 fluorescence imaging scanner system (GE Healthcare) with a 488 nm/520BP40 laser/filter (fluorescence), always according to the manufacturer’s instructions. Bands were quantified by densitometry analysis of images with National Institutes of Health ImageJ software (30).
Microscopy.
Cell cultures at OD750nm = 0.5 were exposed to IPTG for 72 h and then mounted on 1% low-melting point agarose pads for microscopy. Cell autofluorescence confocal microscopy and image analysis were done as described previously (12). For fluorescence microscopy, cells were imaged as described previously (31).
Supplementary Material
Acknowledgments
The authors thank C. V. Racovac for technical and experimental support; J. I. Labella for help with data analysis; and Susan E. Cohen for critical reading of the manuscript. This work was supported by Spanish Ministry of Economy and Competitivity Grant BFU2012-33364 (to A.C.); Generalitat Valenciana Grants ACOMP/2014/144 (to A.C.) and GV/2014/073 (to J.E.); National Institutes of Health Grant R01GM062419 (to S.S.G.); and European Molecular Biology Organization postdoctoral Fellowship ASTF 74–2013 (to J.E.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424632112/-/DCSupplemental.
References
- 1.Ashby MK, Houmard J. Cyanobacterial two-component proteins: Structure, diversity, distribution, and evolution. Microbiol Mol Biol Rev. 2006;70(2):472–509. doi: 10.1128/MMBR.00046-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Mack TR, Stock AM. Bacterial response regulators: Versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32(5):225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 4.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 5.Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22(19):2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-Redondo ML, et al. Environmental control of phosphorylation pathways in a branched two-component system. Mol Microbiol. 2010;78(2):475–489. doi: 10.1111/j.1365-2958.2010.07348.x. [DOI] [PubMed] [Google Scholar]
- 7.Kappell AD, van Waasbergen LG. The response regulator RpaB binds the high light regulatory 1 sequence upstream of the high-light-inducible hliB gene from the cyanobacterium Synechocystis PCC 6803. Arch Microbiol. 2007;187(4):337–342. doi: 10.1007/s00203-007-0213-1. [DOI] [PubMed] [Google Scholar]
- 8.van Waasbergen LG, Dolganov N, Grossman AR. nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942. J Bacteriol. 2002;184(9):2481–2490. doi: 10.1128/JB.184.9.2481-2490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu CJ, Shrager J, Burnap RL, Postier BL, Grossman AR. Consequences of a deletion in dspA on transcript accumulation in Synechocystis sp. strain PCC6803. J Bacteriol. 2004;186(12):3889–3902. doi: 10.1128/JB.186.12.3889-3902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanaoka M, et al. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J Biol Chem. 2012;287(31):26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanaoka M, Tanaka K. Dynamics of RpaB-promoter interaction during high light stress, revealed by chromatin immunoprecipitation (ChIP) analysis in Synechococcus elongatus PCC 7942. Plant J. 2008;56(2):327–335. doi: 10.1111/j.1365-313X.2008.03600.x. [DOI] [PubMed] [Google Scholar]
- 12.Moronta-Barrios F, Espinosa J, Contreras A. In vivo features of signal transduction by the essential response regulator RpaB from Synechococcus elongatus PCC 7942. Microbiology. 2012;158(Pt 5):1229–1237. doi: 10.1099/mic.0.057679-0. [DOI] [PubMed] [Google Scholar]
- 13.Moronta-Barrios F, Espinosa J, Contreras A. Negative control of cell size in the cyanobacterium Synechococcus elongatus PCC 7942 by the essential response regulator RpaB. FEBS Lett. 2013;587(5):504–509. doi: 10.1016/j.febslet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140(4):529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9(12):1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 16.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93(19):10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103(32):12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markson JS, Piechura JR, Puszynska AM, O’Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155(6):1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seki A, et al. Induction of a group 2 sigma factor, RPOD3, by high light and the underlying mechanism in Synechococcus elongatus PCC 7942. J Biol Chem. 2007;282(51):36887–36894. doi: 10.1074/jbc.M707582200. [DOI] [PubMed] [Google Scholar]
- 20.Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50(2):288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H, et al. Exploration of a possible partnership among orphan two-component system proteins in cyanobacterium Synechococcus elongatus PCC 7942. Biosci Biotechnol Biochem. 2012;76(8):1484–1491. doi: 10.1271/bbb.120172. [DOI] [PubMed] [Google Scholar]
- 22.Rippka R, DeReuelles J, Waterbury JB, Herdman M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111(1):1–61. [Google Scholar]
- 23.Golden SS, Sherman LA. Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J Bacteriol. 1984;158(1):36–42. doi: 10.1128/jb.158.1.36-42.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson CR, et al. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 25.Ditty JL, Williams SB, Golden SS. A cyanobacterial circadian timing mechanism. Annu Rev Genet. 2003;37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- 26.Holtman CK, et al. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 2005;12(2):103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 27.Kondo T, Ishiura M. Circadian rhythms of cyanobacteria: Monitoring the biological clocks of individual colonies by bioluminescence. J Bacteriol. 1994;176(7):1881–1885. doi: 10.1128/jb.176.7.1881-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackey SR, Ditty JL, Clerico EM, Golden SS. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol. 2007;362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y-I, Boyd J, Espinosa J, Golden SS. Detecting KaiC phosphorylation rhythms of the cyanobacterial circadian oscillator in vitro and in vivo. Methods Enzymol. 2015 doi: 10.1016/bs.mie.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyd JS, Bordowitz JR, Bree AC, Golden SS. An allele of the crm gene blocks cyanobacterial circadian rhythms. Proc Natl Acad Sci USA. 2013;110(34):13950–13955. doi: 10.1073/pnas.1312793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.