Significance
DNA double-strand breaks (DSBs) are one of the most deleterious types of DNA lesions and may pose a severe threat to genome integrity. Breast cancer type 1 susceptibility protein (BRCA1) is a multifunctional DNA damage response factor that is known to protect the chromosome/genome stability by participating in one of the major DSB repair pathways, homologous recombination (HR). Here we show that in human B cells BRCA1 is also required for another major DSB repair pathway, nonhomologous end-joining (NHEJ) during immunoglobulin class switch recombination (CSR), probably by inhibition of resection and microhomology-mediated end-joining (MMEJ), as well as promotion of long-range recombination. Our study provides previously unrecognized insights into BRCA1’s function in maintaining genome stability and tumor suppression.
Keywords: BRCA1, nonhomologous end-joining, immunoglobulin class switch recombination, alternative end-joining, B cells
Abstract
Breast cancer type 1 susceptibility protein (BRCA1) has a multitude of functions that contribute to genome integrity and tumor suppression. Its participation in the repair of DNA double-strand breaks (DSBs) during homologous recombination (HR) is well recognized, whereas its involvement in the second major DSB repair pathway, nonhomologous end-joining (NHEJ), remains controversial. Here we have studied the role of BRCA1 in the repair of DSBs in switch (S) regions during immunoglobulin class switch recombination, a physiological, deletion/recombination process that relies on the classical NHEJ machinery. A shift to the use of microhomology-based, alternative end-joining (A-EJ) and increased frequencies of intra-S region deletions as well as insertions of inverted S sequences were observed at the recombination junctions amplified from BRCA1-deficient human B cells. Furthermore, increased use of long microhomologies was found at recombination junctions derived from E3 ubiquitin-protein ligase RNF168-deficient, Fanconi anemia group J protein (FACJ, BRIP1)-deficient, or DNA endonuclease RBBP8 (CtIP)-compromised cells, whereas an increased frequency of S-region inversions was observed in breast cancer type 2 susceptibility protein (BRCA2)-deficient cells. Thus, BRCA1, together with its interaction partners, seems to play an important role in repairing DSBs generated during class switch recombination by promoting the classical NHEJ pathway. This may not only provide a general mechanism underlying BRCA1’s function in maintaining genome stability and tumor suppression but may also point to a previously unrecognized role of BRCA1 in B-cell lymphomagenesis.
The genome is under constant threat of DNA damage from external and internal stressors to the cell such as ionizing irradiation, free radicals, and replication fork collapses. One of the most deleterious types of DNA lesions is the DNA double-strand break (DSB), which if unrepaired or misrepaired can result in cell death or genome instability. DSBs are mainly repaired by homologous recombination (HR) or nonhomologous end-joining (NHEJ). HR can only act during the S/G2 phases of the cell cycle, when a sister chromatid is available as a template for repair and is considered to be error-free. NHEJ, on the other hand, can function throughout the cell cycle by joining the broken DNA ends directly without a homologous DNA template, but is also seen as error-prone, as insertions or deletions might be introduced at the recombination junctions. The classical NHEJ (c-NHEJ) pathway requires a number of factors, including X-ray repair cross-complementing protein 6 (XRCC6, Ku70), XRCC5 (Ku80), DNA-PKcs, Artemis, DNA ligase IV, XRCC4, and XLF (Cernunnos), and the repair pattern is characterized by a direct joining of the broken ends or joining based on a few base pairs of sequence homology (microhomology, MH) at the two DNA ends (1). When the c-NHEJ pathway is defective, alternative end-joining (A-EJ) pathway(s), often associated with resections/deletions and longer MHs, may be operating (1–3).
DSBs are not always pathological, but can also be intermediates of physiological processes, such as those that occur during B-cell development. Then, extensive gene rearrangements/modifications at the Ig gene loci may occur, resulting in production of functional antibodies that can recognize and act against an immense number of different pathogens. One of these processes, class switch recombination (CSR), occurs at the mature B-cell stage and exchanges the IgM constant region encoding gene (Cμ) with a downstream constant region gene (Cγ, Cε, or Cα) to generate antibody classes with different effector functions (2). CSR is initiated by the B-cell–specific factor activation-induced cytidine deaminase (AID) (4), which deaminates cytosines into uracils in the switch (S) regions—that is, repetitive intronic DNA sequences that are located upstream of each Ig constant region gene. The uracil/guanine mismatches are processed by the base excision repair (BER) and mismatch repair (MMR) pathways and finally converted into DSBs (5), which are subsequently repaired by NHEJ during the G1 phase of the cell cycle. In addition to the c-NHEJ factors, several DNA damage response (DDR) proteins have been shown to be important for CSR, including histone H2AX, mediator of DNA damage checkpoint protein 1 (MDC1), serine-protein kinase ATM, E3 ubiquitin ligase proteins RING finger (RNF) 8 and RNF168, tumor suppressor p53-binding protein 1 (53BP1), telomere-associated protein RIF1, and the MRN (Mre11, Rad50, Nbs1) complex (2, 6).
The gene encoding the DDR factor BRCA1 was first mapped to chromosome 17 in 1994 as a breast and ovarian cancer susceptibility gene (7). Since then, BRCA1 has been implicated in a vast number of processes, ranging from checkpoint control and chromatin remodeling to transcription and HR (8–10). Its numerous functions could be attributed to its ability to interact with various proteins through its different domains. The N-terminal RNF domain binds to BRCA1-associated RING domain protein 1 (BARD1) and promotes its E3 ubiquitin ligase activity (9). Furthermore, regions within the same domain have been reported to bind, independently of BARD1, to the c-NHEJ–factor Ku80 (11, 12). The BRCA1 C terminus contains two BRCT repeats, which are involved in forming the A-, B-, and C-complexes by interaction with Abraxas, Fanconi anemia group J protein (FACJ, BRIP1), and DNA endonuclease RBBP8 (CtIP), respectively (8, 9). Furthermore, BRCA1 can, through its coiled-coil domain, form a complex with partner and localizer of BRCA2 (PALB2) and breast cancer type 2 susceptibility protein (BRCA2). All of the above-mentioned complexes have, in addition to other functions, been linked to HR. Although the involvement of BRCA1 in HR and cell-cycle checkpoint control seems to be most important for its tumor suppression activities, other functional properties of BRCA1 might also contribute (8–10).
Whether BRCA1 is involved in NHEJ remains controversial (9, 11–18). Notably, BRCA1 forms a large complex, termed the BRCA1-associated genome surveillance complex, with a number of DNA damage repair proteins that have been shown to be involved in CSR, including ATM, the MRN complex, and the MMR proteins MSH2, MSH6, and MLH1 (19). It is thus possible that BRCA1 directly or indirectly regulates NHEJ during CSR, and we therefore tested this hypothesis by analyzing the recombination junctions generated from in vivo switched B cells from individuals carrying mutations within the BRCA1 gene. To further elucidate the mechanisms of its actions, CSR junctions from individuals with defects in the BRCA1 interaction partners BRIP1, CtIP, and BRCA2, as well as RNF168, an ubiquitin ligase that recruits both BRCA1 and 53BP1 to DSBs, were studied.
Results
Longer Microhomologies at Sμ–Sα Junctions in BRCA1-Deficient B Cells.
Complete absence of BRCA1 is most likely not compatible with life, as biallelic, deleterious mutations result in embryonic lethality in mice (20). Sμ–Sα switch fragments were thus amplified from in vivo switched B cells from 15 individuals with heterozygous mutations in BRCA1 (Fig. 1 and SI Appendix, Table S1), using our previously described nested PCR assay (21, 22). Altogether, 227 switch fragments, representing unique CSR events, were characterized from BRCA1-deficient individuals. Of these, 219 contained Sμ–Sα junctions, representing a direct switch from IgM to IgA (illustrated in Fig. 2A), whereas eight showed signs of sequential switching—that is, switching from IgM to IgA via IgG, resulting in a Sμ–Sγ–Sα junction. The Sμ–Sα junctions were subsequently compared with our previously published 155 (23) and 102 newly generated Sμ–Sα junctions derived from healthy adult blood donors. The two sets of controls were largely similar and therefore merged.
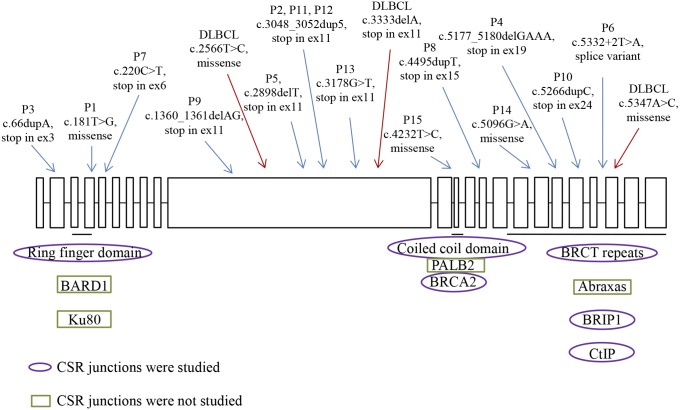
Fig. 1.
Schematic figure of the human BRCA1 gene. The boxes represent the exons (ex), which are numbered from 1 to 24. The positions of the RNF domain, the coiled-coil domain, and the BRCT repeats are underlined. Proteins interacting with these domains are written underneath the domains. The approximate positions of mutations, carried by the individuals included in the CSR junctional analysis, are indicated by arrows (blue, nonlymphoma patients; red, diffused large B-cell lymphoma patients). All mutations are heterozygous.
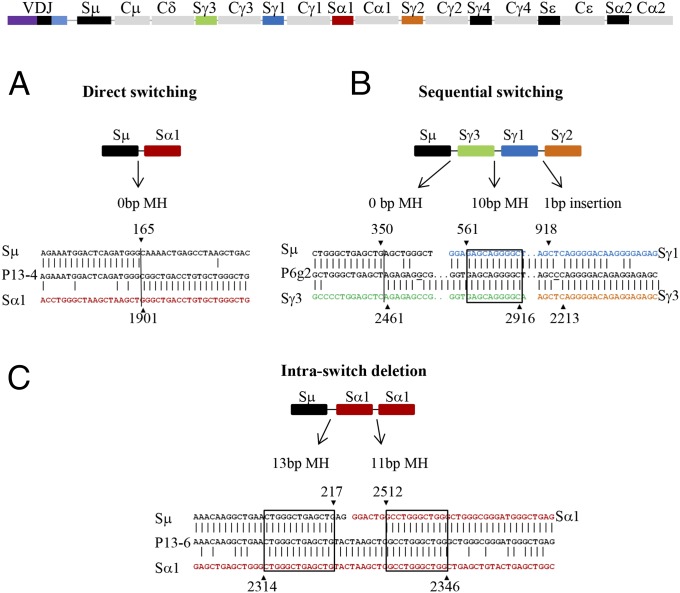
Fig. 2.
Alignment of CSR junctions with germ-line S region sequences. Vertical line indicates direct end-joining, boxes highlight MH, and insertions are underlined. The Sμ, Sα, or Sγ breakpoints are indicated by ▼ or ▲, and positions in germ-line S-region sequences are indicated on top or below the arrowheads. S regions inserted in the reversed direction are designated by (r). Three CSR junctions are shown as examples for different types of end-joining: (A) direct joining; (B) sequential switching from IgM to IgG3, to IgG1, and then to IgG2; and (C) ISD.
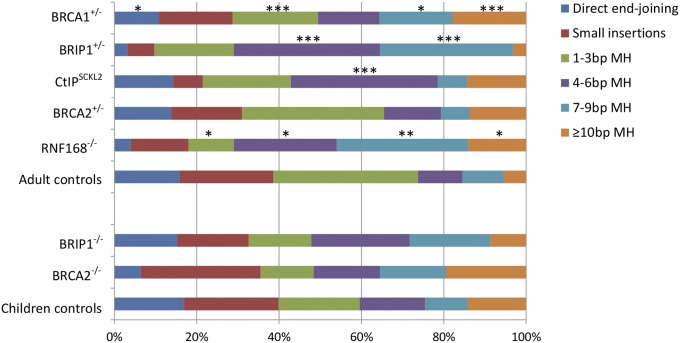
The Sμ–Sα junctions from B cells from BRCA1-deficient individuals showed a preferential use of longer MHs (≥4 bp), a characteristic of A-EJ. Furthermore, 18% of the junctions showed unusually long MHs (≥10 bp), compared with 5% in controls. Conversely, the repair by direct end-joining (no MH, no insertion) or shorter MHs of 1–3 bp, which are typical features for c-NHEJ, was significantly reduced in B cells from the BRCA1-deficient individuals (Table 1 and Fig. 3).
Table 1.
Characterization of Sμ–Sα junctions
| Perfectly matched short homology | |||||||
| 0 bp | |||||||
| Study subjects | Direct end-joining (%) | Small insertions (%) | 1–3 bp (%) | 4–6 bp (%) | 7–9 bp (%) | ≥10 bp (%) | Total no. of S fragments |
| BRCA1+/− | 21 (11)*↓ | 40 (18) | 45 (21)***↓ | 32 (15) | 38 (18)*↑ | 38 (18)***↑ | 214 |
| BRIP1+/− | 1 (3) | 2 (6)*↓ | 6 (19) | 11 (35)***↑ | 10 (32)***↑ | 1 (3) | 31 |
| CtIPSCKL2 | 2 (14) | 1 (7) | 3 (21) | 5 (36)**↑ | 1 (7) | 2 (14) | 14 |
| BRCA2+/− | 4 (14) | 5 (17) | 10 (34) | 4 (14) | 2 (7) | 4 (14) | 29 |
| RNF168−/− | 2 (8) | 2 (8) | 3 (12)*↓ | 7 (27)*↑ | 8 (31)**↑ | 4 (15)*↑ | 26 |
| Controls, adults | 41 (16) | 56 (22) | 91 (36) | 29 (11) | 25 (10) | 14 (5) | 256 |
| BRIP1−/− | 8 (17) | 7 (15) | 7 (15) | 11 (24) | 9 (20) | 4 (9) | 46 |
| BRCA2−/− | 2 (6) | 9 (29) | 4 (13) | 5 (16) | 5 (16) | 6 (19) | 31 |
| Controls, 1-13 y | 31 (17) | 42 (23) | 36 (20) | 29 (16) | 19 (10%) | 26 (14) | 183 |
Statistical analysis was performed using χ2 test, and significant differences are indicated in bold. *P < 0.05, **P < 0.01, ***P < 0.001. All individuals were compared with adult controls (n = 31) for statistic calculations, except the BRCA2−/− and BRIP1−/− patients, who were compared with controls with younger ages (1–13 y, n = 20).
Fig. 3.
Frequencies of different repair patterns at CSR junctions. All groups were compared with adult controls, except for BRCA2−/− and BRIP1−/− patients, who were compared with children controls (1–13 y). Statistical analysis was performed using χ2 test. *P < 0.05, **P < 0.01, ***P < 0.001.
The CSR junctions from the BRCA1-deficient cells were further grouped based on the effect of the mutations carried by the patients (SI Appendix, Table S1). Notably, in the BRCT domain-affected group, the direct end-joining was totally absent. The repair by long MHs (≥10 bp), on the other hand, was strongly increased in both the BRCT and RNF domain-affected groups (SI Appendix, Table S2). The junctions from the BRCA1-deficient individuals with truncating mutations in the middle of the gene, which are likely to cause haploinsufficiency (24), and the patient with a splice site mutation also showed an increased frequency of repair by long MHs (≥10 bp; SI Appendix, Table S2). The Sμ–Sα junctions from the individual who has a mutation affecting the coiled-coil domain, however, showed a largely normal repair pattern (SI Appendix, Table S2). In summary, the c-NHEJ pathway seems to be affected during CSR in BRCA1-deficient cells, and both the RNF domain and BRCT repeats, but not the coiled-coil domain, appear to be involved in the process.
Increased Sequential Switching in Sμ–Sγ Junctions from BRCA1-Deficient B Cells.
Altogether 137 Sμ–Sγ recombination junctions from seven BRCA1-deficient individuals were characterized and compared with our previously published 59 Sμ–Sγ junctions from adult controls (25, 26). The Sμ–Sγ junctions derived from BRCA1-deficient individuals showed a significant reduction in use of 1–3 bp MH and a borderline increased frequency of repair by ≥4 bp MH (SI Appendix, Table S3). Notably, among the Sμ–Sγ junctions derived from P1, who carries a mutation in the RNF domain, two junctions had an unusually long MH of 9 bp. In controls, no Sμ–Sγ junction with ≥6 bp MH was ever observed.
A proportion of Sμ–Sγ junctions (11%) from the BRCA1-deficient individuals also exhibit “footprints” of sequential switching (illustrated in Fig. 2B; see also Table 2). This has previously been detected in Artemis-deficient patients but never in controls and could suggest an impaired repair through c-NHEJ during CSR (23). In conclusion, the c-NHEJ pathway seems to be affected in BRCA1-deficient B cells, during the processes of both IgA and IgG switching.
Table 2.
Frequencies of ISDs, inversions, and sequential switching at CSR junctions
| Study subjects | ISDs (%) | Inversions (%) | Sequential switching (%) | Total no. of S junctions |
| Sμ–Sα | ||||
| BRCA1+/− | 85 (37)*↑ | 9 (4)*↑ | 8 (4) | 227 |
| BRIP1+/− | 11 (34) | 0 (0) | 1 (3) | 32 |
| CtIPSCKL2 | 2 (14) | 0 (0) | 0 (0) | 14 |
| BRCA2+/− | 14 (44) | 2 (6)*↑ | 1 (3) | 32 |
| RNF168−/− | 11 (42) | 0 (0) | 0 (0) | 26 |
| Controls, adults | 78 (29) | 2 (1) | 7 (3) | 268 |
| BRIP1−/− | 15 (31) | 1 (2) | 2 (4) | 48 |
| BRCA2−/− | 8 (25) | 1 (3) | 0 (0) | 32 |
| Controls, 1–13 y | 59 (32) | 1 (1) | 3 (2) | 187 |
| Sμ–Sγ | ||||
| BRCA1+/− | 59 (41)***↑ | 3 (2) | 16 (11)**↑ | 143 |
| RNF168−/− | 8 (24) | 0 (0) | 7 (21)***↑ | 33 |
| Controls, adults | 7 (12) | 0 (0) | 0 (0) | 59 |
Statistical analysis was performed using χ2 test, and significant differences are indicated in bold. *P < 0.05, **P < 0.01, ***P < 0.001. All individuals were compared with adult controls (n = 31 for Sμ–Sα junctions and n = 33 for Sμ–Sγ junctions) for statistical analysis, except the BRIP1−/− and BRCA2−/− patients, who were compared with controls with younger ages (1–13 y, n = 20).
Increased Frequency of Intraswitch Region Recombination in BRCA1-Deficient B Cells.
Aberrant CSR, but normal or enhanced intraswitch recombination (resulting in intraswitch deletions, ISDs), has been observed in mouse cells deficient in several DDR or NHEJ factors (27–30). In contrast to the joining of two heterologous S regions, which could be located several hundreds of kilobases apart, ISDs occur when DSBs are introduced and repaired within the same S region. It may thus be an indication of failed synapsis of distant S regions and long-range recombination (27). Another, nonexclusive, explanation is that ISDs are caused by increased resection and/or use of A-EJ pathway(s), as the probability of finding a homologous template for MH-dependent repair is increased within the same S region, which consists of highly repetitive sequences (31, 32).
As illustrated in Figs. 2C and 4, by analyzing CSR junctions derived from our PCR assay, it is possible to detect ISDs. The proportion of Sμ–Sα and Sμ–Sγ junctions containing ISDs was significantly increased in the BRCA1-deficient group (Table 2). A similar increase was found at CSR junctions from individuals with mutations in the RNF and BRCT BRCA1 domains (SI Appendix, Tables S2 and S3). Taken together, BRCA1 might thus be involved in the synapsis and long-range recombination of S regions and/or in preventing resection and A-EJ during CSR.
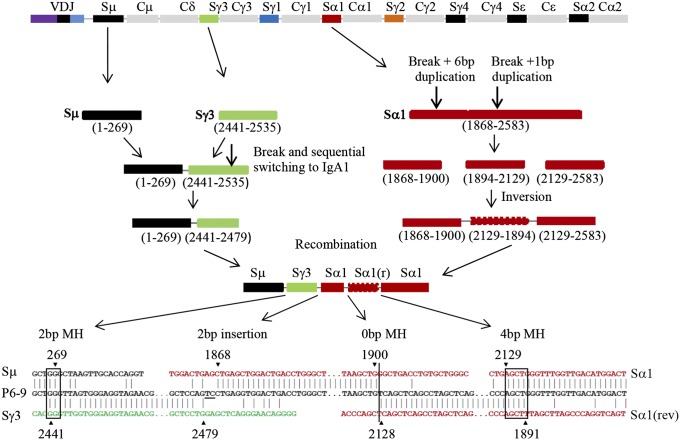
Fig. 4.
An example of CSR junction containing sequential switching, ISDs, and inversion. Symbols are explained in the Fig. 2 legend.
Increased Proportion of CSR Junctions with Inversions in BRCA1-Deficient B Cells.
A small proportion of Sμ–Sα junctions amplified from BRCA1-deficient cells contained insertions of inverted pieces of S regions, which are rarely observed in controls (Table 2 and SI Appendix, Table S2). Most of these junctions comprised only one inverted S region, whereas some harbored several inverted pieces or in combination with sequential switching, as exemplified by the P6–9 and P4–8 junctions in Fig. 4 and SI Appendix, Fig. S1. The generation of these junctions might have occurred through multiple steps of break/inversion/deletion/recombination processes. Few Sμ–Sγ junctions from BRCA1-deficient B cells also comprised inverted pieces of S regions, whereas this has not been observed in those from control cells. Thus, BRCA1 appears to inhibit inversions during recombination of the S regions.
Repair Pattern at CSR Junctions from Patients with Defects in the BRCA1 BRCT Repeat Interaction Partners CtIP or BRIP1.
Sμ–Sα junctions from patients with mutations in either BRIP1 or RBBP8 (SI Appendix, Table S1) were subsequently analyzed. These genes encode the BRIP1 and CtIP proteins that interact with the BRCA1 BRCT repeat through the B and C complexes, respectively (8, 9). As the BRIP1-deficient Fanconi anemia patient was 4.6 y of age at the sampling and we have previously shown that the CSR pattern differs between children and adults [with more MH use in children (23)], the Sμ–Sα junctions from the patient were compared with those derived from healthy children (1–13 y old) (23, 33). There was a slightly increased use of MH at the CSR junctions derived from the BRIP1-deficient patient. More specifically, the changes were significant only when combining the 4–6 bp and 7–9 bp groups (χ2 test, P = 0.022). Surprisingly, the Sμ–Sα junctions from a heterozygous parent showed a more significant change, with increase in the use of long MHs as well as a reduction of small insertions and direct joining (Table 1 and Fig. 3). The frequency of ISDs and inversions, however, were largely normal in both the BRIP1-deficient patient and the heterozygous parent (Table 2).
In total, 14 Sμ–Sα junctions were isolated from the two CtIP-compromised Seckel syndrome patients. A significant increase in the use of 4–6 bp MH was observed (Table 1 and Fig. 3). The proportions of ISDs and inversions were, on the other hand, normal in these patients (Table 2). Taken together, the repair pattern at the Sμ–Sα junctions from the CtIP-compromised and BRIP1-deficient patients and the parent with heterozygous mutations in BRIP1 showed some similarity with the BRCA1-deficient individuals—that is, a shift in using the MH-mediated A-EJ. However, the increased frequency of ISDs and inversions seems to be more specific for BRCA1-deficient individuals.
Repair Pattern at CSR Junctions from BRCA2-Deficient Individuals.
BRCA2 is another breast and ovarian cancer susceptibility gene, which encodes a protein that is known to be important for HR, but has not been implicated in NHEJ (34). It forms a complex with BRCA1 and PALB2 through the BRCA1 coiled-coil domain. Sμ–Sα junctions from a Fanconi anemia patient with compound heterozygous mutations in BRCA2 were analyzed, and these junctions had a slightly elevated MH use and reduced frequency of direct joining compared with the children controls, albeit not to a significant degree (Table 1 and Fig. 3). The Sμ–Sα junctions from the heterozygous mother, who developed breast cancer at 38 y of age, showed a similar repair pattern as adult controls. Thus, it seems that the role of BRCA1 in NHEJ is independent from its interaction with BRCA2. Nevertheless, the frequency of inversions at CSR junctions was significantly elevated in the BRCA2 heterozygous mother (Table 2).
Altered Pattern of CSR Junctions from RNF168-Deficient Cells.
Several studies have shown an involvement of BRCA1 and 53BP1 in DSB repair pathway choice, where 53BP1 promotes NHEJ and BRCA1 facilities end resection and HR (35–37). As 53BP1 has also been implicated in NHEJ during CSR (2, 6, 38), it is somewhat surprising to observe that BRCA1 may have similar rather than opposite function(s) during this process. CSR junctions from B cells derived from an ataxia–telangiectasia-like patient were thus studied, where the recruitments of 53BP1 and BRCA1 were both affected, due to a lack of RNF168 protein in this patient (39). The Sμ–Sα junctions from the RNF168-deficient cells showed a significant shift to the use of longer MHs, even more prominent than that observed in the BRCA1-deficient cells (Table 1 and Fig. 3). The MH use at Sμ–Sγ junctions from patient’s cells was largely similar to controls (SI Appendix, Table S3). However, the frequency of sequential switching was markedly increased in the RNF168-deficient cells, again more prominent than that observed in the BRCA1-deficient cells (Table 2). Thus, in contrast to the previous finding that the HR defect observed in BRCA1-deficient cells can be rescued by knockdown of 53BP1 (36, 37), the combined deficiency in BRCA1 and 53BP1 resulted in a more severe NHEJ defect during CSR (SI Appendix, Fig. S2).
Discussion
The involvement of BRCA1 during NHEJ has been unclear, with studies showing that BRCA1 promotes, inhibits, or has no effect on the process (11–18). All previous experiments on the participation of BRCA1 in NHEJ have been performed in in vitro systems, such as cell lines or cell-free extracts. In this paper, we have analyzed the function of BRCA1 in NHEJ in a more physiological setting, by characterizing the recombination breakpoints derived from in vivo switched B cells in individuals with deleterious mutations in BRCA1. The pattern of these CSR junctions showed a shift from direct end-joining to MH-dependent repair, suggesting that BRCA1 may promote the c-NHEJ pathway and/or inhibit the resection/MH-mediated A-EJ pathway. An indirect role of BRCA1 on CSR through regulation of transcription of genes encoding proteins important for CSR, such as AID, BER, and c-NHEJ factors or DDR proteins including 53BP1, was excluded by mRNA expression analysis in BRCA1-deficient samples (SI Appendix, Figs. S3 and S4). BRCA1 is, however, unlikely to be a component of the core NHEJ machinery, but instead might modulate the NHEJ pathway through a number of mechanisms, as discussed below.
First, BRCA1 may regulate NHEJ through interactions with the key NHEJ component Ku80. Mediated by its N terminus, BRCA1 seems to stabilize the binding of Ku80 to DSBs (11, 12). In the absence of Ku binding, DSBs are preferentially repaired by A-EJ (29).
Second, the functional roles of BRCA1 during NHEJ could be executed through its BRCT repeats that interact with a number of DNA repair factors. The CSR junctions from the heterozygous BRIP1- and CtIP-compromised individuals showed a significant increased use of longer MHs but not as dramatic as those observed from the BRCA1 BRCT-affected individuals. Thus, binding to BRIP1 or CtIP alone does not seem to explain the involvement of the BRCA1 BRCT repeats during NHEJ; instead, there could be a combined effect of impaired formation of several distinct complexes that have varying functions. The BRCT repeats containing A (Abraxas/RAP80/BRCC36) (40) and B (BRIP1) (41) complexes have both been shown to inhibit resection through interaction with MRE11 and/or CtIP, whereas the C (CtIP) complex appears to promote resection and A-EJ (42). Accordingly, one of the functions of BRCA1 during NHEJ could be to modulate the resection process and thus affect the choice between c-NHEJ and A-EJ. This is furthermore supported by the increase in ISDs, which have been suggested to be caused by extensive resection at the DSBs in the S regions, in BRCA1-deficient B cells (32).
Third, BRCA1 may potentially affect NHEJ during CSR through chromatin remodeling (43). It has been proposed that DDR proteins, including H2AX, ATM, and 53BP1, could induce chromatin conformational changes that facilitate synapsis between distant S regions (44–46). This has been supported by the increased frequency of ISDs in B cells from mice deficient in the above-mentioned proteins (27, 28, 30). BRCA1 might thus collaborate with other DDR factors, such as ATM, which is part of BRCA1-associated genome surveillance complex, to induce changes in chromatin that would promote long-range repair during CSR.
Our results from CtIP-compromised patients differ from previous studies, where a reduction or no change in MH use has been observed at CSR junctions derived from a mouse B-cell line treated with CtIP shRNA (42) or from CtIP-depleted mouse B cells (32). It should be noted that the Seckel syndrome patients in our study did not carry null mutations. Instead, cells from our patients expressed normal levels of wild-type CtIP and, in addition, C-terminal truncated protein with an intact BRCA1 interacting site (44). Hence, the CtIP–BRCA1 complex would be expected to form in patients’ cells, but the resection mechanism would be hampered due to the incorporation of truncated CtIP into the CtIP homodimers. Thus, in contrast to the CtIP-depleted mouse models, CtIP-mediated resection might still occur in the cells from the patients, albeit with lower efficiency, which could possibly favor intermediate lengths of MHs at the CSR junctions.
BRCA2 seems to be less important than BRCA1 for c-NHEJ. The balance between the direct end-joining and MH-mediated A-EJ at CSR junctions from the BRCA2-deficient patients and the individual with a mutation affecting the BRCA1 coiled-coil domain, which is necessary for formation of the BRCA1/PALB2/BRCA2 complex, was not significantly altered. However, CSR junctions from these individuals showed a similar increased frequency of inversions as those from the other BRCA1-deficient cells, suggesting that both BRCA1 and BRCA2 might be involved in preventing these events. It is unclear, however, whether an increased frequency of inversions is due to an NHEJ defect or rather due to general chromosomal instability in BRCA1- and BRCA2-deficient cells as a result of impaired HR. Although AID-induced breaks are believed to be repaired during the G1 cell-cycle phase by NHEJ, some of these breaks may persist to the S/G2 phases and are expected to be repaired by HR (45–47). It is therefore possible that an HR defect in these cells contributes to an elevated level of unrepaired DSBs, which might increase the chance for unconventional joining through inversions.
Several recent studies have suggested a central function for BRCA1 in the choice of repair pathways, through promotion of resection and HR during the S/G2 cell-cycle phases (35–37). Thus, it might seem contradictory that BRCA1 could also inhibit resection and promote NHEJ during the G1 cell cycle. However, several DNA repair proteins have opposing functions during HR and NHEJ, including ATM, H2AX, BLM, and RIF1 (48, 49). Furthermore, by studying RNF168-deficient cells, we showed that combined deficiency of 53BP1 and BRCA1 did not rescue the NHEJ defect, as it did for HR. Our results highlight the complexity of the DNA repair machinery. With new cancer therapies, such as poly(ADP ribose) polymerase inhibitors (50), targeting specific DNA repair pathways, it is important to delineate the functional properties of each DDR/repair factor in different contexts, including cell-cycle stages.
The tumor-suppressing activity of BRCA1 has mainly been attributed to its roles in HR and checkpoint control (9, 10). In light of our findings here, the involvement of BRCA1 in regulating the NHEJ pathway may also contribute to its cancer-preventing function. The importance of efficient repair through NHEJ, especially in lymphocytes, becomes evident when studying mice double deficient for p53 and any of the c-NHEJ factors, as most of them develop aggressive lymphomas that often harbor translocations involving the Ig loci (51). Moreover, we have previously shown that DNA repair genes are frequently mutated in human B-cell lymphomas, and NHEJ mutations are associated with IgH translocations in these tumors (52). With the contribution of BRCA1 to both HR and NHEJ, two repair pathways that are important for the maintenance of genome stability and resolution of AID-induced DSBs during CSR, BRCA1 could function as a tumor suppressor in mature B cells. Accordingly, one of the most common tumors observed in BRCA1-deficient mouse models is lymphoma (20). Furthermore, by going through our recently published coding genome sequencing data on 31 diffuse large B-cell lymphomas (53) as well as data from an additional 22 germinal center-related B-cell lymphoma cases, we have observed a number of somatic and germ-line mutations or rare SNPs in BRCA1 in these samples, including a germ-line, pathogenic frame shift mutation (p.Q1111fs) (SI Appendix, Table S4). Notably, CSR junctions from several of these patients showed a similar skewed repair pattern as those from the BRCA1-deficient patients without lymphoma (SI Appendix, Table S5). Furthermore, somatic and germ-line mutations were also found in BRCA2, BRIP1, PALB2, UIMCI (RAP80), and FAM175A (Abraxas) in a number of samples, including nonsense mutations in BRCA2 (p.P3063X) and BRIP1 (p.R162X). BRCA1, BRCA2, and related FA pathway genes might thus also be good candidates for cancer-susceptibility genes in B-cell lymphomas, although further screening of variations in these genes in a larger cohort of patients will be required. Taken together, BRCA1 and its interacting proteins, through their functions in HR and NHEJ, may play an important role in maintaining the chromosome/genome stability and thus in preventing tumorgenesis in mature B lymphocytes.
Materials and Methods
BRCA1-, BRCA2-, BRIP1-, and RNF168-Deficient and CtIP-Compromised Patient Samples.
Fifteen individuals carrying heterozygous mutations in BRCA1 (P1–P15) (Fig. 1), two Fanconi anemia patients (P16 and P17), two previously described Seckel syndrome patients (P18 and P19) (54), and one RNF168-deficient patient (P22) presented with ataxia, microcephaly, and immunodeficiency (39) were included in the study. P16, who carried compound heterozygous mutations in BRCA2, presented with intrauterine growth retardation, short stature, and developed acute myelocytic leukemia at 21 mo of age (55). P17 carried homozygous mutations in BRIP1 (56). P18 and P19 presented with dwarfism, microcephaly, and café au lait spots and carried homozygous SCKL2 mutations, which consist of a splice mutation in the RBBP8 gene, encoding CtIP. In addition, the heterozygous parents of P16 and P17 were also studied (P20 and P21). The details of mutations, age at sampling, and cancer status for P1–P22 are shown in SI Appendix, Table S1. Four lymphoma patients with mutations in BRCA1 (described in SI Appendix, Table S4, and Fig. 1) were furthermore studied. Finally, 14 newly recruited healthy adult blood donors were included in the study as controls. The institutional review board at Karolinska Institutet approved the study.
Characterization of in Vivo Switch Recombination Junctions.
The recombination junctions were characterized as previously described (2, 21). The sequences around the recombination breakpoints (±25 bp) from BRCA1-, BRCA2-, BRIP1-, and RNF168-deficient and CtIP-compromised individuals, as well as from controls, are shown in SI Appendix, Fig. S5.
Supplementary Material
Acknowledgments
We are grateful to all patients and families who have donated blood samples and to the BRCA Laboratory (Lund University) and A. Lindblom (Karolinska Institute) for contributing samples from BRCA1-deficient individuals. We also thank Dr. A. Smogorzewska (The Rockefeller University) and the International Fanconi Anemia Registry for samples from patients with BRCA2 and BRIP1 mutations, and Drs. J.-M. Hertz (University of Southern Denmark) and I. Vogel (Aarhus University Hospital) for samples from members of the SCKL2 family. This work was supported by the European Research Council (242551-ImmunoSwitch), the Swedish Research Council, and the Swedish Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418947112/-/DCSupplemental.
References
- 1.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stavnezer J, Björkman A, Du L, Cagigi A, Pan-Hammarström Q. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv Immunol. 2010;108:45–109. doi: 10.1016/B978-0-12-380995-7.00003-3. [DOI] [PubMed] [Google Scholar]
- 3.Kotnis A, Du L, Liu C, Popov SW, Pan-Hammarström Q. Non-homologous end joining in class switch recombination: The beginning of the end. Philos Trans R Soc Lond B Biol Sci. 2009;364(1517):653–665. doi: 10.1098/rstb.2008.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Chahwan R, Edelmann W, Scharff MD, Roa S. AIDing antibody diversity by error-prone mismatch repair. Semin Immunol. 2012;24(4):293–300. doi: 10.1016/j.smim.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel JA, Nussenzweig A. The AID-induced DNA damage response in chromatin. Mol Cell. 2013;50(3):309–321. doi: 10.1016/j.molcel.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 8.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11(2):138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosen EM. BRCA1 in the DNA damage response and at telomeres. Front Genet. 2013;4:85. doi: 10.3389/fgene.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343(6178):1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- 11.Wei L, et al. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol Cell Biol. 2008;28(24):7380–7393. doi: 10.1128/MCB.01075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang G, et al. BRCA1-Ku80 protein interaction enhances end-joining fidelity of chromosomal double-strand breaks in the G1 phase of the cell cycle. J Biol Chem. 2013;288(13):8966–8976. doi: 10.1074/jbc.M112.412650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, et al. Nonhomologous end-joining of ionizing radiation-induced DNA double-stranded breaks in human tumor cells deficient in BRCA1 or BRCA2. Cancer Res. 2001;61(1):270–277. [PubMed] [Google Scholar]
- 15.Zhong Q, Boyer TG, Chen PL, Lee WH. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 2002;62(14):3966–3970. [PubMed] [Google Scholar]
- 16.Baldeyron C, et al. A single mutated BRCA1 allele leads to impaired fidelity of double strand break end-joining. Oncogene. 2002;21(9):1401–1410. doi: 10.1038/sj.onc.1205200. [DOI] [PubMed] [Google Scholar]
- 17.Thompson EG, Fares H, Dixon K. BRCA1 requirement for the fidelity of plasmid DNA double-strand break repair in cultured breast epithelial cells. Environ Mol Mutagen. 2012;53(1):32–43. doi: 10.1002/em.21674. [DOI] [PubMed] [Google Scholar]
- 18.Dohrn L, Salles D, Siehler SY, Kaufmann J, Wiesmüller L. BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem J. 2012;441(3):919–926. doi: 10.1042/BJ20110314. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14(8):927–939. [PMC free article] [PubMed] [Google Scholar]
- 20.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: Past lessons, current understanding and future prospects. Oncogene. 2006;25(43):5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 21.Pan Q, et al. Regulation of switching and production of IgA in human B cells in donors with duplicated alpha1 genes. Eur J Immunol. 2001;31(12):3622–3630. doi: 10.1002/1521-4141(200112)31:12<3622::aid-immu3622>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 22.Pan Q, et al. Alternative end joining during switch recombination in patients with ataxia-telangiectasia. Eur J Immunol. 2002;32(5):1300–1308. doi: 10.1002/1521-4141(200205)32:5<1300::AID-IMMU1300>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 23.Du L, et al. Involvement of Artemis in nonhomologous end-joining during immunoglobulin class switch recombination. J Exp Med. 2008;205(13):3031–3040. doi: 10.1084/jem.20081915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11(23):2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 25.Pan-Hammarström Q, et al. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J Exp Med. 2005;201(2):189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Q, Rabbani H, Mills FC, Severinson E, Hammarström L. Allotype-associated variation in the human gamma3 switch region as a basis for differences in IgG3 production. J Immunol. 1997;158(12):5849–5859. [PubMed] [Google Scholar]
- 27.Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur J Immunol. 2007;37(1):235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- 28.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197(12):1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boboila C, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107(7):3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200(9):1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 32.Bothmer A, et al. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J Exp Med. 2013;210(1):115–123. doi: 10.1084/jem.20121975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enervald E, et al. A regulatory role for the cohesin loader NIPBL in nonhomologous end joining during immunoglobulin class switch recombination. J Exp Med. 2013;210(12):2503–2513. doi: 10.1084/jem.20130168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia F, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci USA. 2001;98(15):8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escribano-Díaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49(5):872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17(6):688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165(4):459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devgan SS, et al. Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ. 2011;18(9):1500–1506. doi: 10.1038/cdd.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem. 2011;286(15):13669–13680. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suhasini AN, et al. Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol Cell Biol. 2013;33(11):2212–2227. doi: 10.1128/MCB.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18(1):75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye Q, et al. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J Cell Biol. 2001;155(6):911–921. doi: 10.1083/jcb.200108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qvist P, et al. CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet. 2011;7(10):e1002310. doi: 10.1371/journal.pgen.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasham MG, et al. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11(9):820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamane A, et al. RPA accumulation during class switch recombination represents 5′-3′ DNA-end resection during the S-G2/M phase of the cell cycle. Cell Reports. 2013;3(1):138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasham MG, et al. Activation-induced cytidine deaminase-initiated off-target DNA breaks are detected and resolved during S phase. J Immunol. 2012;189(5):2374–2382. doi: 10.4049/jimmunol.1200414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ira G, Nussenzweig A. A new Riff: Rif1 eats its cake and has it too. EMBO Rep. 2014;15(6):622–624. doi: 10.1002/embr.201438825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grabarz A, et al. A role for BLM in double-strand break repair pathway choice: Prevention of CtIP/Mre11-mediated alternative nonhomologous end-joining. Cell Reports. 2013;5(1):21–28. doi: 10.1016/j.celrep.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 50.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Miranda NF, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med. 2013;210(9):1729–1742. doi: 10.1084/jem.20122842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Miranda NF, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124(16):2544–2553. doi: 10.1182/blood-2013-12-546309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Børglum AD, et al. A new locus for Seckel syndrome on chromosome 18p11.31-q11.2. Eur J Hum Genet. 2001;9(10):753–757. doi: 10.1038/sj.ejhg.5200701. [DOI] [PubMed] [Google Scholar]
- 55.Wagner JE, et al. Germline mutations in BRCA2: Shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood. 2004;103(8):3226–3229. doi: 10.1182/blood-2003-09-3138. [DOI] [PubMed] [Google Scholar]
- 56.Levran O, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37(9):931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.