Significance
Transcription termination is an essential regulatory step in gene expression, but its molecular mechanisms remain less understood in comparison with other steps of transcription, owing to the highly dynamic nature of the termination process. Antitermination factors play important regulatory roles in bacteria, viruses, and eukaryotes, but only a few such factors have been studied in molecular detail. We describe a pathway of transcription antitermination by a viral factor that involves suppression of RNA polymerase pausing and stabilization of the transcription complex through interactions with an evolutionary conserved ω subunit of RNA polymerase. The results of the study extend the list of universal RNA polymerase sites able to accept regulatory inputs and provide new insights into the mechanisms of transcription termination.
Keywords: RNA polymerase, transcription antitermination, transcription pausing, omega subunit, NusA
Abstract
Transcription antitermination is a common strategy of gene expression regulation, but only a few transcription antitermination factors have been studied in detail. Here, we dissect the transcription antitermination mechanism of Xanthomonas oryzae virus Xp10 protein p7, which binds host RNA polymerase (RNAP) and regulates both transcription initiation and termination. We show that p7 suppresses intrinsic termination by decreasing RNAP pausing and increasing the transcription complex stability, in cooperation with host-encoded factor NusA. Uniquely, the antitermination activity of p7 depends on the ω subunit of the RNAP core and is modulated by ppGpp. In contrast, the inhibition of transcription initiation by p7 does not require ω but depends on other RNAP sites. Our results suggest that p7, a bifunctional transcription factor, uses distinct mechanisms to control different steps of transcription. We propose that regulatory functions of the ω subunit revealed by our analysis may extend to its homologs in eukaryotic RNAPs.
Transcription promoters and terminators are the main “punctuation marks” that control the expression of individual genes in the genome. In bacteria, transcription termination is an essential regulatory step that determines the relative levels of expression of promoter-proximal and distal genes within operons. Depending on the factors involved, transcription termination in bacteria can be classified as intrinsic (depends on RNA-encoded signals) or factor-dependent (requires additional protein factors such as Rho helicase). Intrinsic terminators consist of a G/C-rich hairpin formed in the nascent RNA followed by a downstream oligo(U) tract. During intrinsic termination, RNA polymerase (RNAP) pauses after synthesizing an oligo(U) sequence, accompanied by hairpin formation, leading to subsequent dissociation of the transcription elongation complex (TEC) (1, 2). Despite the relatively simple overall scenario, the exact mechanisms of pausing and hairpin-induced TEC destabilization remain an issue of debate (1, 3).
Transcription antitermination is a widespread phenomenon involved in regulation of bacterial and phage operons (2). Transcription antitermination is often mediated by specialized protein factors that target host RNAP and can suppress termination at multiple sites in the operon. The well-studied examples of phage antiterminators include proteins N and Q of Escherichia coli phage λ and the gp39 protein of Thermus thermophilus phage P23-45. These factors suppress RNAP pausing, thus inhibiting the first step of the termination pathway (4–6). The antitermination activity of N and Q, but not gp39, is enhanced by cell-encoded proteins, including transcription elongation factor NusA, which by itself stimulates termination but reverses its activity to additionally stabilize the TEC in cooperation with N and Q (5, 6). Curiously, the N, Q, gp39, and NusA proteins all target the flexible β flap domain of RNAP (2, 4, 7–11) that forms a part of the RNA exit channel and has been implicated in intrinsic termination and transcription pausing through direct interaction with RNA hairpins (Fig. 1) (3, 12–14). It therefore seems that unrelated phage-encoded antiterminators use similar strategies to control transcription termination through interactions with the β flap, likely leading to changes in the TEC conformation and stability. Another mechanism is used by RfaH, the best-studied cell-encoded processive antiterminator, which interacts with the coiled-coil motif of the β′ clamp domain and the β gate loop (Fig. 1A) and encloses the RNA–DNA hybrid within the RNAP channel, resulting in suppression of transcription pausing and, probably, TEC stabilization (15).
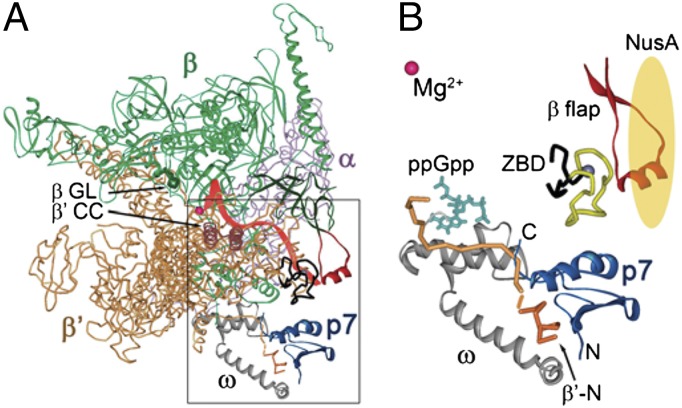
Fig. 1.
Structural model of the RNAP–p7 complex. (A) A model of the RNAP–p7 complex based on the X-ray structure of the E. coli RNAP holoenzyme 4IGC (49) and NMR structure of a complex of p7 with the N-terminal peptide of the X. oryzae β′ subunit (17). Mg2+ ion in the RNAP active center is shown as a red sphere. The σ subunit present in the original holoenzyme structure is not shown; the position of the RNA transcript (red) is drawn based on the T. thermophilus TEC structure (50). The β flap, β′ coiled-coil (β′CC), and β gate loop (βGL) elements are indicated. (B) A close-up view of the p7–RNAP interactions. P7 is shown in blue; the β′ subunit is orange, with the N-terminal peptide [residues 1–10 complexed with p7 (17)] shown in a darker tone. The ω subunit is gray; the β′ ZBD is black, with amino acid residues 70–88 deleted in this work shown in yellow; and the β flap (residues 885–921) is red. The position of ppGpp (turquoise) is superimposed from the E. coli RNAP holoenzyme ppGpp structure (4JK1) (28). NusA is schematically shown as a yellow oval.
The only other phage-encoded antiterminator protein that is known to date is protein p7 of Xp10, a lytic phage of the Siphoviridae family that infects Xanthomonas oryzae, a prominent plant pathogen that causes leaf blight disease in rice (16). Protein p7 likely switches the phage gene expression during infection by affecting several steps of transcription by X. oryzae RNAP. In particular, p7 inhibits recognition of a subset of cellular promoters (belonging to the -10/-35 class) and suppresses intrinsic termination by the host RNAP, which likely ensures efficient expression of early phage genes (transcribed from extended -10 promoters, which are resistant to p7) (16–18). The primary p7 binding site was localized in the N terminus of the β′ subunit, across the RNA exit channel with respect to the β flap and close to the β′ zinc-binding domain (ZBD) and the RNAP ω subunit (Fig. 1A), suggesting that the mechanism of transcription antitermination by p7 may differ from those used by other phage antiterminator proteins (17, 19). Previously, the β′ ZBD, which together with the β flap forms the RNA exit channel, has been implicated in transcription termination and antitermination (20), whereas no termination-specific functions have been reported for the ω subunit. A recent study suggested that p7 can also interact with the β flap domain, which may be important for inhibition of transcription initiation (17). However, the structure of the RNAP–p7 complex, the functional sites involved in p7-dependent antitermination, and the particular step(s) of termination targeted by p7 remain unknown. In this work, we demonstrate that the p7 action depends on its interactions with the RNAP ω subunit, but not the β flap and β′ ZBD, and results from both suppression of RNAP pausing and NusA-dependent TEC stabilization.
Results
p7 Is Active on Intrinsic but Not on Rho-Dependent Terminators.
Previously, it was shown that E. coli RNAP containing a 10-aa segment from the N terminus of the β′ subunit of X. oryzae RNAP instead of corresponding E. coli β′ amino acids binds to and is inhibited by p7 in transcription initiation assays in vitro (17, 19). Because E. coli RNAP does not bind p7 it was concluded that these 10 amino acids comprise the determinant of tight p7 binding. Surprisingly, we observed that p7 did not suppress transcription termination by the hybrid RNAP on several intrinsic terminators tested (Fig. S1 A and C, lanes 1–4). At the same time, p7 acted as an efficient antiterminator of transcription by RNAP purified from X. oryzae (Fig. S1C, lanes 5–8). Thus, transcription antitermination by p7 likely depends on additional elements that distinguish the E. coli and X. oryzae RNAPs and that lie outside the primary binding site in the N terminus of β′. All experiments described below were performed with X. oryzae RNAP.
The paradigmatic antiterminator proteins N and Q of phage λ suppress transcription termination at both intrinsic and Rho-dependent terminator sites (6, 21–23). We therefore tested the effects of p7 on transcription termination at E. coli Rho-dependent terminator trpT′. X. oryzae RNAP responded to the Rho factor at this terminator, resulting in RNA release within the terminator region. However, p7 did not have significant effect on termination when present either alone or together with X. oryzae NusA (Fig. S2).
NusA Stimulates the Antitermination Activity of p7 on Intrinsic Terminators.
To reveal the details of the p7 action on intrinsic termination we performed a series of experiments on the model λ tR2 terminator (Fig. S1A). As measures of transcription termination and antitermination we used the following parameters determined in titration experiments with p7: (i) apparent dissociation constant (Kd) for p7 binding to the TEC, defined as concentration when half of the maximal effect of p7 was observed; (ii) the maximal termination efficiency (Tmax), measured under the standard reaction conditions in the absence of p7; and (iii) the minimal termination efficiency (Tmin), measured at saturating p7 concentrations (see SI Materials and Methods for details). Titration experiments demonstrated that p7 binds the TEC and stimulates full-length RNA synthesis with apparent Kd of ∼270 nM at 37 °C and ∼130 nM at 30 °C (Fig. 2, Table S1, and Fig. S1). The termination efficiency by X. oryzae RNAP in the absence of added factors was higher at 37 °C than at 30 °C (Tmax = 87.6% and 52.7%, respectively). Moreover, a considerable level of termination at 37 °C was observed even at saturating p7 concentrations (Tmin = 40.1% vs. 9.9% at 30 °C). Therefore, most experiments were performed at 30 °C, which is a physiological temperature for X. oryzae.
Fig. 2.
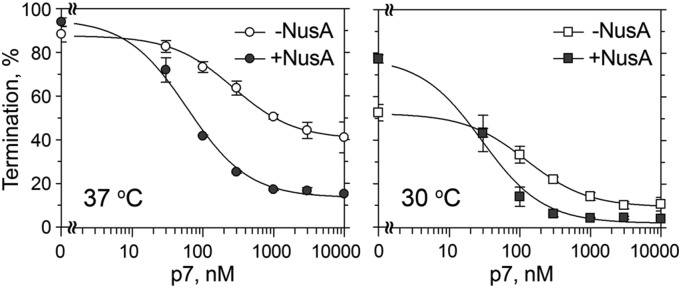
P7-mediated transcription antitermination by X. oryzae RNAP. Termination was analyzed at the λ tR2 terminator at increasing p7 concentrations (from 30 nM to 10 μM) either in the absence or in the presence of NusA. The experiments were performed at 37 °C (Left) or 30 °C (Right). Averages and SDs from three or four independent experiments are shown.
X. oryzae NusA stimulated intrinsic termination by X. oryzae RNAP. This effect was especially visible at 30 °C, because at this temperature termination in the absence of added factors was less efficient (Tmax = 78.1% in comparison with 52.7% in the absence of NusA) (Fig. 2 and Table S1). At the same time, NusA significantly stimulated transcription antitermination by p7. In particular, NusA (i) increased the apparent affinity of p7 to TEC (∼4.5-fold; p7 Kd of ∼60 and ∼30 nM at 37 °C and 30 °C, respectively) and (ii) decreased the termination efficiency at saturating concentrations of p7. The latter effect was especially pronounced at 37 °C, because at this temperature the termination in the absence of the factors was more efficient (Tmin = 13.5% in comparison with 40.1% in the absence of NusA).
p7 Cooperates with NusA to Increase TEC Stability.
The p7 protein could antiterminate transcription by suppressing initial RNAP pausing at the terminator site, preventing RNA hairpin formation, or counteracting hairpin-induced conformational changes and preventing TEC dissociation. We wished to reveal which stage of the termination mechanism is affected by p7.
To reveal possible effects of p7 on transcript release and TEC stability we analyzed oligonucleotide-mediated termination in synthetic TECs assembled on a tR2 terminator-based nucleic acid scaffold (Fig. 3 and Fig. S3). In this assay, the transcription termination in the TEC stalled at a desired template position is induced by the addition of short antisense DNA oligonucleotides complementary to the nascent RNA transcript, thus mimicking the hairpin formation (4, 6). Control experiments demonstrated that the scaffold-based system adequately mimics the properties of a natural terminator (see SI Results and Fig. S3 for details). We then analyzed the kinetics of oligonucleotide-mediated RNA release in the TEC stalled at the termination point. Under the conditions of our experiments, the amount of released RNA increased from ∼30% after 1 min to ∼60% after 10 min of incubation (Fig. 3). The addition of p7 did not inhibit RNA release and even slightly increased it at the 1-min time point. Therefore, p7 by itself does not act by stabilizing the paused termination complexes and does not prevent the termination hairpin formation in the stalled TEC, mimicked by the oligonucleotide annealing.
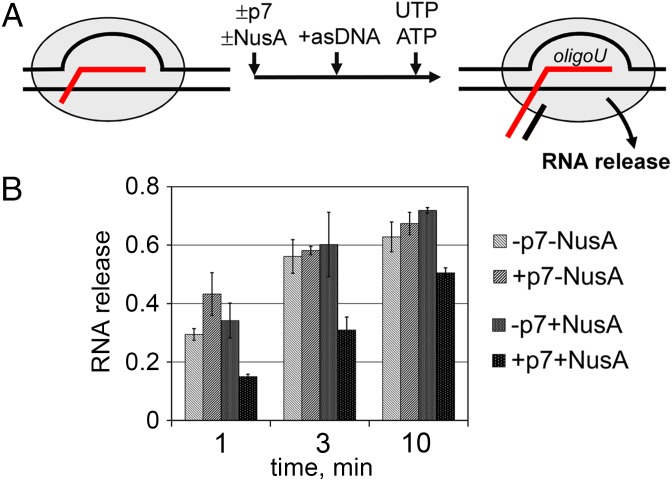
Fig. 3.
Analysis of RNA release in synthetic termination complexes. (A) Outline of experiment. P7, NusA, and antisense oligonucleotides (asDNA) were added to the reconstituted TEC bound to an affinity resin; addition of UTP and ATP resulted in transcription to the end of the U-tract, followed by RNA release into the supernatant fraction. (B) Kinetics of RNA release in TECs stalled at the termination point. The efficiency of RNA release was calculated as a ratio of the amount of released RNA to the sum of the amounts of resin-bound and released RNAs (averages and SDs from three independent experiments are shown). See Fig. S3 for further details.
NusA alone did not affect the kinetics of RNA release in the stalled TEC but significantly slowed it down when added together with p7 (for example, only ∼15% of RNA was released at the 1-min time point; Fig. 3). Therefore, NusA can stimulate p7-mediated transcription antitermination by additionally stabilizing the RNA transcript in the termination complex.
p7 Suppresses Transcription Pausing at a Subset of Pause Sites.
The absence of observable effects of p7 on the dissociation of stalled termination complexes suggested that it may affect the first step of termination; i.e., transcription pausing. Current models of intrinsic termination suggest that the U-tract is a primary cause of pausing (1, 24), although the termination hairpin has also been proposed to contribute (1, 25).
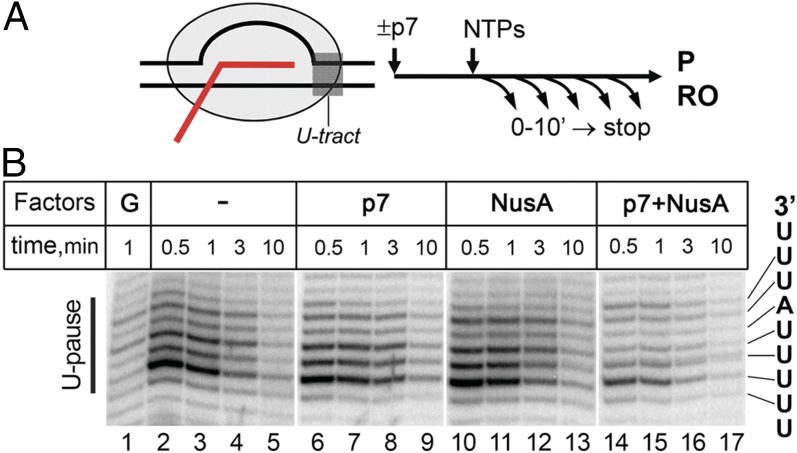
We first tested whether p7 can affect RNAP pausing at a terminator-derived U-tract placed after the initially transcribed sequence of the λ PR promoter (Fig. 4). P7 or NusA alone did not significantly affect the pausing (compare lanes 2–5 and 6–13). At the same time, p7 together with NusA suppressed pausing at most U-tract positions (see Fig. S4 for more details). This effect likely contributes to the observed stimulation of p7 antitermination by NusA. However, pausing was also significantly affected by the nascent transcript cleavage factor GreA (lane 1), suggesting that an isolated U-tract promotes TEC backtracking, which differs from the situation at terminators where hairpin formation likely prevents it. It therefore remains to be established whether p7 could suppress U-tract–dependent pausing in the absence of backtracking.
Fig. 4.
Effects of p7 and NusA on the U-tract-induced pausing. (A) Outline of experiment. (B) Pausing at the U-tract in the absence or in the presence of p7, NusA, and GreA. The sequence of the U-tract is shown on the right. See Fig. S4 for further details.
To reveal whether p7 can affect recognition of other types of pause-inducing signals we analyzed the general effects of p7 on RNA elongation, using a E. coli rpoB-based DNA template. P7 increased the average elongation rate, resulting in a faster appearance of full-length RNA products (Fig. S5). The stimulatory effect of p7 was caused by suppression of some, but not all, transcription pauses observed on this template. At least some of the pause sites that were resistant to p7 disappeared in the presence of GreA (Fig. S5C) and therefore corresponded to backtracked complexes. The result thus suggests that p7 does not act on backtracked TECs. NusA by itself decreased the elongation rate by increasing the RNAP pausing at certain template positions but slightly increased the overall rate of elongation when present together with p7.
In E. coli, the most common type of pausing signals are consensus pauses that likely result from stabilization of the pretranslocated state of the TEC and are defined by several conserved nucleotides within and downstream of the transcription bubble (Fig. S6A) (26, 27). Whereas X. oryzae RNAP responded to this type of pausing signal, p7 not only did not suppress pausing on such a site but actually slightly stimulated it (Fig. S6 B and C).
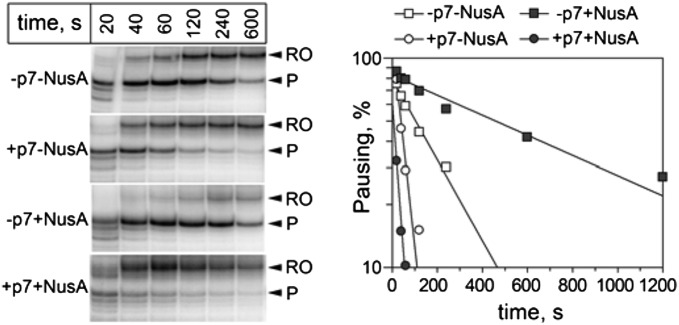
In conrast, p7 efficiently suppressed pausing on a hairpin-dependent hisP pause site (Fig. 5). Whereas NusA by itself increased the pausing, it stimulated the antipausing effect of p7. We therefore propose that the ability of p7 to inhibit hairpin-dependent pausing is related to its antiterminating activity, and that this type of pausing might contribute to intrinsic termination (see Discussion).
Fig. 5.
Effect of p7 and NusA on transcription pausing at the hisP hairpin-dependent pause signal. Positions of the paused (P) and run-off (RO) RNA products are indicated. The plot on the right shows the efficiency of pausing (in logarithmic scale) as a function of time. The data were fit to the single-exponential equation.
The β Flap Domain and the β′ ZBD of X. oryzae RNAP Are Not Essential for p7-Mediated Antitermination.
We further asked which sites of RNAP, in addition to the one in the N-terminal part of the β′ subunit, may be important for the p7-dependent antitermination. To answer this question, we analyzed X. oryzae RNAP variants with changes in the three sites that surround the primary p7-binding site: the β flap domain, the β′ ZBD, and the ω subunit (Fig. 1B).
We reconstituted X. oryzae RNAP with a β−flap deletion (Δ918–954, corresponding to Δ885–921 in E. coli, Fig. 1B) from individual subunits in vitro and analyzed its termination properties. The deletion significantly decreased the termination efficiency (Tmax = 25.5% in comparison with 52.7% for wild-type X. oryzae RNAP) (Fig. S7), suggesting that the β flap is important for intrinsic termination. NusA did not increase the termination efficiency by the Δflap X. oryzae RNAP, similarly to the reported effects of flap mutations on NusA action on E. coli RNAP (8, 13). However, p7 was still able to further decrease termination by the Δflap RNAP to the same level as seen with the wild-type RNAP, although the fold change in the termination efficiency was lower because it was already impaired by the deletion. The deletion also did not decrease the apparent affinity of p7 to RNAP. As expected, NusA did not have any significant effect on the p7 antitermination in the case of the Δflap RNAP (Fig. S7). Therefore, the flap domain is essential for NusA action but is not the primary target for the p7-mediated antitermination.
To test whether the β′ ZBD is important for p7 action we obtained a X. oryzae RNAP variant with deletion of the whole zinc-binding motif (amino acid residues 70–88 in both E. coli and X. oryzae RNAPs) by in vitro reconstitution (Fig. 1B). The mutant ΔZBD RNAP revealed lower efficiency of termination than the wild-type RNAP both in the absence and in the presence of NusA (∼40 and 63%, respectively) (Fig. S8). This corresponds to the previously reported effects of β′ ZBD mutations on intrinsic termination by E. coli RNAP (20). At the same time, p7 suppressed termination by the mutant RNAP to almost the same level as in the case of the control reconstituted wild-type X. oryzae RNAP. Therefore, the β′ ZBD likely plays only a minor role in p7-dependent antitermination.
The Antitermination Activity of p7 Depends on the ω Subunit of Core RNAP and Can Be Modulated by Guanosine Tetraphosphate.
Finally, we tested the transcription termination properties of a RNAP variant lacking the ω subunit, which was obtained by in vitro reconstitution. The ω-less RNAP did not differ significantly from the native wild-type X. oryzae RNAP ω in termination efficiency and was fully responsive to NusA (Tmax = 52.3 and 87.1% in the absence and in the presence of NusA, respectively) (Fig. 6A and Table S1). At the same time, the ω-less RNAP was essentially resistant to the p7 action in termination (apparent p7 Kd ≥10 μM). Remarkably, the addition of purified ω subunit directly to the transcription reaction significantly stimulated the antitermination effect of p7 (p7 Kd ∼800 nM; Tmin = 17.1%) (Fig. 6A and Table S1).
Fig. 6.
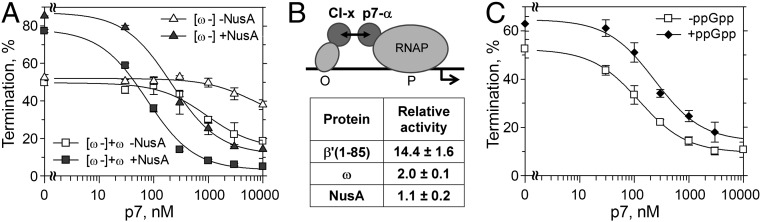
Roles of the ω subunit and ppGpp in p7-mediated antitermination. (A) Termination at the λ tR2 terminator at different p7 concentrations by reconstituted ω-less X. oryzae RNAP without ([ω-], triangles) or with addition of purified ω subunit ([ω-]+ω, squares), either in the absence (open symbols) or in the presence (filled symbols) of NusA. (B) Bacterial two-hybrid analysis of p7 interactions. P7 was fused to the α subunit of E. coli RNAP (interacting with a promoter, P, placed before the reporter lacZ gene) whereas the other interacting partner (x) was fused to the λ CI repressor (interacting with its operator sequence, O). Relative β-galactosidase activities for various p7 partners (in comparison with control noninteracting protein pairs) are shown in the table. (C) Effect of ppGpp on intrinsic termination and p7-dependent antitermination at the λ tR2 terminator. The experiment was performed at 30 °C with native wild-type X. oryzae RNAP in the absence (open symbols) or in the presence (filled symbols) of 100 µM ppGpp.
NusA also dramatically stimulated p7-dependent antitermination by ω-less RNAP, by increasing the apparent p7 affinity (p7 Kd ∼200 nM) and decreasing the Tmin (11.5%) (Fig. 6A and Table S1). The addition of purified ω further decreased the termination efficiency at saturation (Tmin ∼3.8%). Therefore, the ω subunit and NusA likely participate in p7 antitermination by acting on parallel pathways, rather than subsequent steps of the same pathway.
To test whether p7 may directly interact with the ω subunit, we performed a bacterial two-hybrid assay (Fig. 6B; see the figure legend for experimental details). As expected, p7 strongly interacted with the N terminus of the X. oryzae β′ subunit, resulting in significant transcription activation of a reporter gene (14.4-fold increase in the reporter activity). In contrast, p7 did not interact with NusA in this assay. At the same time, we observed a weak but reproducible interaction between p7 and the ω subunit (twofold increase in the reporter activity), suggesting that p7 may indeed directly interact with ω in the TEC.
Recent structural and biochemical work indicated that the ω subunit constitutes a part of the binding site for guanosine tetraphosphate (ppGpp), an alarmone that has been implicated in the regulation of both transcription initiation and elongation (Fig. 1B) (28–30). We therefore tested whether ppGpp will have any effects on antitermination by p7. The addition of ppGpp increased the termination efficiency by X. oryzae RNAP (from 52.7 to 62.6%) (Fig. 6C and Table S1), which is similar to its effects on termination by E. coli RNAP (31, 32). Furthermore, the p7 titration experiments demonstrated that ppGpp decreased the antitermination efficiency at most p7 concentrations and decreased the apparent affinity of p7 to the TEC (Kd ∼265 nM in comparison with ∼130 nM in the absence of ppGpp). Therefore, ppGpp and p7 may use the same pathway to control transcription termination (see Discussion).
The β′ ZBD but Not the ω Subunit Is Important for Inhibition of Transcription Initiation by p7.
To reveal whether removal of the ω subunit has a general effect on p7 interactions with RNAP we analyzed p7-dependent inhibition of transcription initiation by ω-less RNAP, by measuring the efficiency of abortive RNA synthesis from the T7 A1 promoter (a model -10/-35 class promoter) in the presence of increasing p7 concentrations. Remarkably, the absence of the ω subunit did not decrease the efficiency of transcription inhibition by p7 (Fig. S9). This suggests that the observed effect of removal of ω on transcription antitermination cannot be explained by a simple loss of p7 binding to RNAP. In contrast, the β′ ZBD deletion severely affected the efficiency of transcription initiation inhibition, even at the highest p7 concentrations tested (Fig. S9), indicating that this domain is essential for p7 function during transcription initiation. Thus, the studied RNAP mutations differentially affect regulation of transcription initiation and termination by p7, suggesting that this bifunctional protein uses distinct subsets of contacts with RNAP to exert its partial functions and control different steps of the transcription cycle.
Discussion
In this study we investigated transcription antitermination by the p7 protein of X. oryzae phage Xp10. A two-tier mechanism of the p7 action emerges from our analysis.
First, p7 can suppress intrinsic termination in the absence of any cofactors, without changing TEC stability. This activity crucially and uniquely depends on the RNAP core ω subunit with which p7 seems to interact but does not require the β flap and β′ ZBD. P7 also suppresses RNAP pausing at some sites, including hairpin-dependent pause sites, but not at the U-tract or consensus pause sequences, thus revealing mechanistic differences between various types of pause-inducing signals.
Second, NusA modulates p7 antitermination by several mechanisms. NusA increases apparent p7 affinity to the TEC, enables it to suppress the U-tract pausing, enhances the effect of p7 on hairpin-dependent pausing, and helps to prevent RNA release from the termination complexes. These NusA effects do not require the ω subunit but are strictly dependent on the β flap domain of RNAP, in agreement with the proposed role of the β flap in NusA binding. Because our two-hybrid data suggest that p7 and NusA do not interact directly, the stimulation of p7 binding likely results from NusA-induced conformational changes in the TEC structure. NusA may also stimulate the antipausing and antitermination activities of p7 by affecting the interaction of the β flap with the nascent RNA transcript, which has been implicated in both transcription pausing and termination (8, 33).
The NusA-independent antitermination activity of p7 is likely related to its ability to suppress hairpin-dependent pausing, because both types of signals depend on RNA hairpins that are formed in the RNA exit channel of RNAP. The pausing hairpin formation was shown to induce the β′ clamp opening that further inhibits RNAP translocation and prevents the trigger loop folding required for catalysis in the RNAP active site. These effects depend on hairpin interactions with the β flap domain, which is essential for pausing and associated conformational changes but not for hairpin formation per se (14, 34, 35). Although the contribution of hairpin-dependent pausing to termination remains controversial (reviewed in ref. 1), the termination hairpin may induce similar changes in the TEC structure, at least at the initial steps of its folding. The p7 action on both pausing and termination could therefore result from its effects on the hairpin formation and/or subsequent TEC rearrangements. P7 likely does not prevent hairpin formation because it does not affect oligonucleotide-mediated RNA release in stalled TECs. Furthermore, deletions in the β flap tip decrease transcription termination (refs. 4, 12, and 14 and this work) but do not have additive effects with p7 antitermination, which could have been expected if p7 disrupted the hairpin folding. P7 could in principle affect the kinetics of RNA folding, a hypothesis that can be further tested using a recently developed technique that uses quenching of fluorophore-labeled nascent transcript with antisense RNA (34). Alternatively, p7 binding at the bottom of the β′ clamp may inhibit hairpin-induced conformational changes, including clamp opening and associated effects on the trigger loop folding and/or RNAP translocation, the pathway that can also be probed with recently described cystein-pair reporters that fix various RNAP conformations (34, 35). Based on available evidence, we hypothesize that hairpin-induced pausing might play a role in intrinsic termination and its regulation by various factors, including antiterminators that bind in the vicinity of the RNA exit channel (discussed below). P7-dependent antitermination may provide a useful model to reveal the intricate details of TEC conformational changes that occur during these processes.
The ω subunit is one of the universally conserved core subunits present in all cellular RNAPs (36), but its functions remain only partially understood (37). We provide the first evidence, to our knowledge, that ω is involved in the regulation of transcription elongation and termination. Recently, certain mutations in the ω subunit were shown to affect the properties of the RNAP active site, revealing its potential role in the regulation of RNAP activity (38). The ω subunit was also shown to participate in the binding of ppGpp, an alarmone that plays a key role in the stringent response in bacteria. The binding of ppGpp was demonstrated to affect the active site conformation through changes in the relative positions and mobility of the core and shelf/clamp RNAP modules, resulting in destabilization of promoter complexes and, probably, modification of the elongation properties of RNAP (28–30). Importantly, ppGpp stimulates intrinsic transcription termination and transcription pausing (refs. 31, 32, and 39 and this study), likely as a result of conformational changes in the RNAP active site [although a direct competition with NTP substrates was also proposed to contribute to the transcription effects of ppGpp (ref. 40 and references therein)]. Furthermore, ppGpp counteracts the p7 effects on transcription termination by X. oryzae RNAP, probably by interfering with p7-induced conformational changes and/or p7 binding (but its independent effect on RNA elongation cannot also be excluded). We therefore propose that p7, which binds RNAP from the opposite site of the ω subunit with respect to ppGpp (Fig. 1B), may use the same allosteric pathway to suppress pausing and stimulate RNA elongation.
The eukaryotic ω ortholog, the Rpb6 subunit, which is shared by all three cellular RNAPs, was shown to genetically interact with TFIIS, a factor that directly accesses the active site through the secondary RNAP channel, suggesting that Rpb6 may also allosterically regulate RNAP II activity (41). Furthermore, Rpb6 was proposed to affect the clamp opening and DNA/RNA binding, depending on its phosphorylation state, and to interact with accessory RNAP II subunits (42). Therefore, the regulatory functions of the ω subunit likely extend to its Rpb6 counterpart in eukaryotic RNAPs.
Comparison of various classes of processive antiterminator proteins studied to date reveals three major RNAP effector sites involved in transcription antitermination (Fig. 1A and Table S2): (i) the β′ coiled-coil/β gate loop, targeted by RfaH and its paralogs (15); (ii) the β flap/RNA exit channel, targeted by N, Q and gp39 (4, 7, 9–11); and (iii) the β′ N terminus and adjacent regions, involved in antitermination by p7. In addition, p7 seems to partially share its target site with an RNA-based antiterminator, the put hairpin encoded by phage HK022, the function of which was shown to depend on the β′ ZBD (20, 43, 44). Despite the different binding sites, all classes of factors suppress transcription pausing by bacterial RNAP, which therefore is likely a universal part of all antitermination mechanisms (Table S2). Most of the studied factors (with a notable exclusion of RfaH and gp39) also rely on cell-encoded proteins, primarily NusA, to strengthen their intrinsic antipausing activity and additionally stabilize the TEC against dissociation (refs. 5, 6, and 45 and this work). Therefore, NusA may function as a general switch to enhance the termination/antitermination properties of RNAP in response to various regulatory stimuli. Overall, analysis of known phage and cellular factors suggests that they apparently use a limited number of strategies to control transcription termination through suppression of the transcription pausing and prevention of the RNA hairpin-dependent TEC dissociation. Because eukaryotic RNAPs can respond to the hairpin termination signals in vitro (46, 47), one can expect that similar termination and antitermination pathways may be discovered in eukaryotic gene expression.
Intriguingly, the RNAP sites targeted by p7 during antitermination seem to be different from those targeted for the inhibition of initiation. In particular, the ω subunit, which is critical for p7-dependent antitermination, is not important for p7 inhibition of transcription initiation, and the β flap and β′ ZBD are not absolutely required for antitermination but play crucial roles in the inhibition of initiation (ref. 17 and this work). The combination of two apparently antagonistic activities in the same small protein beautifully illustrates the exquisite ability of phages to manipulate the cell gene expression apparatus by targeting key steps of the transcription cycle by compact multifunctional proteins.
Materials and Methods
Proteins and Bacterial Two-Hybrid Assay.
Native X. oryzae RNAP was purified from X. oryzae strain XO604 as described in ref. 18. The p7 protein, X. oryzae σA, and core RNAP subunits, NusA and Rho were cloned, expressed in E. coli, and purified as described in SI Materials and Methods. Mutant Δflap, ΔZBD, and ω-less X. oryzae RNAPs were obtained by site-directed mutagenesis and in vitro RNAP reconstitution from individual subunits (48). The bacterial two-hybrid assay was performed as described in ref. 4. See SI Materials and Methods for details.
In Vitro Transcription Assays.
Analysis of intrinsic transcription termination was performed as previously described (4). The antitermination parameters were calculated from the p7 titration experiments as described in SI Materials and Methods. Rho-dependent termination was analyzed on a DNA template containing the E. coli trp T′ terminator. Analysis of the elongation rates was performed on a DNA template containing a 500-bp fragment of the E. coli rpoB gene. Analysis of transcription pausing was performed on PCR templates or scaffolds assembled from synthetic oligonucleotides. Analysis of transcription termination in reconstituted TECs was performed essentially as described in ref. 4. Analysis of transcription inhibition by p7 was performed on the T7A1 promoter template. See SI Materials and Methods and Figs. S1–S9 for further details.
Supplementary Material
Acknowledgments
We thank I. Artsimovitch, Y. Yuzenkova, and N. Zenkin for plasmids and helpful discussions; S. Wigneshweraraj for sharing the RNAP-p7 model and discussions; and B. Nickels for plasmids used in the two-hybrid assay. This work was supported in part by the Russian Academy of Sciences Presidium Program in Molecular and Cellular Biology grant (to A.K.) and by Russian Foundation for Basic Research Grants 14-04-01696 and 14-04-31994. Work in K.S. laboratory is supported by an NIH Grant GM59295.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416330112/-/DCSupplemental.
References
- 1.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: The RNA 3′-end chronicles. J Mol Biol. 2011;412(5):793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat Rev Microbiol. 2011;9(5):319–329. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28(6):991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Berdygulova Z, et al. A novel phage-encoded transcription antiterminator acts by suppressing bacterial RNA polymerase pausing. Nucleic Acids Res. 2012;40(9):4052–4063. doi: 10.1093/nar/gkr1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees WA, Weitzel SE, Das A, von Hippel PH. Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage lambda. J Mol Biol. 1997;273(4):797–813. doi: 10.1006/jmbi.1997.1327. [DOI] [PubMed] [Google Scholar]
- 6.Shankar S, Hatoum A, Roberts JW. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol Cell. 2007;27(6):914–927. doi: 10.1016/j.molcel.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deighan P, Diez CM, Leibman M, Hochschild A, Nickels BE. The bacteriophage lambda Q antiterminator protein contacts the beta-flap domain of RNA polymerase. Proc Natl Acad Sci USA. 2008;105(40):15305–15310. doi: 10.1073/pnas.0805757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401(5):708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, et al. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 2009;10(9):997–1002. doi: 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tagami S, et al. Structural basis for promoter specificity switching of RNA polymerase by a phage factor. Genes Dev. 2014;28(5):521–531. doi: 10.1101/gad.233916.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuznedelov KD, Komissarova NV, Severinov KV. The role of the bacterial RNA polymerase beta subunit flexible flap domain in transcription termination. Dokl Biochem Biophys. 2006;410:263–266. doi: 10.1134/s1607672906050036. [DOI] [PubMed] [Google Scholar]
- 13.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292(5517):730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 14.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12(5):1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 15.Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43(2):253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuzenkova J, et al. Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J Mol Biol. 2003;330(4):735–748. doi: 10.1016/s0022-2836(03)00634-x. [DOI] [PubMed] [Google Scholar]
- 17.Liu B, et al. A bacteriophage transcription regulator inhibits bacterial transcription initiation by σ-factor displacement. Nucleic Acids Res. 2014;42(7):4294–4305. doi: 10.1093/nar/gku080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nechaev S, Yuzenkova Y, Niedziela-Majka A, Heyduk T, Severinov K. A novel bacteriophage-encoded RNA polymerase binding protein inhibits transcription initiation and abolishes transcription termination by host RNA polymerase. J Mol Biol. 2002;320(1):11–22. doi: 10.1016/S0022-2836(02)00420-5. [DOI] [PubMed] [Google Scholar]
- 19.Yuzenkova Y, Zenkin N, Severinov K. Mapping of RNA polymerase residues that interact with bacteriophage Xp10 transcription antitermination factor p7. J Mol Biol. 2008;375(1):29–35. doi: 10.1016/j.jmb.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King RA, Markov D, Sen R, Severinov K, Weisberg RA. A conserved zinc binding domain in the largest subunit of DNA-dependent RNA polymerase modulates intrinsic transcription termination and antitermination but does not stabilize the elongation complex. J Mol Biol. 2004;342(4):1143–1154. doi: 10.1016/j.jmb.2004.07.072. [DOI] [PubMed] [Google Scholar]
- 21.Mason SW, Li J, Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J Biol Chem. 1992;267(27):19418–19426. [PubMed] [Google Scholar]
- 22.Muteeb G, Dey D, Mishra S, Sen R. A multipronged strategy of an anti-terminator protein to overcome Rho-dependent transcription termination. Nucleic Acids Res. 2012;40(22):11213–11228. doi: 10.1093/nar/gks872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang XJ, Roberts JW. Gene Q antiterminator proteins of Escherichia coli phages 82 and lambda suppress pausing by RNA polymerase at a rho-dependent terminator and at other sites. Proc Natl Acad Sci USA. 1989;86(14):5301–5305. doi: 10.1073/pnas.86.14.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3(4):495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 25.Chan CL, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3′ end spacing distinguishes pause and termination pathways. J Mol Biol. 1997;268(1):54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 26.Larson MH, et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344(6187):1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vvedenskaya IO, et al. Transcription. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344(6189):1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41(12):6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50(3):420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50(3):430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furman R, Sevostyanova A, Artsimovitch I. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012;40(8):3392–3402. doi: 10.1093/nar/gkr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Artsimovitch I, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117(3):299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 33.Kolb KE, Hein PP, Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J Biol Chem. 2014;289(2):1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hein PP, et al. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol. 2014;21(9):794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol Cell. 2013;50(6):882–893. doi: 10.1016/j.molcel.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minakhin L, et al. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci USA. 2001;98(3):892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew R, Chatterji D. The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol. 2006;14(10):450–455. doi: 10.1016/j.tim.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar P, Sardesai AA, Murakami KS, Chatterji D. Inactivation of the bacterial RNA polymerase due to acquisition of secondary structure by the ω subunit. J Biol Chem. 2013;288(35):25076–25087. doi: 10.1074/jbc.M113.468520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krohn M, Wagner R. Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. J Biol Chem. 1996;271(39):23884–23894. doi: 10.1074/jbc.271.39.23884. [DOI] [PubMed] [Google Scholar]
- 40.Jöres L, Wagner R. Essential steps in the ppGpp-dependent regulation of bacterial ribosomal RNA promoters can be explained by substrate competition. J Biol Chem. 2003;278(19):16834–16843. doi: 10.1074/jbc.M300196200. [DOI] [PubMed] [Google Scholar]
- 41.Ishiguro A, Nogi Y, Hisatake K, Muramatsu M, Ishihama A. The Rpb6 subunit of fission yeast RNA polymerase II is a contact target of the transcription elongation factor TFIIS. Mol Cell Biol. 2000;20(4):1263–1270. doi: 10.1128/mcb.20.4.1263-1270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan Q, Prysak MH, Woychik NA. Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II, and III. Mol Cell Biol. 2003;23(9):3329–3338. doi: 10.1128/MCB.23.9.3329-3338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komissarova N, et al. Inhibition of a transcriptional pause by RNA anchoring to RNA polymerase. Mol Cell. 2008;31(5):683–694. doi: 10.1016/j.molcel.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sen R, King RA, Mzhavia N, Madsen PL, Weisberg RA. Sequence-specific interaction of nascent antiterminator RNA with the zinc-finger motif of Escherichia coli RNA polymerase. Mol Microbiol. 2002;46(1):215–222. doi: 10.1046/j.1365-2958.2002.03154.x. [DOI] [PubMed] [Google Scholar]
- 45.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: The basic mechanisms. Cell. 2001;107(4):437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 46.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol Cell. 2002;10(5):1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 47.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340(6140):1577–1580. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borukhov S, Goldfarb A. Recombinant Escherichia coli RNA polymerase: Purification of individually overexpressed subunits and in vitro assembly. Protein Expr Purif. 1993;4(6):503–511. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 49.Murakami KS. X-ray crystal structure of Escherichia coli RNA polymerase σ70 holoenzyme. J Biol Chem. 2013;288(13):9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448(7150):157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.