Significance
Stem cells hold great promise in the field of regenerative medicine because of their capability both to self-renew and to differentiate. Regulation of these processes by molecular players is crucial to maintain stem cells' unique functions. We previously reported the generation of a library of embryonic stem cells (ESCs) with engineered chromosomal deletions, in an effort to identify novel genes or elements essential for ESC differentiation. We now reveal that ESCs heterozygous for ribosomal protein (RP)-coding genes show strong defects in embryoid body (EB) differentiation but not in self-renewal. We also identify p53-dependent and -independent mechanisms that mediate the defects in EB formation of RP-deleted clones. Together, our results highlight previously unidentified roles for RP genes in ESC fate regulation.
Keywords: embryonic stem cells, differentiation, embryoid body, ribosomal proteins
Abstract
In a functional genomics screen of mouse embryonic stem cells (ESCs) with nested hemizygous chromosomal deletions, we reveal that ribosomal protein (RP) genes are the most significant haploinsufficient determinants for embryoid body (EB) formation. Hemizygocity for three RP genes (Rps5, Rps14, or Rps28), distinguished by the proximity of their corresponding protein to the ribosome's mRNA exit site, is associated with the most profound phenotype. This EB phenotype was fully rescued by BAC or cDNA complementation but not by the reduction of p53 levels, although such reduction was effective with most other RP-deleted clones corresponding to non-mRNA exit-site proteins. RNA-sequencing studies further revealed that undifferentiated ESCs hemizygous for Rps5 showed reduced expression levels of several mesoderm-specific genes as compared with wild-type counterparts. Together, these results reveal that RP gene dosage limits the differentiation, not the self-renewal, of mouse ESCs. They also highlight two separate mechanisms underlying this process, one of which is p53 independent.
Embryonic stem cell (ESC) fate is intricately controlled at the transcriptional level by factors such as Oct4 (Pou5f1), Sox2 and Nanog (1–3), Dax1, Rex1, and SalI (reviewed in ref. 4), and the Mediator complex (3, 5, 6). Although much progress has been made on the identification of the transcriptional circuitry that defines pluripotency, the regulation of mRNA translation and posttranslational protein modifications currently are considered in this context only as fine-tuning to balance protein production and activity (7).
ESC self-renewal can be assessed readily by their ability to give rise to undifferentiated progeny in a clonal fashion. Similarly, in the absence of leukemia inhibitory factor (LIF) and bone morphogenetic protein signaling, ESC pluripotent potential can be investigated in vitro by initiating differentiation of the three germ layers using an embryoid body (EB) formation assay (8, 9).
To identify novel regulators of ESC fate, we developed a retrovirus-based methodology that randomly generates hemizygous chromosomal deletions with increasing size (nested) (Fig. 1A and detailed in ref. 10). Using this method, we reported the generation of a library of 1,307 ESC clones covering more than 25% of the mouse genome (the DelES library) along with an improved methodology for BAC recombineering and complementation (11). Phenotypical characteristics and a genetic map of all clones in our DelES collection are available at www.bioinfo.iric.ca/deles.
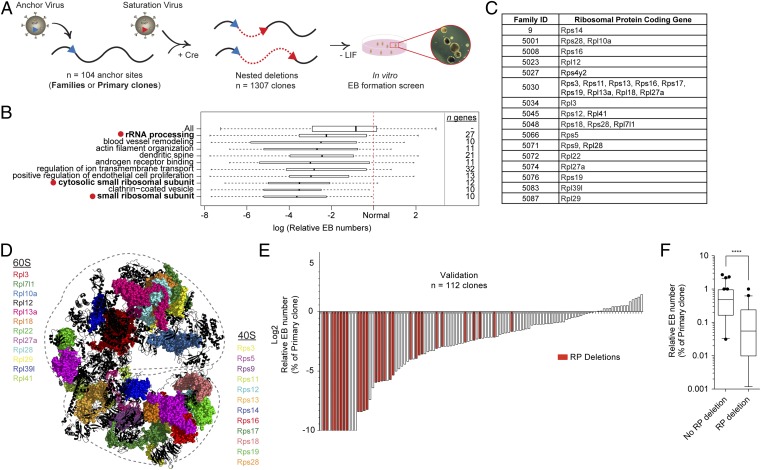
Fig. 1.
The EB-formation phenotype correlates with RP gene deletions. (A) Schematic overview of DelES library generation. For more details, see ref. 11. (B) GO analysis of deleted regions with respect to EB-formation phenotypes obtained in our high-throughput screen. The 10 GO terms present in at least 20 deletions with the most significant (P < 0.0005) negative effect on EB formation are shown. Bar thickness indicates the square root of the number of clones that hit a given GO term. See Dataset S1 for complete analysis of GO-term clustering. Relative EB numbers are calculated by dividing EB numbers of hemizygous clones by those of primary (wild-type) clones. (C) Rps and Rpl genes deleted in the first 100 families of the DelES library. (D) Location of deleted RP-coding genes highlighted on the 80S eukaryotic ribosome 3D structure (PDB ID codes 3U5E, 3U5F, and 3U5G) (26). (E) Waterfall-plot representation of the validation experiment in which 112 randomly selected DelES ESC clones from 35 unique anchor points were tested for the EB-formation phenotype in 60-mm dishes. Bars with values of −10 represent absolute EB numbers of 0 and should be considered as a negative infinite. Red shading indicates clones with RP gene deletions. (F) Box plot of EB-formation validation experiments; error bars indicate 95% confidence intervals. ****P < 0.0001.
Here, we report the integrative analysis linking the physical mapping to phenotype for all 1,307 clones in our library, documenting that genes involved in ribosome biogenesis and function are key players in ESC fate. We tested this hypothesis extensively in functional studies and identified a p53-independent response to ribosomal stress for mRNA exit-site ribosomal proteins (RPs).
Results
Gene Ontology Term Analysis Identifies RP Genes as Prime Candidates for EB Formation.
Using our collection of 1,307 previously described ESC clones, we first tested the contribution to overall fitness of the hemizygous deletion of gene sets extracted from the Gene Ontology (GO) database to determine underlying biological processes with the most reproducible and extreme effect on embryoid body (EB) development. GO term clustering (n = 7,083 genes out of 1,307 clones) showed that deletion of genes linked with the small ribosomal subunit (n = 10 genes) strongly correlated with a negative EB-formation outcome. Consistent with this finding, other categories such as rRNA processing (n = 27 genes) and cytosolic small ribosomal subunit (n = 12 genes) also are found in the 10 most significant categories of genes negatively affecting EB formation (P < 0.0005; red dots in Fig. 1B and Dataset S1). Within our collection, RP gene deletions are present in 16 families (Fig. 1C and Table S1), representing members of both the small (Rps, n = 12) and large (Rpl, n = 12) ribosome subunits (Fig. 1D). Because the DelES collection is made up of clones with hemizygous deletions, the abnormal EB-formation phenotype shared by this group of clones suggests that although RP gene dosage has no clear impact on self-renewing ESCs, it appears to be crucial for ESC differentiation.
To confirm this observation, we randomly selected 112 clones from DelES and validated their phenotype in standard in vitro assays (60-mm dish format). These validation experiments revealed that clones with the hemizygous deletion of at least one RP gene invariably exhibit defects in EB formation (Fig. 1E). Fig. 1F shows that the median relative EB number is 0.48 for non–RP-deletion clones versus 0.055 for RP-deletion clones (P < 0.0001). Importantly, no significant correlation was found between EB formation and chromosome deletion size (Fig. S1).
Physiological Consequences of RP Haploinsufficiency.
Under self-renewal conditions, wild-type and RP-hemizygous ESCs (hereafter called “ΔRP-ESCs”) could be maintained undifferentiated and were undistinguishable based on colony morphology and proliferation and apoptosis rates. Importantly, although cell-cycle progression was not affected (Fig. S2A), we detected an increased tendency of ΔRP-ESCs to undergo cell death following LIF removal (Fig. S2 B and C).
RP Genes Are Critical for EB Formation: Complementation Studies.
To confirm the requirement of RP genes for EB formation, we performed BAC complementation as described (11) and using the experimental design detailed in Fig. 2A. A total of 17 BACs were modified to complement clones from 11 different families, each containing one distinct RP gene deletion, i.e., Rps5, Rps12, Rps14, Rps16, Rps19, Rps28, Rpl12, Rpl22, Rpl27a, Rpl29, and Rpl39. Of these 11 families, five (Rps5, Rps14, Rps28, Rpl12, and Rpl27a) were functionally rescued (indicated by asterisks in Fig. 2B and Fig. S3). Projection of these RP proteins onto a 3D ribosomal structure revealed a consistent localization around the mRNA exit site for the three identified Rps proteins (Fig. 2C, Right). Interestingly, these three clones, which showed the strongest EB-formation defects (no background and early disaggregation), also exhibited the highest level of complementation (Fig. 2B), suggesting that RPs located in this region may be critical for EB formation.
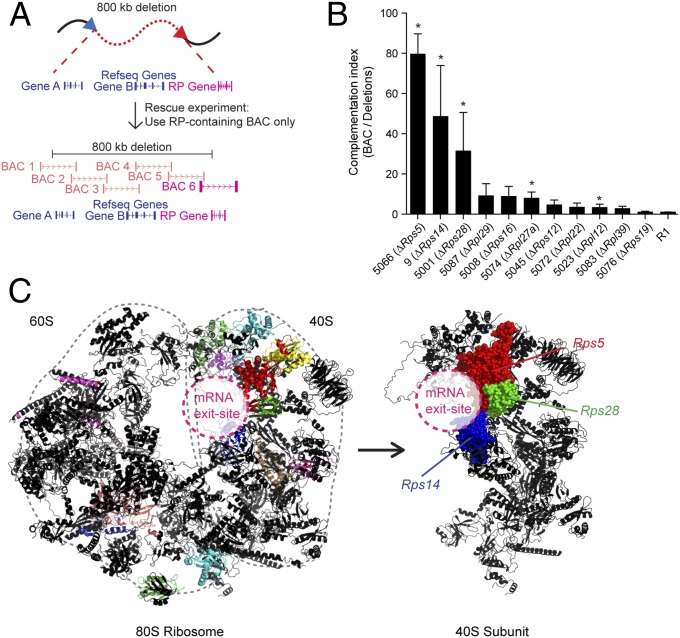
Fig. 2.
Complementation experiments reveal a site-specific role for RP genes. (A) Design of BAC complementation experiments. The backbone of a BAC vector containing the affected RP gene was modified using puromycin-, hygromycin-, or zeocin-resistance genes and was transfected into DelES clones of interest lacking one copy of the corresponding RP gene. (B) Complementation experiments of RP-deleted clones. Clones with a single RP gene deletion are shown. (See Fig. S3 for all values in the bar graph.) The complementation index was obtained by dividing relative EB numbers (based on primary clone values) of BAC-transfected clones by those of mock-transfected clones. Error bars show SEM of at least two independent experiments. (C) The RP-deleted clones that showed the best BAC complementation (farthest left columns in B) are colocalized at the mRNA exit site of the 40S ribosomal subunit (PDB ID codes 3U5E, 3U5F, and 3U5G) (26, 27).
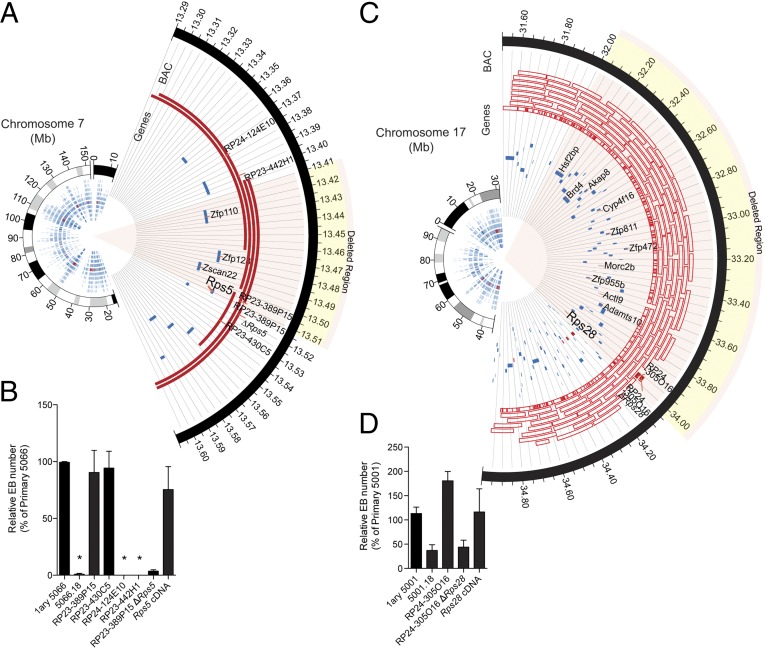
Additional studies were performed to confirm that RP genes—not other elements— are responsible for this complementation. To do so, we focused our studies on the two best complemented families, namely those in which Rps5 and Rps28 are deleted [families 5066 and 5001 in Fig. 2B; Rps14 haploinsufficiency for EB formation already has been described with the characterization of DelES library (11)]. Family 5066 (Rps5 heterozygote) was complemented only by the two BACs that included the Rps5 gene (BAC coverage is indicated by red lines in Fig. 3A; complementation results are shown in Fig. 3B). Importantly, a modified version of BAC RP23-389P15 engineered to lack the coding region of Rps5 and all other BACs lacking this gene failed to complement (Fig. 3B). In line with these results, transfection of a Rps5 cDNA expression vector effectively rescued EB formation to levels observed in primary clones (those containing no deletion but only an anchor virus; Fig. 3B). Similar data were obtained for clone 5001 (which has a large hemizygous deletion including the Rps28 gene), in which BAC complementation was dependent on the presence of Rps28, and cDNA complementation was sufficient to rescue EB formation to the levels seen in undeleted cells (Fig. 3 C and D).
Fig. 3.
Rps5 and Rps28 are essential for EB formation. (A) Circos (28) plots representing the genomic context of clone 5066.18 (Rps5Δ/wt) ∼90-kb deletion (mm9 mouse assembly). (B) Relative EB numbers (expressed as percentage of primary clone numbers) following transfection of BAC DNA identified in A. Error bars represent the SEM for at least two independent experiments performed in duplicate. Asterisks denote null values. (C) Circos plots representing the genomic context of clone 5001.18 (Rps28Δ/wt) ∼2-Mb deletion (mm9 mouse assembly). Genes have been partially annotated to simplify visualization. (D) Relative EB numbers (expressed as percentage of primary clone numbers) following transfection of BAC DNA identified in C. Error bars represent the SEM for at least two independent experiments performed in duplicate.
p53-Independent Ribosomal Stress in a Subset of RP-Deleted Clones.
RP imbalance affects ribosome biogenesis, leading to human diseases commonly referred to as “ribosomopathies” (reviewed in ref. 12). These syndromes, often causing hematological and skeletal anomalies, are characterized by ribosomal or nucleolar stress, a condition most frequently associated with p53 activation. Mechanistically, a subset of free RPs from destabilized ribosomal subunits physically associates with mouse double minute 2 (Mdm2), thus preventing p53 degradation (summarized in Fig. 4A) (13). This p53 response has been hypothesized to represent the cornerstone of ribosomopathies, given that many symptoms of these conditions can be corrected by a simple attenuation of p53 activity (12, 14, 15).
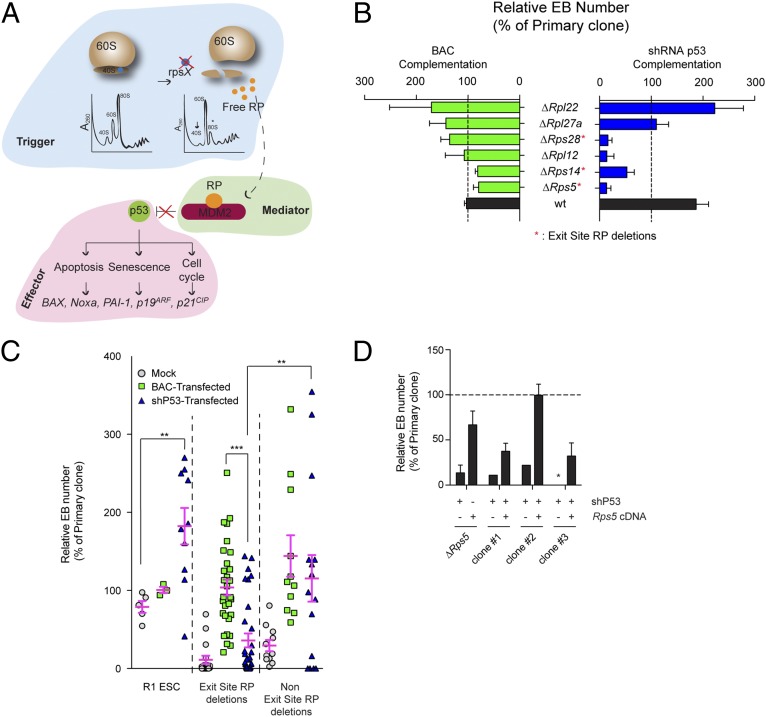
Fig. 4.
p53-independent role of exit-site–specific RP deletions in EB formation. (A) Schematic representation of ribosomal (nucleolar) stress pathway. An RP imbalance can lead to the formation of specific RP–Mdm2 complexes, resulting in p53 stabilization and downstream signaling. (B) Relative EB numbers (expressed as a percentage of primary clone numbers) following BAC or shRNA p53 transfection. Results from clones in which BAC complementation was at least 50% are shown. See Fig. S4D for results of all clones. Error bars show the SEM of at least two (BAC transfections) or four (shp53 transfections) independent experiments performed in duplicate. (C) Comparison of BAC vs. shRNA p53 complementation in wild-type R1 ESCs, exit-site–specific deletions (n = 3 clones), and other RP deletions (n = 3 clones). An shRNA to luciferase has been used in mock conditions. Pink bars represent mean values ± SEM. p53 knockdown levels can be found in Fig. S4 B and C. (D) Reintroduction of Rps5 cDNA following unsuccessful shRNA p53 rescue experiments. Three clones refractory for shRNA p53 complementation were subsequently transfected with the Rps5 cDNA to test for EB-formation capability and p53 independency. Error bars show the SEM of at least two independent experiments performed in duplicate.
Using an shRNA vector against p53, we first noted a twofold increase in EB numbers in wild-type R1 ESCs compared with controls (black bar in Fig. 4B, Right), likely indicating that p53 normally limits EB formation. Reduction of p53 levels in “non–exit-site” Rpl22Δ/wt and Rpl27Δ/wt ESC clones produced a complete rescue of EB formation (Fig. 4B, Right). In contrast, treatment with p53 shRNA was unable to complement “exit-site” clones hemizygous for Rps5Δ/wt, Rps14Δ/wt, or Rps28Δ/wt (Fig. 4B, Right), despite evidence of ribosomal stress (reduced 40S subunit abundance and/or increased p53 protein levels) (Fig. S4A) and successful p53 protein reduction upon shRNA expression (Fig. S4 B and C). However, p53 levels were not significantly higher in Rps5Δ/wt than in control cells, although polysome profiles clearly showed decreased levels of the 40S subunit. Interestingly, although not localized at the mRNA exit site, Rpl12Δ/wt ESCs were not rescued by reduction in p53 levels.
These studies thus reveal two different types of complementation, one that involves attenuation of p53 activity and one that does not (see Fig. S4D for all clones studied herein). Indeed, p53 shRNA treatment and BAC complementation were comparable in rescuing EB formation in clones hemizygous for non–exit-site RP genes (Fig. 4C, Right). In contrast, none of the three mRNA exit-site RP-deletion clones could be rescued by p53 shRNA (blue triangles in in Fig. 4C), although BAC complementation (green squares in Fig. 4C) or cDNA efficiently rescued these clones. To gain further insights into the nature of this phenotype, we isolated three subclones from the p53 shRNA-transduced Rps5Δ/wt ESC clone and reintroduced Rps5 cDNA separately in each clone. Again, and confirming the results shown in Fig. 4C, cDNA transfection was very effective even when p53 levels were experimentally decreased (Fig. 4D).
Expression of Mesoderm Genes Is Impaired in Rps5Δ/wt Mutants.
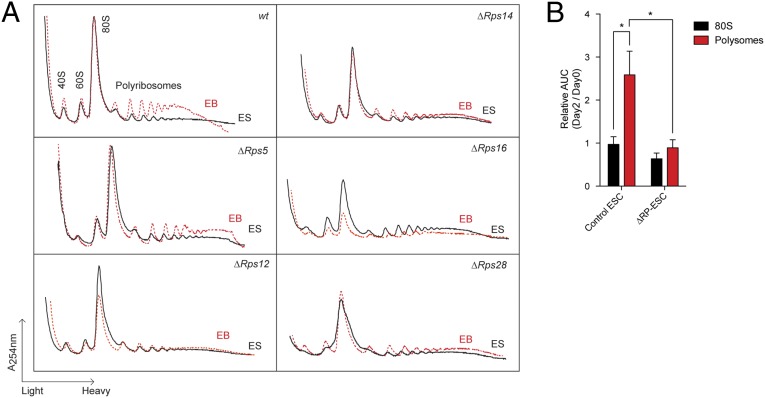
Next, we analyzed the polyribosome profiles of wild-type ESCs in self-renewing and differentiating conditions. Using area under the curve (AUC) measurement (Fig. S5A) (7), we observed a 2.5-fold increase in polyribosome fractions with the onset of EB formation. (Ribosome profiles are shown in Fig. 5A, and a summary is given in Fig. 5B.) In contrast to this polyribosome response, the 80S fraction was constant between the two states (Fig. 5).
Fig. 5.
The polyribosomal AUC is increased upon EB formation. (A) Polysome profiles obtained from wild-type and ΔRP-ESCs (ES, black lines) and EBs (red dashed lines). Profiles are representative of at least two independent experiments. For a full description of the experimental setup, see Materials and Methods. (B) Quantification of the AUC of polysomal profiles. Values represent relative area units (day 2/day 0). Error bars show SEM values of six wild-type (various ESC lines) and seven RP deletions. *P < 0.05. See Fig. S5A and Materials and Methods for detailed AUC quantification methodology.
Although self-renewing ΔRP-ESCs showed nearly normal polysome profiles, their polyribosome response was almost absent upon LIF removal (Fig. 5A; quantification in Fig. 5B). As expected of a ribosomal stress condition, the small and large ribosome subunits, and hence the mature 80S ribosome fractions, often were perturbed in these cells (Fig. S5B).
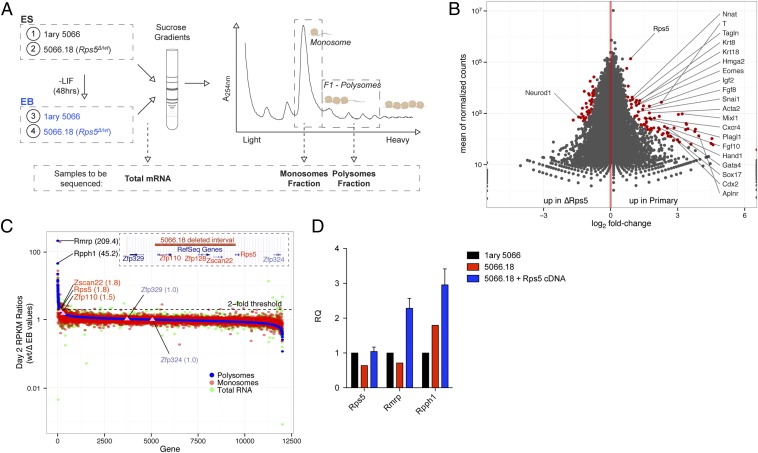
To gain further insights into the impact of this failure in polyribosome response, we performed RNA sequencing (RNAseq) using mRNA isolated from different ribosome fractions from wild-type and Rps5Δ/wt cells, in both self-renewing (ESCs) and differentiation onset (EBs, 48 h) conditions (Fig. 6A), before the appearance of disaggregation to avoid potential contamination by cell death signals. Analyses conducted from all EB fractions (total mRNA and monosomal or polysomal fractions) showed a significant enrichment of genes linked to mesodermal lineage specification in wild-type clones as compared with clones with Rps5 hemizygous deletion (Fig. 6B; see also the network representation in Fig. S6A, the comprehensive list of all differentially expressed genes in Dataset S2, and GO term clustering in Dataset S3). Interestingly, similar results were obtained when we compared the read counts of undifferentiated samples (see the representative DESeq graph in Fig. S6B), suggesting that a defect in the expression of mesodermal-specific genes already is established in self-renewing Rps5Δ/wt ESCs, even before LIF removal and EB formation.
Fig. 6.
RNAseq analysis of ribosomal fractions. (A) Experimental design. Wild-type (primary 5066) and Rps5Δ/wt lysates (undifferentiated ESCs and EBs) were fractionated on sucrose gradients, and RNA was extracted from monosome and polysome fractions. Twelve samples were hybridized to three lanes of an Illumina array and subjected to next-generation sequencing. (B) Differential expression (DESeq package) (29) analysis obtained from EB polysomal fractions. Genes that are differentially expressed (adjusted P value < 0.1) are shown in red. Genes found in the mesodermal differentiation network are labeled (Fig. S6). Full analysis data are presented in Dataset S2. (C) Enrichment curve showing RPKM ratios (wild-type/Rps5Δ/wt EBs) of genes found in polysomal (blue track), monosomal (red track), and total RNA (green track). Genes showing the greatest differential expression based on RPKM values of polysomal fractions are labeled in black. Genes present in the deletion (red) and those located just outside the deletion (blue) are identified also. (D) qRT-PCR assessment of relative Rmrp and Rpph1 expression (RQ) in wild-type, Rps5Δ/wt, and Rps5 cDNA-complemented ESCs. Error bars are representative of eight independent cDNA clones and are expressed as mean ± SEM.
We next compared differentially expressed genes from monosomal and polysomal fractions of the Rps5 hemizygous clone which undergo EB formation. We found that the expression level of genes deleted in these cells, namely Rps5, Zfp110, and Zscan22, was reduced by approximately twofold in all fractions examined (Fig. 6C; see Fig. S7 A–C for fraction-specific enrichment curves). As expected, the expression level of genes outside the deleted region, Zfp329 and Zfp324, was unchanged (Fig. 6C). We also found that very few genes are enriched in wild-type and Rps5Δ/wt clones. Apart from genes related to the previously identified mesodermal differentiation network, no significant differentially expressed candidate could be observed in monosomal fractions. However, in polysomal fractions, two noncoding genes, RNA component of mitochondrial RNA processing (Rmrp) and ribonuclease P RNA component H1 (Rpph1), are the most enriched genes, having reads per kilobase of transcript per million reads mapped (RPKM) ratios of 209.4 and 45.2, respectively (Fig. 6C). Reinforcing this observation, we observed that ESCs transduced with Rps5 expression vector (Fig. 3B) showed increased levels of Rmrp and Rpph1 by RNAseq [38.2- and 10.4-fold, respectively; see Fig. 6D and Table S2 for confirmation by quantitative RT-PCR (qRT-PCR) studies].
Discussion
The idea that RP-coding genes could have more than a merely structural function has evolved recently. Several RP-coding genes were shown to be essential in a species- and tissue-specific fashion, and mutations of these genes have been identified as genetic markers of human malignancies (12, 16). In the present study, we report a critical role for RP genes in EB formation from mouse ESCs. Of interest, this approach allowed us to highlight fundamental differences by which self-renewing pluripotent ESCs are less sensitive to nucleolar stress (RP hemizygous) than their differentiation-engaged progenies. Complementation experiments revealed two subsets of RPs regulating EB formation, one of which acts independently of p53. Of interest, these RPs are localized to the mRNA exit site, and their reduction conferred the most severe phenotypes.
Rmrp and Rpph1 were identified as depleted in the Rps5Δ/wt polysomal fraction. Of interest, these two genes are the most positively correlated ones (based on Pearson's correlation coefficient derived from expression values) in all publicly available RNAseq datasets analyzed to date (Fig. S7D). Although currently there is no known link between these two noncoding transcripts, their high degree of correlation suggests their participation in a network that might be involved in mesodermal differentiation, given that Rmrp mutations in humans give rise to dysplasia-related syndromes such as cartilage hair hypoplasia syndrome (17). Future experiments will be needed to determine the nature of this Rmrp–Rpph1 coexpression and their regulation by Rps5.
The transition of ESCs to EBs is not a lineage-specific assay and thus allows the generation of the three germ-layer derivatives (8, 9). Therefore, it is difficult to reconcile the “mesodermal signature” observed in Rps5Δ/wt ESCs and the severe EB phenotype observed in these clones. Previous studies have shown hematopoietic defects, but not defects in EB formation, upon deletion of mesodermal genes (18–20). However, it is possible that the combined deficiencies of several mesodermal genes as observed in Rps5Δ/wt ESCs contribute to this severe EB phenotype. Further studies are needed to clarify these observations, but the early onset of this phenotype makes it difficult to set a directed differentiation approach, because those techniques usually require EB formation before cytokine induction.
The ongoing cell differentiation occurring within EBs is a highly demanding metabolic process and most likely triggers an important increase in protein synthesis, which could be impossible to achieve in RP hemizygous cells. Along these lines, we observed that ribosomal loading (AUC of polyribosomal profiles; Fig. 5) was higher in differentiating than in self-renewing wild-type ESCs. Sampath et al. (7) also observed this “polyribosome response” in EBs and further demonstrated an enhanced protein production in these cells compared with ESCs.
It is well known that RP gene imbalance activates p53 signaling. qRT-PCR analysis of a panel of p53 target genes showed no significant expression changes in the context of RP hemizygous deletions. Imbalance in RP-coding genes also affects other cellular functions, such as transcription (21, 22). In line with this finding, our results document a p53-independent function for at least a subset of RP genes, those associated with the ribosome mRNA exit site.
The essential role of RP-coding genes reported here highlights fundamental differences between self-renewing ESCs and their differentiated counterparts, as well as a unique p53-independent role in this transition step for the mRNA exit-site Rps5, Rps14, and Rps28 genes. To our knowledge, this is the first time that a p53-independent response to nucleolar stress has been identified in a model that assesses self-renewal and EB formation from pluripotent ESCs.
Materials and Methods
ESC Maintenance.
Male R1 ES cells (23) were grown on irradiated mouse embryonic fibroblasts or on gelatin-coated dishes in DMEM medium (high glucose with l-glutamine and pyruvate) (Invitrogen) containing 15% (vol/vol) FCS (Invitrogen), 1.5 × 10−4 M α-monothioglycerol (Sigma), and 1 × 10−4 M nonessential amino acids (Invitrogen), supplemented with 1,000 U/mL of LIF-conditioned medium from transfected COS cells. The generation of the DelES library and clone maintenance have been described previously (11).
EB Formation.
ESC differentiation into EBs was performed in LIF-deprived semisolid medium, as previously described (8). See SI Materials and Methods for more details.
Ribosome Profiling.
Sucrose gradient velocity sedimentation was used to isolate polysomal fractions, as described (24). See SI Materials and Methods for more details.
GO Analysis.
To determine common mechanisms and pathways underlying the measured EB-formation phenotypes, GO terms were tested for a significant association with EB formation. See SI Materials and Methods for a complete description.
RNAseq.
RNAseq was performed on RNA samples purified from sucrose gradient fractions of 1.5 mL. Briefly, read count data were generated using the htseq-count module and were mapped to the mm9 .gtf file from the University of California, Santa Cruz. Reads mapping to introns were excluded from the analysis. The full methodology can be provided on request. Transcriptome sequencing was done as described for our previously reported T-ALL collection (25). Four samples were hybridized per lane of an Illumina HiSeq2000 (coverage of ∼87–113 million reads per sample). RNAseq data have been deposited in the Gene Expression Omnibus database under accession no. GSE57648.
Transfections, flow cytometry, and complementary methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tatiana Traboulsi, who helped with qRT-PCR studies and cDNA generation as a summer student in our laboratory; Geneviève Boucher and Jean-Philippe Laverdure for RNAseq data processing and help with bioinformatic analyses; Joseph Tcherkezian for his expertise with polysome experiments; Raphaëlle Lambert and Marianne Arteau of the Institute for Research in Immunology and Cancer genomic platform for expertise with qRT-PCR and RNAseq; and Dr. Bernhard Lehnertz for scientific comments. This work was supported by Canadian Institute of Health Research (CHIR) Grant MOP 82814. G.S. holds a Canada Research Chair in Molecular Genetics of Stem Cells. S.F. was supported in part by a studentship from CHIR. T.S.’s contribution was made possible through Victorian State Government Operational Infrastructure Support, the Australian Government National Health and Medical Research Council (NHMRC) Independent Research Institutes Infrastructure Support Scheme, and NHMRC Program Grant 1016647.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.H. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession no. GSE57648).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418845112/-/DCSupplemental.
References
- 1.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 2.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 3.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145(6):835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampath P, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993;13(1):473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7(6):862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 10.Bilodeau M, Girard S, Hébert J, Sauvageau G. A retroviral strategy that efficiently creates chromosomal deletions in mammalian cells. Nat Methods. 2007;4(3):263–268. doi: 10.1038/nmeth1011. [DOI] [PubMed] [Google Scholar]
- 11.Fortier S, et al. Genome-wide interrogation of Mammalian stem cell fate determinants by nested chromosome deletions. PLoS Genet. 2010;6(12):e1001241. doi: 10.1371/journal.pgen.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23(1):63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Liao J-M, Liao W-J, Lu H. Scission of the p53-MDM2 Loop by Ribosomal Proteins. Genes Cancer. 2012;3(3-4):298–310. doi: 10.1177/1947601912455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fumagalli S, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11(4):501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGowan KA, et al. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood. 2011;118(13):3622–3633. doi: 10.1182/blood-2010-11-318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narla A, Ebert BL. Ribosomopathies: Human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridanpää M, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104(2):195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 18.Keller G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes Dev. 2005;19(10):1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 19.Lacaud G, Kouskoff V, Trumble A, Schwantz S, Keller G. Haploinsufficiency of Runx1 results in the acceleration of mesodermal development and hemangioblast specification upon in vitro differentiation of ES cells. Blood. 2004;103(3):886–889. doi: 10.1182/blood-2003-06-2149. [DOI] [PubMed] [Google Scholar]
- 20.Fehling HJ, et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130(17):4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 21.Lindström MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun. 2009;379(2):167–170. doi: 10.1016/j.bbrc.2008.12.083. [DOI] [PubMed] [Google Scholar]
- 22.Provost E, et al. Ribosomal biogenesis genes play an essential and p53-independent role in zebrafish pancreas development. Development. 2012;139(17):3232–3241. doi: 10.1242/dev.077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90(18):8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cargnello M, et al. Phosphorylation of the eukaryotic translation initiation factor 4E-transporter (4E-T) by c-Jun N-terminal kinase promotes stress-dependent P-body assembly. Mol Cell Biol. 2012;32(22):4572–4584. doi: 10.1128/MCB.00544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon C, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26(7):651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330(6008):1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 28.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.