Significance
Therapeutic antibodies represent the largest and fastest growing class of biopharmaceuticals. There is a trend in moving from intact antibodies toward “armed” antibody products, in which the antibody moiety serves as pharmacodelivery vehicle. The impact of glycosylation on the targeting performance of armed antibodies is still largely unknown. Our article sheds light on the surprising finding that relatively small variations in glycostructures and sialic acid content can have dramatic effects on therapeutic agent performance. A better understanding of the impact of glycosylation on pharmaceutical activity is likely to be relevant not only for future antibody development activities, but also for changes in current manufacturing processes and for the development of biosimilar products.
Keywords: tumor targeting, glycosylation, armed antibody, interleukin-9, site-specific glycan analysis
Abstract
The ability of antibodies to extravasate out of blood vessels is critical for therapeutic activity, because molecular targets for most diseases are located outside of the endothelial lining. By performing detailed biodistribution studies with a novel IL9-armed cancer-specific antibody, we identified a clear correlation between N-linked glycan structures and tumor-targeting efficiencies. Site-specific glycan analysis provided a detailed view of the glycan microheterogeneity present on the IL9 portion of the recombinant protein. Nonsialylated glycan structures have a negative impact on disease-homing activity, highlighting the importance of glycosylation control and characterization during process development.
Monoclonal antibodies represent the largest and fastest growing class of pharmaceutical biotechnology products (1). Most biopharmaceuticals, including antibody-based therapeutics, feature posttranslational modifications such as protein N-glycosylation and therefore rely on mammalian cell expression systems (2). For example, monoclonal antibodies, used in a human IgG format, contain structurally distinct N-linked glycans at conserved positions within the Fc region. Depending on the IgG subtype, Fc-glycosylation has been recognized to have a profound effect on the activation of immune cells (3, 4). Indeed, the first glyco-engineered antibody product, obinutuzumab (Gazyva), has recently been approved for chronic lymphocytic leukemia. The impact of protein glycosylation on pharmacokinetics has been extensively studied for glycoprotein hormones, including the prominent examples of recombinant erythropoietin and its glyco-engineered derivative Darbepoetin alfa (5). The introduction of additional N-glycosylation motifs into the peptide sequence of erythropoietin can result in increased serum half-lives (6). Glycosylation also dictates the serum half-life of glycoprotein drugs, capable of neonatal Fc-receptor–mediated recycling, as shown for the systemic TNF inhibitor Lenercept, a fusion protein consisting of an IgG1 Fc portion and the extracellular p55 TNF receptor domain (7). However, quantitative studies investigating the impact of protein glycosylation on disease-homing properties of therapeutic proteins are rare.
Exploiting their exquisite target selectivity and ability to localize at sites of disease, there is an emerging trend to use monoclonal antibodies as pharmacodelivery vehicles, thus moving from intact antibodies toward armed antibody products (8, 9). The attachment of therapeutic payloads to a targeting antibody can be accomplished either by chemical conjugation, in the case of small molecules, or by genetic fusion of protein domains (9). The fusion of bioactive protein payloads (e.g., cytokines) may lead to additional O- or N-glycans in the resulting armed antibody when expressed in eukaryotic cell expression systems.
Most antibody-mediated pharmacodelivery approaches rely on extravasation of the biopharmaceutical product to diffuse into tissues and reach the site of disease. Glycans present on the therapeutic protein can modulate this process by different glycan–receptor interactions in the bloodstream. For example, hepatocytes express the asialoglyoprotein receptor with specificity for nonsialylated proteins with terminally exposed galactose residues (10). Another receptor involved in glycoprotein homeostasis, primarily expressed by macrophages and dendritic cells, is the mannose receptor, recognizing terminal mannose or N-acetylglucosamine (11). Certain glycan epitopes are also known to be immunogenic and can lead to antidrug antibody responses in humans (12).
Our group has worked extensively on the production and in vivo characterization of armed antibody products directed against splice isoforms of extracellular matrix components, which are undetectable in normal adult tissues, but abundantly expressed at sites of cancer and other inflammatory conditions (13). In particular, we have studied the high-affinity human monoclonal antibody F8, which is specific to the alternatively spliced extradomain A (EDA) of fibronectin, a marker of angiogenesis expressed in the subendothelial extracellular matrix of tumor blood vessels (13, 14). The F8 antibody has been used for the pharmacodelivery of drugs, radionuclides, and cytokines to various types of disease lesions (9, 15). Small bivalent antibody fragments without Fc portion may be preferred for the delivery of highly potent payloads, because they are rapidly cleared from circulation while exhibiting favorable biodistribution profiles (8). In this study, we found that variations in N-linked glycan structures, present on the interleukin-9 (IL9) moiety of different F8-based diabody fusion protein preparation, led to dramatic changes in tumor targeting efficiencies, as revealed by quantitative biodistribution analysis.
Results and Discussion
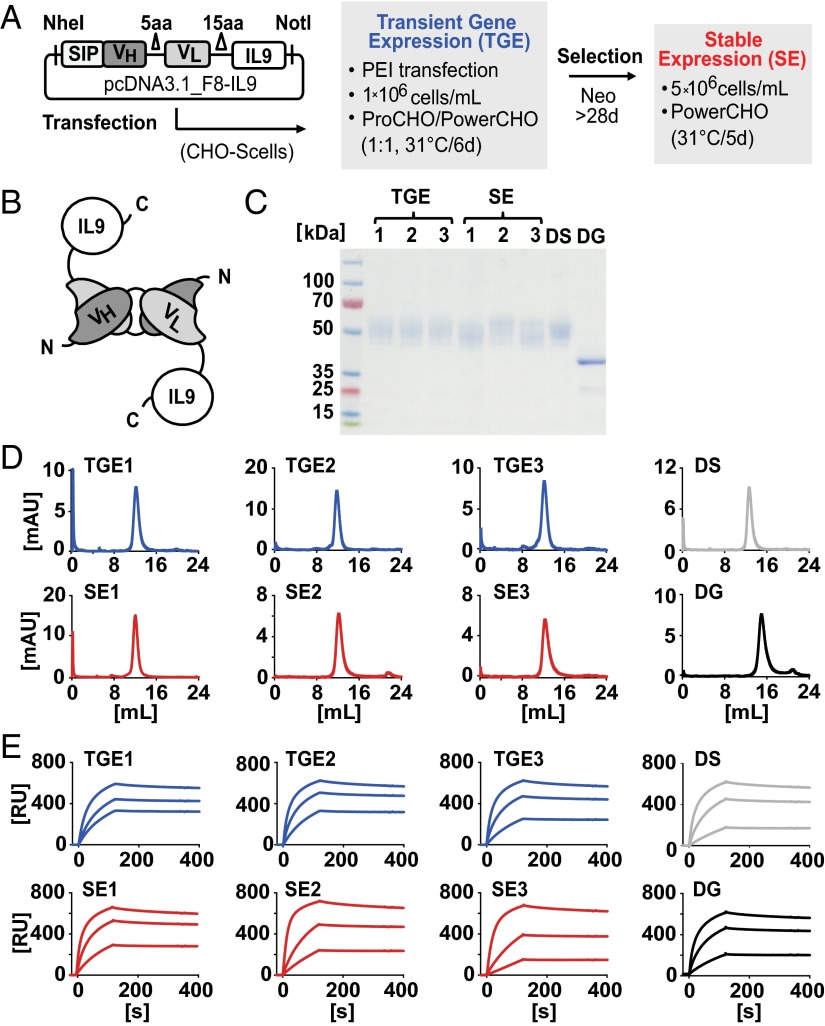
We focused our attention on IL9 as a therapeutic payload, based on reports on its potent T-cell–mediated antitumor activities (16, 17). IL9 is a special cytokine because it contains four distinct N-glycosylation sites, while being devoid of O-linked glycans (Fig. S1). In this study, we genetically fused IL9 to the C terminus of a nonglycosylated F8-based diabody (Fig. S2) and expressed the recombinant immunocytokine either by transient gene expression (TGE) or by stable expression (SE) in stably transfected Chinese hamster ovary (CHO) cells (Fig. 1 A and B) (18). Stably transfected polyclonal CHO cells were cultured at a higher cell density and modified medium composition. SDS/PAGE analysis showed similar patterns for protein preparations obtained in various experimental conditions. The mass difference of ∼10 kDa observed for glycosylated F8-IL9 samples in SDS/PAGE suggested similar glycosylation site occupancies, irrespective of the production method. F8-IL9 preparations tested in vivo eluted as a single peak in gel filtration and displayed comparable EDA-binding kinetics in surface plasmon resonance (SPR) analysis (Fig. 1 D and E) . The products could be converted into a fully deglycosylated form upon peptide-N-glycosidase F (PNGase F) treatment (Fig. 1C and Figs. S1 and S3). Ex vivo, all F8-IL9 preparations selectively stained the subendothelial extracellular matrix of blood vessels in murine F9 teratocarcinoma, regardless of their production method and enzymatic modification (Fig. S4).
Fig. 1.
Production methods and protein characterization data of F8-IL9 preparations investigated in vivo. Data from TGE (blue) and SE (red) F8-IL9 batches (1–3) are displayed. Desialylated SE (DS; gray) and deglycosylated TGE (DG; black) F8-IL9 were compared. (A) Expression vector and production methods. (B) Schematic of homobivalent diabody format. (C) Analytical SDS/PAGE analysis. (D) Gel-filtration chromatograms. (E) EDA-binding sensograms (SPR analysis).
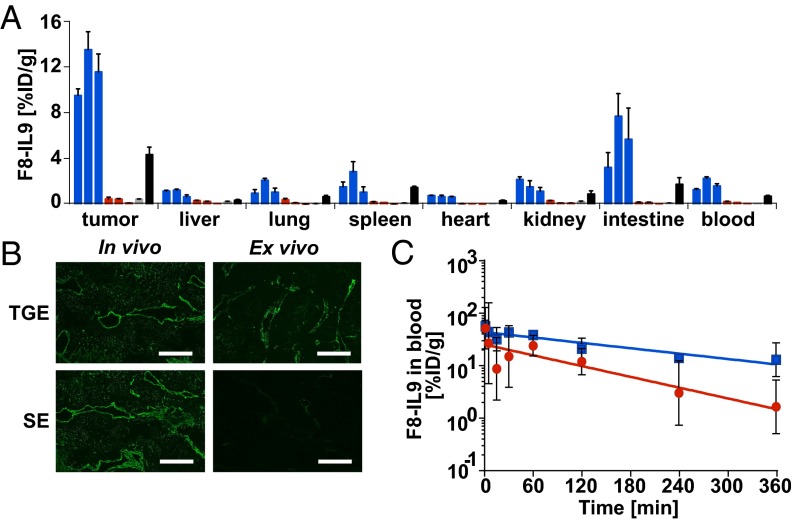
When various batches of F8-IL9 were studied by quantitative biodistribution analysis in immunocompetent 129/Sv mice bearing s.c. F9 tumors, a strikingly different tissue distribution profile was observed for proteins produced by using either TGE or SE methodologies (Fig. 2A). F8-IL9 derived from TGE cultures was able to efficiently and selectively localize to tumors 24 h after i.v. injection with 11.53 ± 0.71% injected dose per gram (%ID/g) in the neoplastic lesions while exhibiting favorable tumor-to-organ ratios (Fig. 2A). By contrast, batches of the same protein derived from SE cultures failed to target tumors in vivo, with only 0.36 ± 0.05%ID/g in the tumor and 0.16 ± 0.01%ID/g in blood. In line with the biodistribution data, immunofluorescence microscopy revealed that only TGE-derived F8-IL9 could successfully localize to the perivascular space of tumor blood vessels in vivo (Fig. 2B). These biodistribution data were surprising, because the stable polyclonal expression cultures were directly derived from preceding TGE cultures via antibiotic selection. Our finding proved to be highly reproducible because the data originate from three independent experiments for each type of sample. Additional biodistribution experiments, performed after treating F8-IL9 with PNGase F, revealed that deglycosylated F8-IL9 from SE cultures had regained its tumor-targeting ability (Fig. 2A). Importantly, TGE- and SE-produced F8-IL9 displayed close to identical biodistribution profiles after enzymatic deglycosylation with PNGase F (Fig. 2A and Fig. S5), indicating that the protein components were of equivalent quality. F8-IL9 produced by TGE also failed to target the tumor neovasculature of F9 tumors upon enzymatic removal of terminal sialic acids by α2-3,6,8,9 neuraminidase (Fig. S5), pointing out the special role of this carbohydrate residue.
Fig. 2.
In vivo biodistribution profiles, microscopic analysis, and pharmacokinetic data. (A) Quantitative biodistribution profiles 24 h after i.v. administration to F9-tumor–bearing mice. TGE (blue), SE (red), desialylated TGE (DS; gray), and deglycosylated SE (DG; black) derived F8-IL9 products are shown. (B) Juxtaposition of immunofluorescence detection images of F8-IL9 at 24 h after i.v. administration or after ex vivo application onto F9 tumor sections. (Scale bar: 100 µm.) (C) Pharmacokinetic data of F8-IL9 from SE (red) and TGE (blue) during the first 6 h after injection.
The quantitative biodistribution profiles shown in Fig. 2A provide a global view of potential glycan-mediated F8-IL9 interactions in vivo. A potential lectin-trapping mechanism (e.g., by immune cells) would be detectable by elevated radioactivity levels in blood and in the spleen. However, only low levels of radiolabeled F8-IL9 were found in normal organs, with the exception of intestinal uptake, which is often observed using anti-EDA antibody products. We therefore assumed that changes in glycostructures could have an impact both on drug clearance and on extravasation. To support this conclusion, a formal pharmacokinetic analysis comparing radiolabeled F8-IL9 samples from either TGE or SE cultures was performed (Fig. 2C). However, F8-IL9 produced by the SE method displayed only slightly faster blood clearance, suggesting that the two products could also differ in their ability to cross the tumor endothelium.
The model system used is appropriate to study extravasation and vascular targeting because of the specific site of expression of the target antigen EDA in the subendothelial matrix of the tumor-associated vasculature (Fig. 2B and Fig. S4). High expression levels of EDA at this site prevent saturation effects over a wide dose range (19). Additionally, the turnover rate for this type of antigens is considered to be very low because bound antibody can be detected up to 5 d after injection (10). Targeting of solid tumors and metastases is often limited by the buildup of an antigen barrier in proximity to the neovasculature (20). Hence, our findings may also be relevant for other antibody-based pharmacodelivery approaches.
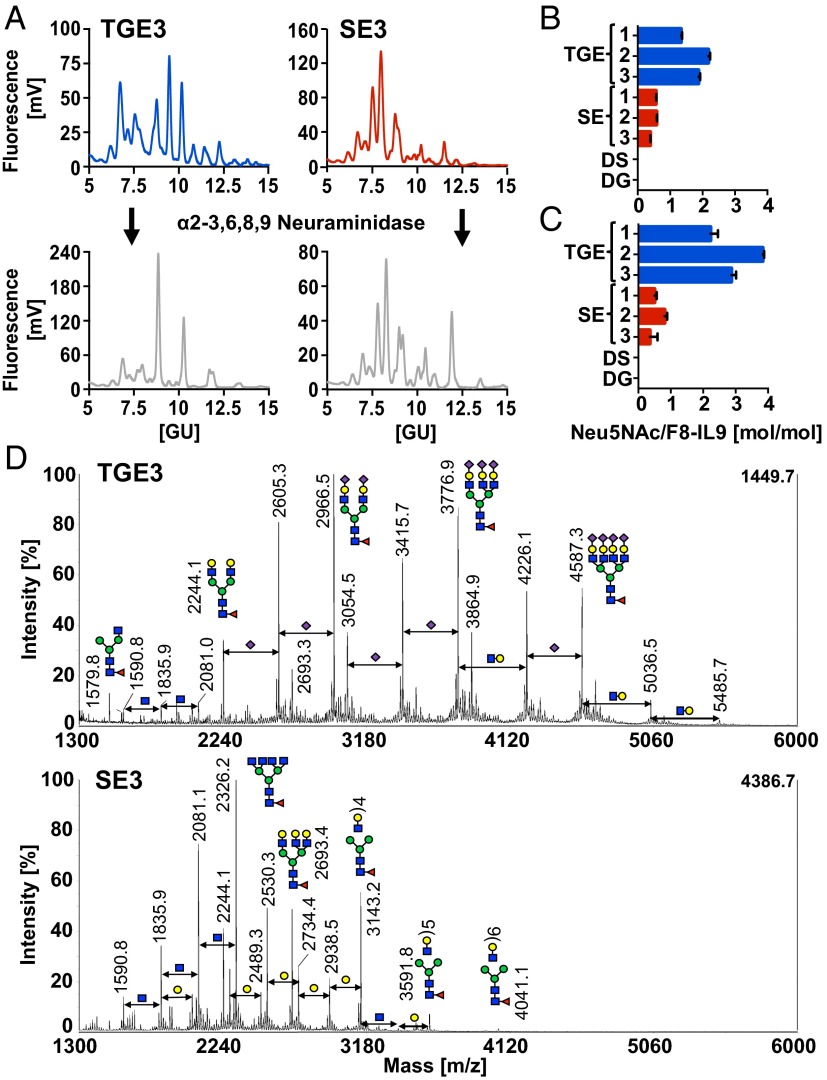
F8-IL9 glycoforms produced by either SE or TGE were extensively characterized by using several complementary methods. Hydrophilic interaction chromatography (HILIC)-HPLC–based glycoprofiling of fluorescently labeled glycan pools after PNGase F treatment showed substantial differences between TGE and SE samples (Fig. 3A). We also quantified the amount of terminal N-acetylneuraminic acid (Neu5Ac) moieties in various protein preparations, using two orthogonal assays based either on chemical or enzymatic sialic acid release, followed by different fluorescent labeling and detection methods as described in Methods. The TGE-produced batches of F8-IL9 exhibited significantly higher Neu5Ac-to-protein ratios, compared with the same protein derived from stably transfected cells in both assays (Fig. 3 B and C). By comparison, the glycan profiles before and after α2-3,6,8,9 neuraminidase treatment revealed characteristic peak shifts for the TGE samples, again indicating terminally sialylated N-glycans (Fig. 3A). Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/TOF MS) analysis of permethylated glycans further confirmed this finding (Fig. 3D). The major N-glycan structures from TGE-derived F8-IL9 were sialylated N-glycans with core fucosylation. On the contrary, MS profiling showed that incomplete galactosylated and neutral N-glycans were the main species present on SE-derived F8-IL9. Further, we have predominantly found α-2,3–linked sialic acids on TGE product as confirmed by enzymatic treatment with α-2,3-neuraminidase. Surprisingly, we have also observed antennary N-acetyllactosamine (LacNAc) repeats on the glycans of SE F8-IL9. Such poly-LacNAc structures represent ligands for galectins, which are known to be involved in cell adhesion and tumor progression (21, 22). A detailed list of all observed N-glycan structures is given in Table S1.
Fig. 3.
Characterization data of N-linked glycan pools. F8-IL9 samples from TGE (blue), SE (red), TGE after desialylation (DS; gray), and SE after deglycosylation (DG; black) were analyzed. (A) Representative HILIC-HPLC profiles of 2-AB–labeled N-glycan pools after PNGase F release and neuraminidase treatment. (B) Quantification of Neu5Ac by reversed-phase HPLC upon mild acid hydrolysis. (C) Fluorimetric quantification of terminal Neu5Ac after enzymatic release with α2-3,6,8,9 neuraminidase. (D) MALD-TOF/TOF spectra of permethylated glycan pools. All color schemes followed the guidelines of the Consortium for Functional Glycomics (33).
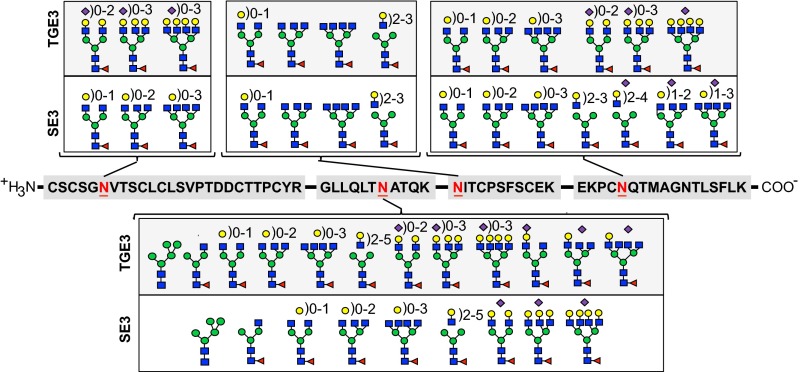
IL9 features four putative glycosylation sites. To obtain a detailed view of the glycan microheterogeneity, site-specific glycosylation analysis was performed. After proteolytic cleavage, the four sets of F8-IL9 derived glycopeptides were analyzed by nanoLC–higher-energy collisional dissociation (HCD) tandem mass spectrometry (MS/MS). Sialylated glycans were again identified with the help of α2-3,6,8,9 neuraminidase. Site-specific N-linked glycan structures for representative SE and TGE preparations are summarized in Fig. 4. All annotated glycan structures and corresponding glycopeptides are summarized in Table S2. MS and MS/MS spectra for glycosylation site 2 are exemplified in Fig. S6. Consistent with the data mentioned above, sialylated N-glycans were again predominantly found in the TGE preparations but were also present in the SE product. Notably, sialylation differences were site-specific; i.e., terminal sialic acids as well as galactose residues at the penultimate position were absent at glycosylation site 3 in both types of products (Fig. 4).
Fig. 4.
Site-specific glycan microheterogeneity. Representative TGE- and SE-derived F8-IL9 samples were subjected to nanoLC-HCD-MS/MS glycopeptide analysis. Peptide sequences upon cleavage by Trypsin, Glu-C endopeptidase, and AspN endopeptidase are shown in the center. Glycosylation sites are numbered from left to right (sites 1–4).
Together, the absence of terminal sialic acids and the exposure of terminal galactose or N-acetylglucosamine residues control both the elimination of the fusion protein from blood and its extravasation properties. Low levels of terminal sialic acids can lead to unacceptably fast clearance, as previously described (5). A contribution to protein clearance of either the asialoglycoprotein receptor and of the mannose receptor would be compatible with our analytical data.
Changes in bioavailability and extravasation of biopharmaceuticals, including therapeutic antibodies, can have a profound impact on safety and therapeutic action (23). Indeed, substantial differences in pharmacokinetics and biological activities have been reported for products undergoing changes in manufacturing processes (24). However, variations in extravasation rates and disease-homing properties may often go unnoticed, unless quantitative biodistribution studies are performed. The findings of this study suggest that the engineering and development of biopharmaceuticals may favor either a complete absence of glycostructures or, alternatively, the engineering of well-defined sialylated carbohydrates (25–27). Pharmacokinetic and biodistribution studies combined with thorough glycan profiling appear mandatory for therapeutic glycoproteins, especially when the production process is modified or when biosimilar products are developed.
Methods
Cloning of F8-IL9 Fusion Proteins.
Mouse IL9 (mIL9) cDNA (Sino Biological) was amplified by PCR using the forward primer link15-mIL9_for (5′-TCAGGCGGAGGTGGCTCTGGCG GTGGCGGATCACAGAGATGCAGCACCACATGGGGC-3′) that appends the C-terminal part of the flexible (Gly4Ser)3-linker sequence and the reverse primer mIL9-NotI_rev (5′-TTTTCCTTTTG CGGCCGCTCACTATGGTCGGCTTTTCTGCCTTTGCATCTC-3′) encoding two stop codons and a NotI restriction site. The F8 diabody gene was PCR-amplified by using the primers SIP-F8_for (5′-CCTGTTCCTCGTCGCTGTGGCTACAGGTGTGCACTCGGAGGTGCAGCTGTTGGAGTCTGGGG-3′) that appends part of the signal peptide (SIP) sequence and F8-link15_rev (5′-CCGCCA-GAGCCACCTCCGCCTGAACCGCCTCCACCTTTGATTTCCACCTTGGT CCCTTGG-3′), which encodes the N-terminal part of the (Gly4Ser)3-linker. The mIL9 and F8-diabody DNA fragments were assembled by PCR and amplified by using the primers NheI-SIP_for (5′-CCCGCTAGCGTCGACCATGGGCTGGAGCCTGATCCTCCTGTTCCTCGTCGCTGTGGC-3′), containing an NheI restriction site followed by the N-terminal part of the SIP sequence, and mIL9-NotI_rev. The PCR-assembled full-length F8-IL9 immunocytokine gene was double-digested with NheI and NotI restriction endonucleases (New England Biolabs) and ligated into the mammalian cell expression vector pcDNA3.1(+) (Life Technologies) with T4 ligase (New England Biolabs).
Cell Lines and Protein Expression.
Percentages (%) are based on vol/vol unless stated differently. Transiently expressed F8-IL9 fusion proteins were produced in suspension cultures of Freestyle CHO (CHO-S) cells (Life Technologies). The medium was composed of a 1:1 mixture of ProCHO-4 (Lonza) and PowerCHO-2 CD (Lonza), both supplemented with HT supplement containing 100 µM hypoxanthine and 16 µM thymidine (Life Technologies), 2 mM Ultraglutamine (Lonza), and 1% antibiotic–antimycotic solution (Life Technologies). Protein expression occurred in a shaking incubator at 31 °C for 6 d, starting at an initial cell density of 1 × 106 cells per mL.
To generate stably transfected cells, 10 mL of the TGE culture were taken 24 h after transfection, centrifuged (at 1,000 × g for 4 min), and resuspended in RPMI medium (Life Technologies) supplemented with 10% FCS (Life Technologies), 1% antibiotic–antimycotic solution, and 0.5 mg/mL Geneticin (Life Technologies). Stable integration into the CHO-S genome was achieved after cultivation for >28 d at 37 °C and 5% CO2 under antibiotic selection. Polyclonal stably transfected cells were then grown in suspension at 37 °C in PowerCHO-2 CD medium supplemented as described above with HT supplement, Ultraglutamine, and antibiotic–antimycotic solution. As soon as the cells reached a density of 4.5–5 × 106 cells per mL, cultures were transferred to a 31 °C shaking incubator for protein expression until day 5. F9 teratocarcinoma cells (ATCC no. CRL-1720) were grown according to supplier’s protocol in 0.1% gelatin-coated tissue culture flasks in DMEM (Life Technologies) supplemented with 10% FCS and 1% antibiotic–antimycotic solution.
Protein Purification and Characterization.
Both stably and transiently expressed fusion protein preparations were purified from the supernatant to homogeneity by protein-A (Sino Biological) affinity chromatography and further analyzed by SDS/PAGE (NuPAGE system; Life Technologies), size-exclusion chromatography (gel-filtration) on a Superdex S200 10/300GL column (GE Healthcare), and SPR analysis with a Biacore 3000 system (GE Healthcare) on a CM5 sensor chip coated with ∼1,500 resonance units of EDA antigen performed as described (13).
Immunofluorescence Detection.
Ex vivo immunofluorescence staining with F8-IL9 preparations was performed with F9 teratocarcinoma tumors from 129/Sv mice (Charles River), which served as control mice in previous therapy experiments. In vivo immunofluorescence analysis was performed with F9 tumors, which were excised 24 h after i.v. injection of F8-IL9 samples and embedded in NEG-50 cryo-embedding medium (Thermo Scientific). Tumor sections of 10 µm were stained by using the following primary antibodies: PECAM1 goat anti-mCD31 (M-20; Santa Cruz Biotechnology) and RM4A9 rat anti-mouse IL9 antibody (BioLegend). Detection was accomplished with anti-goat Alexa Fluor 488 and anti-rat Alexa Fluor 594-coupled secondary antibodies (Life Technologies). The slides were analyzed with an Axioskop 2 Plus microscope (Zeiss) and processed with Adobe Photoshop.
Biodistribution and Pharmacokinetic Analysis.
Twelve-week-old female 129/Sv mice were s.c. injected in the flank with 2.5 × 107 F9 teratocarcinoma cells. As soon as tumor sizes were >50 mm3, mice were grouped (n = 4), and ∼10 µg of 125I-labeled protein was injected into the lateral tail vein as described (13). Mice were killed 24 h after injection, organs were excised and weighed, and radioactivity was quantified with a Packard Cobra γ-counter. Values are given in %ID/g ± SD (wt/wt). The pharmacokinetic analysis was performed as follows: The radiolabeling of F8-IL9 samples and i.v. administration into healthy 129/Sv mice (n = 4) was carried out as described for the quantitative biodistribution analysis. Blood samples at suitable time points were taken by puncturing the lateral tail vein and withdrawing 1–5 µL of blood, which was analyzed by scintillation counting. One mouse per group was killed after 20 min to obtain short-time biodistribution profiles. Experiments were performed under the project license granted by the local authority Verterinäramt des Kantons Zürich, Switzerland (License No. 42/2012).
Sialic Acid-Release and Quantification.
Sialic acids were released enzymatically via α2-3,6,8,9 neuraminidase (New England Biolabs). Specifically, 3 U of enzyme per µg of F8-IL9 was incubated at 37 °C for 16 h. The samples for subsequent in vivo experiments were then purified via protein-A affinity chromatography. Alternatively, sialic acids were released by mild hydrolysis in 0.5 M NaHSO4 (Sigma Aldrich), requiring an incubation period of 40 min at 80 °C (28). Enzymatically released Neu5Ac was quantified by using a fluorimetric Neu5Ac assay kit (BioVision). Neu5Ac (Sigma Aldrich) represents the most common sialic acid species and was used as a standard for calibration for both assays. Fluorescence was measured in a SpectraMax Paradigm plate reader (Molecular Devices) at excitation/emission (Ex/Em) wavelengths of 535/587 nm. Hydrolyzed sialic acids were fluorescently labeled with o-phenylenediamine (Sigma Aldrich) in 0.5 M NaHSO4 for 2 h at 80 °C. RP-HPLC was performed with a Hitachi Lachrom D-7000 HPLC-system (Merck) equipped with an Xterra 5-µm, 4.6- × 150-mm C18 column (Waters). Sialic acid derivatives were eluted by using an isocratic buffer system and detected at Ex/Em 280/425 nm as described (28). Data points were normalized per F8-IL9 monomer (average of triplicates ± SD).
Enzymatic Release of N-Glycan Pools.
N-linked oligosaccharides were released by glycerol-free PNGase F (New England Biolabs). Specifically, 10 U of enzyme per µg of protein were incubated for 24 h at 37 °C. Samples for biodistribution experiments were deglycosylated in PBS buffer, followed by an additional protein A purification step to remove the enzyme and contaminants. All other samples were dialyzed against 50 mM ammonium bicarbonate (Sigma Aldrich) (pH 8.0) buffer before deglycosylation reactions for a prolonged incubation time of up to 24 h. Released glycans were then separated form proteins by Vivaspin 500 centrifugal filter units (Sartorius Stedim) with a 10-kDa cutoff and washed with 4 × 400 µL of deionized water. The flow-through was collected and vacuum-dried.
HILIC-HPLC–Based Glycoprofiling.
Vacuum-dried N-glycan samples were fluorescently labeled with 2-aminobenzamide (2-AB) (29), by using a Glycoprofile labeling kit (Sigma Aldrich) and purified via GlycoClean S cartridges (Prozyme), according to the providers’ instructions. Samples were again reduced to dryness in a vacuum centrifuge before they were dissolved in 200 µL of 50% acetonitrile/water. HPLC-HILIC was performed with the Hitachi Lachrom D-7000 HPLC system equipped with a TSKgel Amide-80 column (TOSOH Bioscience). The injected sample volume was 12 µL. Gradient elution was achieved by using a buffer system consisting of acetonitrile (Sigma Aldrich) and 0.1 M ammonium formate (ARCOS Chemicals) (pH 4.5) solution. A gradient of 0.3%/min starting with 24% 0.1 M ammonium formate provided the highest resolution. A dextran calibration ladder (Waters) was run after each sample as external standard to calibrate the system for subsequent conversion of retention times (minutes) into glucose units (GU).
MALDI-TOF/TOF Analysis for Permethylated Glycans.
N-glycans from 50 μg of purified F8-IL9 proteins were released by PNGase F (Promega), as described above. To verify glycosidic α-2,3–linkage of terminal sialic acids, samples were incubated with 20 U of α-2,3-neuraminidase (New England Biolabs) per µg of F8-IL9 for 16 h at 37 °C. The released glycans from all samples were isolated by C18 Sep-Pak cartridge (Waters) and permethylated as described by Dell et al. (30). Permethylated glycans samples were mixed 1:1 with dihydroxy-benzonic acid matrix (15 mg/mL in 75% acetonitrile in water with 0.1% formic acid) and then spotted onto a MALDI-TOF/TOF MS target plate. Data acquisition was performed manually on a Model 4800 Proteomics Analyzer (Applied Biosystems) with a Nd:YAG laser, and 1,000 shots were accumulated in the reflectron positive ion mode. MALDI-TOF/TOF mass spectrometer was calibrated externally by permethylated N-glycans from RNase B (Sigma). N-glycan structures of m/z 2081.1 and 2,362.2 from SE F8-IL9 were further confirmed by MALDI-TOF/TOF MS/MS. All other annotations are based on the current knowledge for the N-glycosylation synthesis pathway, and the m/z value of each peak was labeled as monoisotopic mass for spectra acquired in reflectron mode. Data interpretation was processed manually or with the help of GlycoWorkbench (Version 2.0; ref. 31).
Sample Preparation for Glycopeptides.
A total of 50-μg purified F8-IL9 protein samples were digested by filter-assisted sample preparation procedure (32) before MS measurement. Briefly, proteins were reduced by 50 mM DTT in 50 mM ammonium bicarbonate buffer (pH 8.5) at 37 °C for 1 h, following by alkylation by 65 mM iodoacetamide at 37 °C in the dark for 1 h. After washing the filter device with ammonium bicarbonate buffer four times, proteins were digested by sequencing-grade modified trypsin (Promega) at a ratio of 50:1 at 37 °C overnight. All digested peptides and glycopeptides were collected by centrifugation and dried with a vacuum centrifuge. Additionally, two-thirds of the samples were further treated with Glu-C endopeptidase (Promega), AspN endopeptidase (Promega), neuraminidase (Calbiochem), and PNGase F (Roche) individually. All samples were desalted by Zip-Tip C18 (Millipore) before nanoLC-MS/MS analysis.
Glycopeptide Analysis by NanoLC-HCD-MS/MS.
Samples were analyzed on a calibrated LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific) coupled to an Eksigent-Nano-HPLC system (Eksigent Technologies). Peptides were resuspended in 2.5% acetonitrile and 0.1% formic acid and loaded on a self-made fritted column (75 µm × 150 mm) packed with reverse-phase C18 material (AQ, 1.9 μm 200 Å; Bischoff GmbH) and eluted with a flow rate of 300 nL/min by a gradient from 3% to 30% of B in 22 min, 50% of B in 25 min, and 97% of B in 27 min. One scan cycle comprised a full-scan MS survey spectrum, followed by up to 10 sequential HCD MS/MS on the most intense signals above a threshold of 2,000. Full-scan MS spectra (700–2,000 m/z) were acquired in the FT-Orbitrap at a resolution of 60,000 at 400 m/z, and HCD MS/MS spectra were recorded in the FT-Orbitrap at a resolution of 15,000 at 400 m/z. HCD was performed with a target value of 1e5, and stepped collision energy rolling from 35, 40, and 45 V was applied. AGC target values were 5e5 for full Fourier transform MS. For all experiments, dynamic exclusion was used with one repeat count, 15-s repeat duration, and 60-s exclusion duration.
Database Search and Site-Specific Glycosylation Analysis.
MS and MS/MS data were processed into the Mascot generic format files and searched against the Swissprot database (Version 201408) through Mascot engine (Version 2.4) with the consideration of carbamidomethylation at cysteine and oxidation at methionine. For PNGase F digestions, deamination was considered as a variable modification. The monoisotopic masses of 2+ or more charged peptides were searched with a peptide tolerance of 10 ppm and a MS/MS tolerance of 0.25 Da for fragment ions. Only peptides with a maximum of two missing cleavage sites were allowed in database searches. Positive identification of deaminated peptides was performed by using a variety of strict criteria, including manual inspection of spectra. For site-specific glycosylation analysis, all data were interpreted manually. Here, XCalibur (Version 2.2 sp1.48) was used for data analysis.
Supplementary Material
Acknowledgments
We thank Dr. Danilo Ritz, Dr. Tim Fugmann, and Dr. Vivianne Otto for useful discussions. We give special thanks to Dr. Peter Gehrig (Functional Genomics Center Zürich) for help with the mass spectrometer and to Ruth Alder and Prof. Dr. Irmgard Werner for providing the HPLC system. This research was supported by the ETH Zürich, the Swiss National Science Foundation, and Philochem AG.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416694112/-/DCSupplemental.
References
- 1.Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol. 2014;32(10):992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 2.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol. 2006;24(10):1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 3.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313(5787):670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara C, et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci USA. 2011;108(31):12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinclair AM, Elliott S. Glycoengineering: The effect of glycosylation on the properties of therapeutic proteins. J Pharm Sci. 2005;94(8):1626–1635. doi: 10.1002/jps.20319. [DOI] [PubMed] [Google Scholar]
- 6.Elliott S, et al. Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat Biotechnol. 2003;21(4):414–421. doi: 10.1038/nbt799. [DOI] [PubMed] [Google Scholar]
- 7.Jones AJ, et al. Selective clearance of glycoforms of a complex glycoprotein pharmaceutical caused by terminal N-acetylglucosamine is similar in humans and cynomolgus monkeys. Glycobiology. 2007;17(5):529–540. doi: 10.1093/glycob/cwm017. [DOI] [PubMed] [Google Scholar]
- 8.Wu AM, Senter PD. Arming antibodies: Prospects and challenges for immunoconjugates. Nat Biotechnol. 2005;23(9):1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 9.Hess C, Venetz D, Neri D. Emerging classes of armed antibody therapeutics against cancer. Medchemcomm. 2014;5(4):408–431. [Google Scholar]
- 10.Morell AG, Gregoriadis G, Scheinberg IH, Hickman J, Ashwell G. The role of sialic acid in determining the survival of glycoproteins in the circulation. J Biol Chem. 1971;246(5):1461–1467. [PubMed] [Google Scholar]
- 11.Lee SJ, et al. Mannose receptor-mediated regulation of serum glycoprotein homeostasis. Science. 2002;295(5561):1898–1901. doi: 10.1126/science.1069540. [DOI] [PubMed] [Google Scholar]
- 12.Ghaderi D, Taylor RE, Padler-Karavani V, Diaz S, Varki A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat Biotechnol. 2010;28(8):863–867. doi: 10.1038/nbt.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villa A, et al. A high-affinity human monoclonal antibody specific to the alternatively spliced EDA domain of fibronectin efficiently targets tumor neo-vasculature in vivo. Int J Cancer. 2008;122(11):2405–2413. doi: 10.1002/ijc.23408. [DOI] [PubMed] [Google Scholar]
- 14.Rybak JN, Roesli C, Kaspar M, Villa A, Neri D. The extra-domain A of fibronectin is a vascular marker of solid tumors and metastases. Cancer Res. 2007;67(22):10948–10957. doi: 10.1158/0008-5472.CAN-07-1436. [DOI] [PubMed] [Google Scholar]
- 15.Perrino E, et al. Curative properties of noninternalizing antibody-drug conjugates based on maytansinoids. Cancer Res. 2014;74(9):2569–2578. doi: 10.1158/0008-5472.CAN-13-2990. [DOI] [PubMed] [Google Scholar]
- 16.Purwar R, et al. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat Med. 2012;18(8):1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, et al. Tumor-specific IL-9-producing CD8+ Tc9 cells are superior effector than type-I cytotoxic Tc1 cells for adoptive immunotherapy of cancers. Proc Natl Acad Sci USA. 2014;111(6):2265–2270. doi: 10.1073/pnas.1317431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajendra Y, Kiseljak D, Baldi L, Hacker DL, Wurm FM. A simple high-yielding process for transient gene expression in CHO cells. J Biotechnol. 2011;153(1-2):22–26. doi: 10.1016/j.jbiotec.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Gutbrodt KL, Casi G, Neri D. Antibody-based delivery of IL2 and cytotoxics eradicates tumors in immunocompetent mice. Mol Cancer Ther. 2014;13(7):1772–1776. doi: 10.1158/1535-7163.MCT-14-0105. [DOI] [PubMed] [Google Scholar]
- 20.Graff CP, Wittrup KD. Theoretical analysis of antibody targeting of tumor spheroids: Importance of dosage for penetration, and affinity for retention. Cancer Res. 2003;63(6):1288–1296. [PubMed] [Google Scholar]
- 21.Hughes RC. Galectins as modulators of cell adhesion. Biochimie. 2001;83(7):667–676. doi: 10.1016/s0300-9084(01)01289-5. [DOI] [PubMed] [Google Scholar]
- 22.Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5(1):29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 23.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6(5):343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 24.Schiestl M, et al. Acceptable changes in quality attributes of glycosylated biopharmaceuticals. Nat Biotechnol. 2011;29(4):310–312. doi: 10.1038/nbt.1839. [DOI] [PubMed] [Google Scholar]
- 25.Brik A, Ficht S, Wong CH. Strategies for the preparation of homogenous glycoproteins. Curr Opin Chem Biol. 2006;10(6):638–644. doi: 10.1016/j.cbpa.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz F, et al. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat Chem Biol. 2010;6(4):264–266. doi: 10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meuris L, et al. GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat Biotechnol. 2014;32(5):485–489. doi: 10.1038/nbt.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anumula KR. Rapid quantitative determination of sialic acids in glycoproteins by high-performance liquid chromatography with a sensitive fluorescence detection. Anal Biochem. 1995;230(1):24–30. doi: 10.1006/abio.1995.1432. [DOI] [PubMed] [Google Scholar]
- 29.Bigge JC, et al. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230(2):229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 30.Dell A, et al. Mass spectrometry of carbohydrate-containing biopolymers. Methods Enzymol. 1994;230:108–132. doi: 10.1016/0076-6879(94)30010-0. [DOI] [PubMed] [Google Scholar]
- 31.Ceroni A, et al. GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7(4):1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 32.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 33.Varki A, et al. Symbol nomenclature for glycan representation. Proteomics. 2009;9(24):5398–5399. doi: 10.1002/pmic.200900708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.