Significance
Insulin is key for control of glucose homeostasis in vertebrates. Insufficient insulin secretion relative to metabolic needs causes diabetes. Pancreatic beta cells store insulin into secretory granules (SGs), which release insulin extracellularly upon fusion with the plasma membrane. SGs exist in different functional pools, with newly generated SGs being preferentially secreted. Here we show that aged SGs display reduced competence for glucose-stimulated microtubule-mediated transport and are disposed within actin-positive multigranular bodies. These data highlight the link between SG age and mobility and thus are relevant for better understanding insulin secretion in health and diabetes.
Keywords: islets, diabetes, secretion, Bayesian probability theory, processivity
Abstract
Insulin secretion is key for glucose homeostasis. Insulin secretory granules (SGs) exist in different functional pools, with young SGs being more mobile and preferentially secreted. However, the principles governing the mobility of age-distinct SGs remain undefined. Using the time-reporter insulin-SNAP to track age-distinct SGs we now show that their dynamics can be classified into three components: highly dynamic, restricted, and nearly immobile. Young SGs display all three components, whereas old SGs are either restricted or nearly immobile. Both glucose stimulation and F-actin depolymerization recruit a fraction of nearly immobile young, but not old, SGs for highly dynamic, microtubule-dependent transport. Moreover, F-actin marks multigranular bodies/lysosomes containing aged SGs. These data demonstrate that SGs lose their responsiveness to glucose stimulation and competence for microtubule-mediated transport over time while changing their relationship with F-actin.
Peptide hormones, neuropeptides, and bioactive amines are stored into secretory granules (SGs), also referred to as large dense-core vesicles (1, 2). Upon appropriate cell stimulation SGs undergo exocytosis and release their cargoes. Pancreatic beta cells are suitable models to study regulated secretion because they are responsible for production and release of insulin, a key hormone for maintenance of glucose homeostasis that affects a wide range of metabolic processes (3). Glucose-stimulated insulin secretion exhibits a biphasic pattern (4–6). The first phase, which develops rapidly after stimulation of beta cells by hyperglycemia, only lasts a few minutes, whereas the second, slowly rising phase persists until euglycemia is restored and should last <2 h in a healthy subject (7, 8). Because lack of the first phase and a blunted second phase of glucose-induced insulin release are hallmarks of glucose intolerance and diabetes (9), a detailed characterization of the SG pools contributing to insulin secretion is relevant for understanding insulin secretion in health and disease.
A beta cell contains on average 5 × 103 insulin SGs (10). Ultrastructural and biochemical analyses combined with electrophysiological measurements have revealed that insulin SGs can be mainly divided into two distinct populations, named readily releasable pool and reserve pool. These two pools differ in their size and release competence. The readily releasable pool consists of ∼50 SGs that are already docked at the plasma membrane (PM) before stimulation (11, 12). Exocytosis of these SGs has been considered to primarily account for the first phase of glucose-stimulated insulin release. After consumption of the readily releasable pool, the mobilization of more distal SGs in the reserve pool would give rise and sustain the second phase of insulin release (7, 8). This classical model has been recently challenged following the observation of living beta cells by total internal reflection fluorescence microscopy (TIRFM) that “newcomer” SGs suddenly appearing at the PM can undergo exocytosis without virtually any time delay (13, 14). According to some reports newcomer SGs contribute to both phases of insulin release (15–19). Additional evidence suggests that docking of SGs to the PM can constrain SG exocytosis (20, 21). Hence, mobility seems to be a critical factor for assessing the probability of SGs to undergo exocytosis (22, 23).
Notably, neither the classical nor the revised model of glucose-stimulated insulin secretion takes into account the preferential release of newly synthesized insulin (24–26). Preferential release of newly generated SGs is a shared feature among peptide-secreting endocrine cells (27, 28). Using SG cargoes fused to fluorescent timer proteins, which change their emission spectra over time, it was shown that SGs of bovine chromaffin cells (29) and PC12 cells (30) segregate into functionally and spatially distinct pools, with newer SGs being preferentially released upon stimulation. The stochastic nature and restricted temporal window of its spectral transition limit, however, the reliability and flexibility of fluorescent timer proteins for accurate determination of SG age. To overcome this hurdle, we developed a time-resolved protocol for SG labeling by tagging insulin with SNAP (Ins-SNAP) (31). SNAP is derived from alkyl guanine DNA alkyl transferase, a DNA repair enzyme that irreversibly transfers an alkyl group from its substrate to a reactive cysteine residue (32–36). The availability of cell-permeable nonfluorescent and fluorescent SNAP substrates, such as TMR-Star, allows for a broad variety of applications. Ins-SNAP labeled with TMR-Star (Ins-SNAPTMR-Star) is properly sorted into SGs and secreted upon glucose stimulation (31). Importantly, high-content image analyses formally demonstrated that the preferential release of young Ins-SNAPTMR-Star+ SGs is a homogenous response to stimulation rather than being primarily sustained by a more biosynthetically and secretory active subpopulation of cells. In this study we exploited the same protocol to investigate in mechanistic and quantitative terms the dynamics of time-resolved insulin SGs and their relationship with the cytoskeleton.
Results
Three Dynamic Components Account for the Collective Mobility of Insulin SGs.
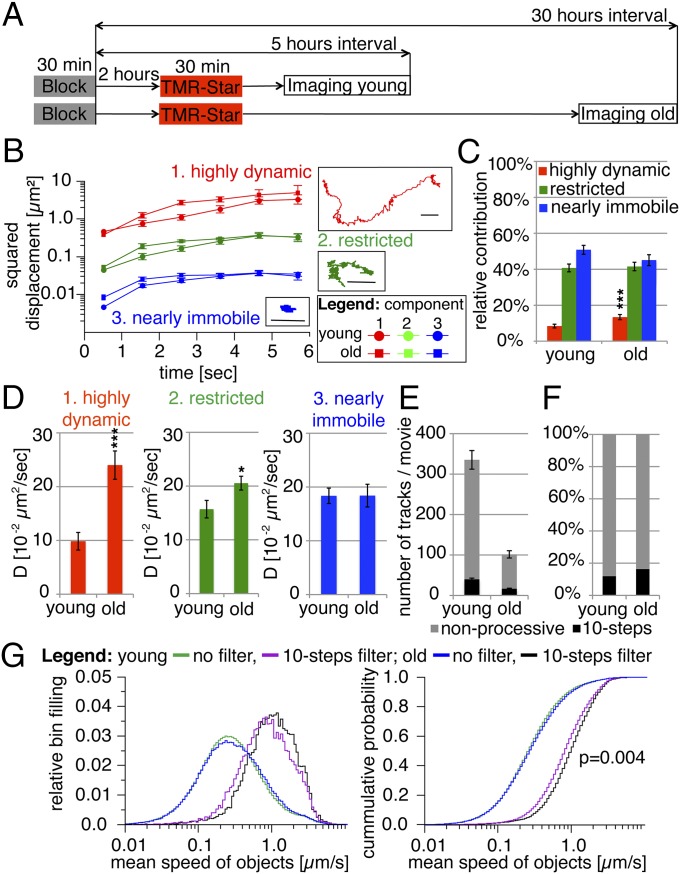
Fluorescent labeling of insulinoma INS-1 cells expressing Ins-SNAP with the SNAP substrate TMR-Star allows for the detection of age-distinct Ins-SNAPTMR-Star+ SGs (31). We imaged cortical 3- to 5- or 28- to 30-h-old Ins-SNAPTMR-Star+ (henceforth defined as young and old) SGs using TIRFM and analyzed their dynamics with automated particle tracking and statistical analysis based on the Motion Tracking software (Fig. 1A and Movie S1). These time intervals were close to those applied in previous studies on insulin turnover (31, 37). Our initial analyses indicated that young and old SGs did not significantly differ in regards to their collective diffusion coefficient (Fig. S1). The power of this analysis was restricted, however, because it did not provide information about the different components contributing to the collective dynamics of SGs, which are highly heterogeneous in their mobility (Movie S1). Therefore, we performed analysis based on Bayesian probability theory (38). This approach identified the number of dynamic components for which there was most evidence in the experimental data, as well as their relative contribution and diffusion coefficients. Three components, defined as “highly dynamic” [diffusion coefficient (D) ∼10−2 µm2/s], “restricted” (D ∼10−3 µm2/s), and “nearly immobile” (D ∼10−4 µm2/s), were found to be necessary and sufficient to account for the collective dynamics of both young and old SG pools (Fig. 1B and Table S1). The highly dynamic component accounted for the minority of events in the case of both SG pools, whereas most young and old SGs were either restricted or nearly immobile (Fig. 1C). The relative contribution of highly dynamic old SGs (Fig. 1C) as well as the D of highly dynamic and restricted old SGs (Fig. 1D), as calculated by regression analysis of squared displacements plotted over time (Fig. S1), were higher than those of young SGs.
Fig. 1.
Three dynamic components account for the collective mobility of SGs. (A) Schematic representation of the labeling protocol for fluorescent visualization of 3- to 5- (young) and 28- to 30-h-old (old) Ins-SNAP+ SGs by TMR-Star labeling. For details see Material and Methods. (B) Squared displacements (log-scale) as a function of time for each dynamic component contributing to the collective dynamics of young and old Ins-SNAPTMR-Star+ SGs. Representative tracks for each component are shown. (Scale bars, 1 µm.) (C) Relative contribution of the dynamic components shown in B to the collective dynamics of young and old Ins-SNAPTMR-Star+ SGs. (D) Diffusion coefficients of young and old Ins-SNAPTMR-Star+ SGs as calculated by regression analysis of the squared displacement of each dynamic component plotted over time (see Fig. S1). (E) Mean number of the total and processive (see Fig. S1) tracks of Ins-SNAPTMR-Star+ SGs detected per movie. (F) Relative representation of the processive tracks of Ins-SNAPTMR-Star+ SGs. (G) Collective and processive mean speeds of young and old Ins-SNAPTMR-Star+ SGs shown as normalized distribution (Left) and cumulative probability (Right). The figure presents the data obtained from 12 independent experiments counting 33,984 tracks in 111 cells for young SGs and 12 independent experiments counting 11,486 tracks in 102 cells for old SGs.
Next, we evaluated the collective mean speed of young and old SGs as well as the mean speed in both age groups of processive SGs, which moved for ≥10 steps without changes in direction >30° in between each step (Fig. S1). Although the average number of tracks per movie was higher in the case of young SGs (Fig. 1E), the percentage of processive tracks was higher for old SGs (Fig. 1F). Young and old SGs did not differ in their collective mean speed, but processive old SGs were faster (Fig. 1G). Hence, using two independent statistical approaches we identified a fraction of highly mobile SGs, which in the case of old SGs was larger and displayed greater D and processive speed. Notably, these findings were in contrast with previous analyses, including our own, suggesting that SG motility decreases over time (29, 31).
Glucose Stimulation Mobilizes Young but Not Old SGs.
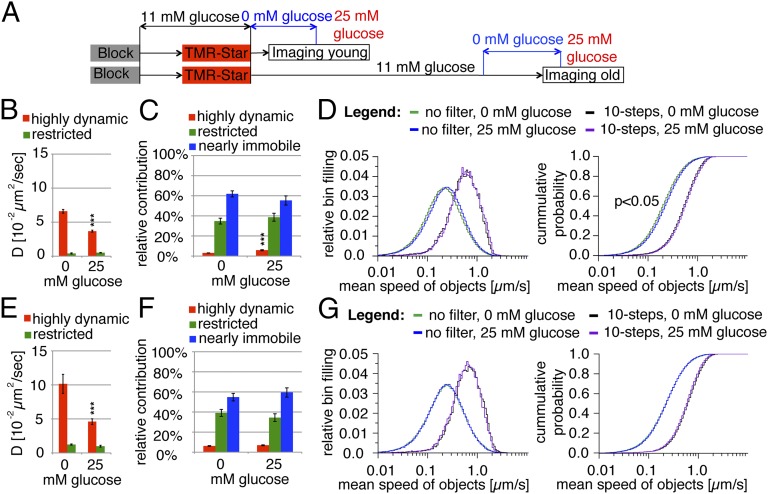
Newly synthesized insulin is released preferentially in response to glucose stimulation of beta cells (24–26), which mobilizes SGs (39). Therefore, we analyzed the impact of glucose stimulation on the mobility of young and old SGs (Fig. 2A). Upon glucose stimulation the D of both highly dynamic young (Fig. 2B) and old (Fig. 2E) SGs decreased to ∼50%, whereas the relative contribution of the first (Fig. 2C), but not of the second (Fig. 2F), to the collective SG mobility increased approximately twofold. Moreover, glucose stimulation increased the collective, but not the processive, speed of young SGs (Fig. 2D), whereas in the case of old SGs both speeds were unchanged (Fig. 2G). Thus, glucose recruits more young SGs to the highly dynamic pool, although their reduced D indicates more frequent changes in directionality, as also reflected by the increase in the collective, but not in the processive, speed.
Fig. 2.
Glucose stimulation increases the frequency of young but not old highly dynamic SGs. (A) Schematic representation of the labeling (as in Fig. 1A) and resting (0 mM glucose) and stimulation (25 mM glucose) protocol. (B) Diffusion coefficients of young highly dynamic and restricted Ins-SNAPTMR-Star+ SGs in resting and stimulated cells. (C) Relative contribution of the dynamic components to the collective dynamics of young Ins-SNAPTMR-Star+ SGs in resting and stimulated cells. (D) The collective and processive mean speed of young Ins-SNAPTMR-Star+ SGs shown as normalized distribution (Left) and cumulative probability (Right). (E) Diffusion coefficients of young highly dynamic and restricted old Ins-SNAPTMR-Star+ SGs in resting and stimulated cells. (F) Relative contribution of the dynamic components to the collective dynamics of old Ins-SNAPTMR-Star+ SGs in resting and stimulated cells. (G) Collective and processive mean speeds of old Ins-SNAPTMR-Star+ SGs shown as normalized distribution (Left) and cumulative probability (Right). The figure presents the data obtained from three independent experiments counting 19,176 tracks in 57 resting cells and 24,868 tracks in 32 stimulated cells for young SGs and three independent experiments counting 6,917 tracks in 35 resting cells and 5,608 tracks in 25 stimulated cells for old SGs.
Different Dynamics of Age-Distinct SGs Is Regulated by the Cytoskeleton.
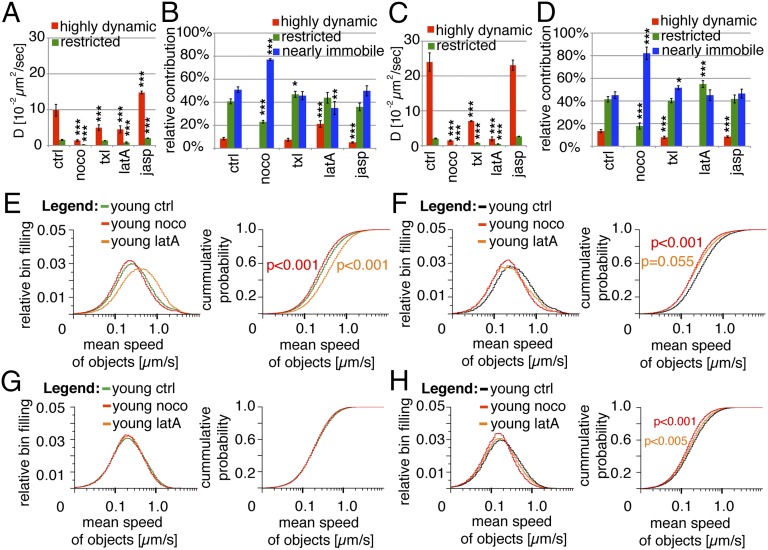
Microtubules (MTs) are implicated in the transport of SGs (40–45) (Movie S2). Therefore, we analyzed the role of MTs in regards to the different dynamics of young and old SGs. Nocodozole treatment, which causes MT depolymerization (Fig. S2), reduced the D of the highly dynamic young (Fig. 3A) and old (Fig. 3C) SGs to that of restricted SGs in untreated cells. Thus, in this condition SGs in both age groups were either restricted or nearly immobile, with the latter being predominant (Fig. 3 B and D). Following MT disruption the collective mean speeds of young (Fig. 3E) and old (Fig. 3G) SGs were also decreased. Stabilization of MTs with taxol also reduced the D of highly dynamic young and old SGs (Fig. 3 A and C). Moreover, it decreased the pool of highly dynamic old (Fig. 3D), but not young (Fig. 3B), SGs. This effect was mirrored by the decreased frequency of highly dynamic and increased frequency of nearly immobile old SGs (Fig. 3D) and the reduced collective mean speed of old (Fig. 3H), but not young (Fig. 3F), SGs. Hence, dynamic MTs are required for the excursions of both highly mobile young and old SGs but seem to be more critical for the latter.
Fig. 3.
Disruption of FA, but not MTs, differently affects the dynamics of young and old SGs. (A) Impact of nocodozole (noco), taxol (txl), latrunculin A (latA), or jasplakinolide (jasp) treatments on the mean diffusion coefficients of the highly dynamic and restricted young Ins-SNAPTMR-Star+ SGs. (B) Impact of noco, txl, latA, or jasp treatments on the relative contribution of the three dynamic components to the collective dynamics of young Ins-SNAPTMR-Star+ SGs. (C) Impact of noco, txl, latA, or jasp treatments on the mean diffusion coefficients of the highly dynamic and restricted old Ins-SNAPTMR-Star+ SGs. (D) Impact of noco, txl, latA, or jasp treatments on the relative contribution of the three dynamic components to the collective dynamics of old Ins-SNAPTMR-Star+ SGs. (E) Impact of noco or latA treatments on the collective mean speed of young Ins-SNAPTMR-Star+ SGs shown as normalized distribution (Left) and cumulative probability (Right). (F) Impact of txl or jasp treatments on the collective mean speed of young Ins-SNAPTMR-Star+ SGs shown as normalized distribution (Left) and cumulative probability (Right). (G) Impact of noco or latA treatments on the collective mean speed of old Ins-SNAPTMR-Star+ SGs shown as normalized distribution (Left) and cumulative probability (Right). (H) Impact of txl or jasp treatments on the collective mean speed of old Ins-SNAPTMR-Star+ SGs shown as normalized distribution (Left) and cumulative probability (Right). Data for ctrls are the same as in Fig. 1. Other data are from two independent experiments counting 7,149 tracks of young SGs in 24 cells treated with noco; six experiments counting 18,343 tracks of young SGs in 50 cells treated with txl; four experiments counting 8,767 tracks in 24 cells treated with latA; five experiments counting 18,513 tracks of young SGs in 49 cells treated with jasp; two experiments counting 7,735 tracks of old SGs in 23 cells treated with noco; five experiments counting 3,721 tracks of old SGs in 39 cells treated with txl; four experiments counting 3,362 tracks of old SGs in 30 cells treated with latA; and five experiments counting 5,116 tracks of old SGs in 42 cells treated with txl.
Filamentous actin (FA) regulates the access of SGs to the PM (46–53). Therefore, its role in relationship to the different dynamics of young and old SGs was also investigated. Upon latrunculin A treatment, which depolymerizes FA (Fig. S2), the D of highly dynamic young SGs was reduced by ∼50% (Fig. 3A), whereas their number increased by ∼1.5-fold (Fig. 3B). This increase came at the expense of the nearly immobile SGs, which were comparably fewer than in control cells (Fig. 3B). Conversely, disruption of FA abolished the highly dynamic pool of old SGs such that only restricted and nearly immobile old SGs remained (Fig. 3 C and D). Moreover, latrunculin A treatment increased the collective mean speed of young SGs (Fig. 3E), whereas it decreased that of old SGs (Fig. 3G). Opposite to latrunculin A, jasplakinolide treatment, which stabilizes FA, increased the D of highly dynamic and restricted young SGs, but it did not alter that of the corresponding pools of old SGs (Fig. 3 A and C). It also reduced the frequency of highly dynamic young and old SGs (Fig. 3 B and D), as well as the collective mean speed of old, but not of young, SGs. Hence, opposite to MTs, FA negatively regulates the mobility of highly dynamic young SGs, whereas it supports that of highly dynamic old SGs. Notably, its pharmacological disruption mimicked the effects of glucose stimulation on highly dynamic young SGs.
Old SGs Are More Frequently Associated with F-Actin.
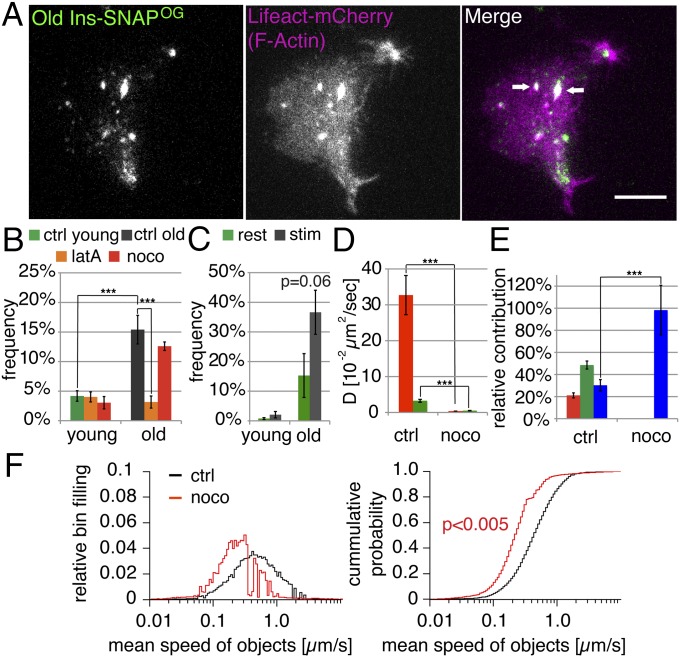
To further investigate the differential involvement of FA in the mobility of young and old SGs we performed dual-color TIRFM in INS-1 cells coexpressing Ins-SNAP labeled with Oregon Green (OG) (hence Ins-SNAPOG+ objects) and Lifeact-mCherry (54), which specifically stains FA (Fig. S3). In addition to labeling filamentous structures Lifeact-mCherry localized to mobile punctuate organelles, including some Ins-SNAPOG+ objects (Fig. 4A and Movie S3). Old Ins-SNAPOG+, Lifeact-mCherry+ objects were threefold more frequent than the corresponding young objects (Fig. 4B) and they were reduced upon latrunculin A treatment (Fig. 4B), whereas their number tended to increase upon glucose stimulation (Fig. 4C). In control cells we detected highly mobile, restricted, and nearly immobile old Ins-SNAPOG+, Lifeact-mCherry+ objects, whereas in nocodozole-treated cells only the nearly immobile component was present (Fig. 4 D and E and Movies S4 and S5). This treatment decreased also the collective mean speed of Ins-SNAPOG+, Lifeact-mCherry+ SGs (Fig. 4F). Thus, the association of FA with SGs increases with their age, it is enhanced by glucose, and FA+ SGs move on MTs.
Fig. 4.
Old SGs are more frequently FA+ and move on MTs. (A) Dual-color TIRFM images showing the overlap of some old Ins-SNAPOG+ SGs with Lifeact-mCherry. See Movie S3. (Scale bar, 10 µm.) (B) Percentage of young or old Ins-SNAPOG+ tracks positive for Lifeact-mCherry in control (ctrl), nocodazole- (noco), or latrunculin A- (latA) treated cells. (C) Percentage of young or old Ins-SNAPOG+ tracks positive for Lifeact-mCherry in resting (0 mM glucose; rest) and stimulated (25 mM glucose; stim) cells. (D) Impact of noco treatment on the mean diffusion coefficients of old highly dynamic and restricted Ins-SNAPOG+, Lifeact-mCherry+ SGs. (E) Impact of noco treatment on the relative contribution of the three dynamic components to the collective dynamics of old Ins-SNAPOG+, Lifeact-mCherry+ SGs. (F) Impact of noco treatment on the mean speed of old Ins-SNAPOG+, Lifeact-mCherry+ SGs shown as normalized distribution (Left) and cumulative probability (Right). See also Movies S4 and S5. The figure presents the data obtained from three (for young SGs) and four (for old SGs) independent experiments counting 15,379 tracks of young SGs in 19 ctrl cells, 11,113 tracks of young SGs in 17 latA-treated cells, and 8,149 tracks of young SGs in 17 noco treated cells; 7,476 tracks of old SGs in 35 ctrl cells, 2,921 tracks of old SGs in 19 latA-treated cells, and 3,695 tracks of old SGs in 22 noco-treated cells. Data in C are derived from three independent experiments where 21,950 tracks of young SGs in 45 resting cells, 27,632 tracks of young SGs in 58 stimulated cells, 4,462 tracks of old SGs in 47 resting cells, and 5,716 tracks in 58 stimulated cells were counted.
Aged SGs Are Disposed in Actin-Positive Multigranular Bodies.
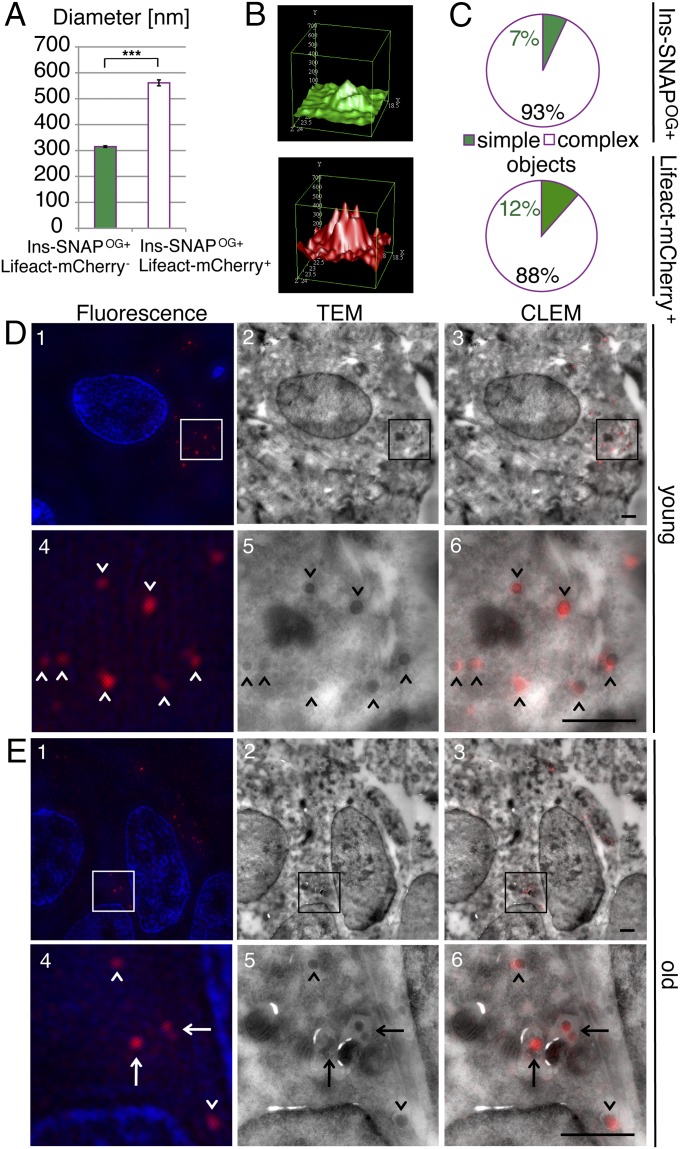
By electron microscopy insulin SGs are rather uniform in regards to their spherical appearance and size (10). However, the shape of old Ins-SNAPOG+, Lifeact-mCherry+ SGs was pleiomorphic (Fig. 4A, arrow). Moreover, the mean diameter of Ins-SNAPOG+, Lifeact-mCherry+ SGs was twice that of Ins-SNAPOG+, Lifeact-mCherry− SGs (Fig. 5A). Indeed, two classes of Ins-SNAPOG+ SGs could be distinguished based on the intensity profile of their fluorescence: “simple objects” and “complex objects.” Simple objects had one intensity peak whereas complex objects consisted of several simple objects, which in time-lapse movies moved together (Fig. 4A, arrow, and Fig. S4). Ins-SNAPOG+, Lifeact-mCherry+ represented mostly complex objects, including on average 3.4 ± 0.4 and 3.2 ± 0.6 fluorescence peaks for Ins-SNAPOG and Lifeact-mCherry, respectively (Fig. 5 B and C). To gain further insight into the ultrastructure of young and old Ins-SNAP+ SGs we performed correlative-light electron microscopy (Fig. 5 D and E). Most Ins-SNAPTMR-Star+ objects detected by super resolution microscopy (Fig. 5 D and E, Left, arrowheads) displayed by electron microscopy the typical ultrastructure of SGs (Fig. 5 D and E, Center and Right, arrowheads). However, 9.2 ± 1.3% of young and 44.3 ± 6.5% of old Ins-SNAPTMR-Star+ objects correlated by electron microscopy with multigranular bodies (MGBs) (Fig. 5E, arrows), that is, pleiomorphic organelles delimited by a membrane enclosing two or more electrondense cores (55). The higher proportion of old Ins-SNAPTMR-Star in MGBs (P < 0.05) is consistent with a greater fraction of old Ins-SNAP OG being in complex objects larger than bona fide SGs.
Fig. 5.
A fraction of old SGs is found in multigranular bodies. (A) Mean diameter of Ins-SNAPOG+, Lifeact-mCherry− SGs, and of Ins-SNAPOG+, Lifeact-mCherry+ SGs. Data are derived from the same dataset as in Fig. 4B. (B) Representative intensity profiles of a single complex Ins-SNAPOG+, Lifeact-mCherry+ object showing several overlapping fluorescence peaks in the green (Top) and red (Bottom) channels (possibility of bleed-through between channels was ruled out as described in Materials and Methods). (C) Frequency of the complex Ins-SNAPOG+, Lifeact-mCherry+ objects in the green (Top) and red (Bottom) channels as a result of counting 40 Ins-SNAPOG+ and 23 Lifeact-mCherry+ objects. (D) Representative images of young Ins-SNAPTMR-Star+ SGs visualized by (1) structured illumination microscopy (fluorescence). (2) TEM of the same region as in 1. (3) Overlay (CLEM) of images in 1 (red channel) and 2. (4–6) High magnification of the area boxed in 1–3. Arrowheads point to young Ins-SNAPTMR-Star+ SGs. (Scale bar, 1 µm.) (E) Representative images of old Ins-SNAPTMR-Star+ SGs visualized by (1) structured illumination microscopy (fluorescence). (2) TEM of the same region as in 1. (3) Overlay (CLEM) of images in 1 (red channel) and 2. (4–6) High magnification of the area boxed in 1–3. Arrowheads point to old Ins-SNAPTMR-Star+ SGs; arrows point to several old Ins-SNAPTMR-Star+ SGs within a MGB. In D and E, two independent CLEM experiments were performed counting in total in 19 cells 169 young Ins-SNAPTMR-Star+ SGs, out of which 16 were found in MGBs, and in 31 cells counting 197 Ins-SNAPTMR-Star+ SGs, out of which 76 were found in MGBs. (Scale bar, 1 µm.)
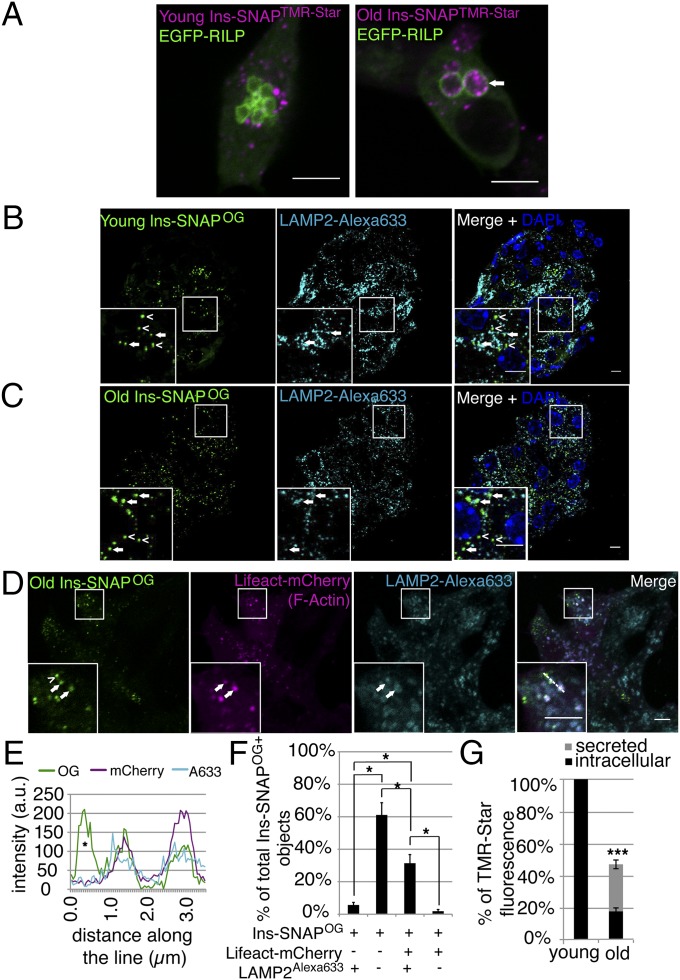
MGBs represent lysosomes at an early stage of SG disposal (56). To investigate in more detail the relationship between age of SGs and their degradation we analyzed the colocalization of young and old SGs with lysosomes. In INS-1 cells 14.7 ± 2.6% of the young Ins-SNAPTMR-Star+ SGs colocalized with the lysosomal reporter EGFP-RILP compared with 33.5 ± 5.4% (P < 0.05) of the old Ins-SNAPTMR-Star+ SGs (Fig. 6A). A similar trend was observed in primary beta cells of SOFIA mice, which express mINS2-SNAP (31) and in which 12.1 ± 0.7% of the young Ins-SNAPOG+ SGs colocalized with the lysosomal marker lysosome-associated membrane protein 2 (LAMP2) compared with 21.3 ± 1.2% of the old Ins-SNAPOG+ SGs (P < 0.05) (Fig. 6 B and C). Moreover, old Ins-SNAPOG+, Lifeact-mCherry+, but not Ins-SNAPOG+ Lifeact-mCherry−, organelles colocalized with LAMP2 in INS-1 cells. (Fig. 6 D–F and Fig. S5). The intracellular levels of young and old Ins-SNAPTMR-Star as well as the amount of Ins-SNAPTMR-Star released in the media during the two time points (i.e., between 5 and 30 h postlabeling) was further measured by fluorimetry. Combined, intracellular, and secreted old Ins-SNAPTMR-Star only accounted to approximately half of young Ins-SNAPTMR-Star (Fig. 6G), indicating that during this time interval half of Ins-SNAPTMR-Star was degraded. This half-life is close to that of 22.5 h estimated for insulin in mouse insulinoma MIN6 cells (57). Taken together, these data show that a significant fraction of old SGs is marked by FA and destined for intracellular degradation.
Fig. 6.
MGBs represent FA+ lysosomes involved in degradation of aged SGs. (A) Confocal images of EGFP-RILP and young or old Ins-SNAPTMR-Star+ objects in fixed INS-1 cells. An arrow points to old Ins-SNAPTMR-Star+ SGs within an EGFP-RILP+ compartment. Quantification was performed on the data obtained from three independent experiments. In total 23 cells containing 1,157 young Ins-SNAPTMR-Star+ SGs, out of which 170 were EGFP-RILP+ and 19 cells containing 684 old Ins-SNAPTMR-Star+ SGs, out of which 196 were EGFP-RILP+. (Scale bars, 5 µm.) (B) Confocal images of young Ins-SNAPOG+ SGs (open arrowheads) and LAMP2+ lysosomes (filled arrowhead) in beta cells of SOFIA mice. (Scale bar, 5 µm.) (C) Confocal images of old Ins-SNAPOG+ SGs (empty arrowheads) and LAMP2+ lysosomes (full arrowhead) in beta cells of SOFIA mice. In B and C, two independent islet isolations were performed from two SOFIA mice each time. In total was counted 100 cells with 1,975 young Ins-SNAPOG+ SGs, out of which 240 were LAMP2+ and 218 cells with 1,615 old Ins-SNAPOG+ SGs, out of which 335 were LAMP2+. (Scale bar, 5 µm.) (D) Colocalization of old Ins-SNAPOG+, Lifeact-mCherry+ (arrow) SGs with LAMP2 in INS-1 cells. Ins-SNAPOG+, Lifeact-mCherry− SG (arrowhead) does not colocalize with LAMP2. Quantification (shown in F) was performed on the data obtained from threee independent experiments counting in total 44 old Ins-SNAPOG+/LAMP2+/Lifeact-mCherry− SGs, 458 old Ins-SNAPOG+/LAMP2−/Lifeact-mCherry− SGs, 244 old Ins-SNAPOG+/LAMP2+/Lifeact-mCherry+ SGs, and 14 old Ins-SNAPOG+/LAMP2−/Lifeact-mCherry+ SGs. (Scale bar, 5 µm.) (E) Fluorescence intensity profile across the line for OG, mCherry, and Alexa633. An asterisk marks the intensity profile of the SG indicated in B by the arrowhead. (F) Percentage of LAMP2+ and/or Lifeact-mCherry+ old Ins-SNAPOG+ SGs in INS-1 cells. (G) Fluorimetric analysis of Ins-SNAPTMR-Star turnover. Fluorescence of 28- to 30-h-old Ins-SNAPTMR-Star in the cells and in media compared with the 3- to 5-h-old Ins-SNAPTMR-Star in the cells. The difference between 3- to 5-h-old Ins-SNAPTMR-Star in the cells and 28- to 30-h-old Ins-SNAPTMR-Star in the cells plus in media equals the amount of degraded Ins-SNAPTMR-Star. Data were obtained from three independent fluorimetric experiments, each containing biological triplicate for young and old Ins-SNAPTMR-Star and measured in loading triplicates. For all measurements the same number of cells was seeded. To confirm that the loss of Ins-SNAPTMR-Star signal is not due to the loss of the cells over the time course we measured total protein concentration in the samples. The total protein concentration increased over time (from 0.271 µg/mL to 0.357 µg/mL), which reflects the increase in the cell number during incubation time. Because we measured the signal of Ins-SNAPTMR-Star signal originating from the labeling at the same starting point, increase in the cell number in the culture in the absence of TMR-Star substrate could not affect the fluorimetric analysis.
Young Bona Fide SGs Are More Mobile Than Their Older Counterparts.
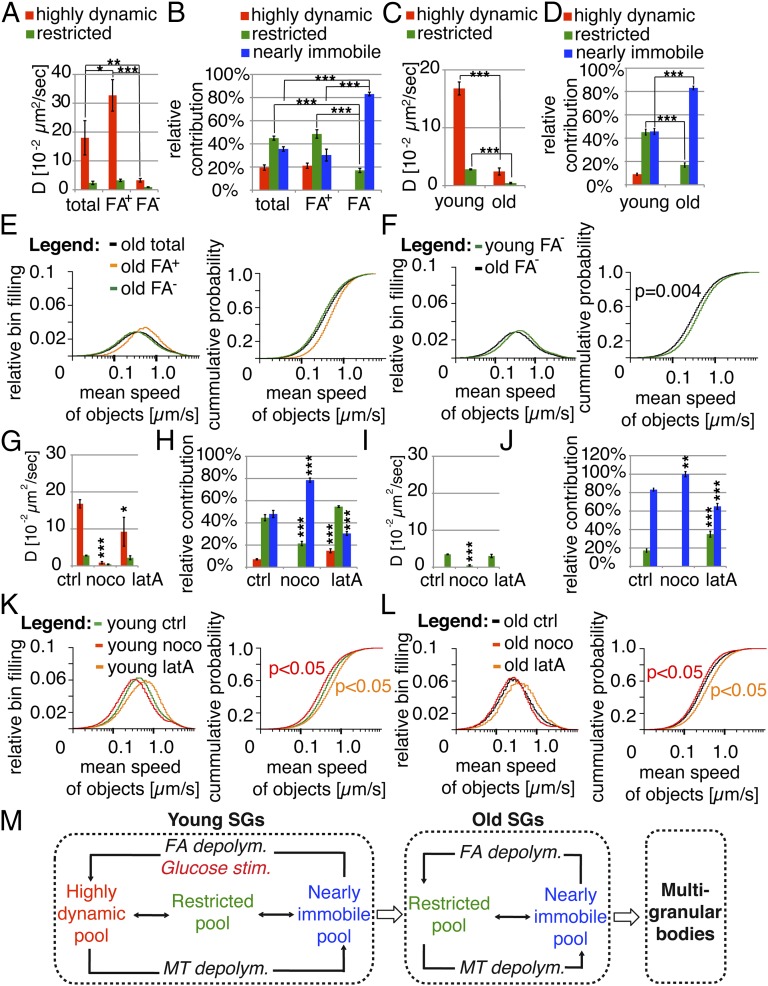
Identification of FA as a marker associated with aged SGs localized within MGBs suggests that only Ins-SNAPOG+, Lifeact-mCherry− objects represent bona fide SGs. Therefore, we evaluated separately the dynamics of old Ins-SNAPOG+ objects either positive or negative for Lifeact-mCherry. Ins-SNAPOG+, Lifeact-mCherry+ objects displayed a higher D and were faster than the whole set of highly dynamic old Ins-SNAPOG+ objects (Fig. 7 A and E), whereas the D of the Lifeact-mCherry− subset (i.e., of bona fide old SGs) was equivalent to that of restricted SGs. Hence, in the case of bona fide old SGs the highly dynamic component was absent and most of them were nearly immobile (Fig. 7B), whereas Lifeact-mCherry+ SGs accounted for the high mobility of the whole set of old Ins-SNAPOG+ objects.
Fig. 7.
Young SGs are more mobile than their older counterparts. (A) Diffusion coefficients of the whole set of highly dynamic and restricted old Ins-SNAPOG+ objects and of the corresponding Lifeact-mCherry+ (FA+) or Lifeact-mCherry− (FA−) subsets. (B) Relative contribution of the three dynamic components to the collective motion of old Ins-SNAPOG+ objects and of the corresponding Lifeact-mCherry+ (FA+) or Lifeact-mCherry− (FA−) subsets. (C) Diffusion coefficients of young and old highly dynamic and restricted Ins-SNAPOG+, Lifeact-mCherry− objects. (D) Relative contribution of the three dynamic components to the collective dynamics of young and old Ins-SNAPOG+, Lifeact-mCherry− objects. (E) Collective mean speeds of young and old Ins-SNAPOG+, Lifeact-mCherry− objects. (F) Collective mean speeds of old Ins-SNAPOG+ objects and of the corresponding Lifeact-mCherry+ (FA+) or Lifeact-mCherry− (FA−) subsets. (G) Impact of nocodazole (noco) or latrunculin A (latA) treatment on the mean diffusion coefficients of highly dynamic and restricted Ins-SNAPOG+, Lifeact-mCherry− (i.e., bona fide young SGs). (H) Impact of noco or latA treatment on the relative contribution of the three dynamic components to the collective dynamics of young bona fide SGs. (I) Impact of noco or latA treatment on the mean diffusion coefficients of restricted old Ins-SNAPOG+, Lifeact-mCherry− objects. (J) Impact of noco or latA treatment on the relative contribution of the restricted and immobile components to the collective dynamics of old Ins-SNAPOG+, Lifeact-mCherry− objects. (K) Impact of noco or latA treatment on the collective mean speed of young bona fide SGs shown as normalized distribution (Left) and cumulative probability (Right). (L) Impact of latA or noco treatments on the collective mean speed of old Ins-SNAPOG+, Lifeact-mCherry− objects shown as normalized distribution (Left) and cumulative probability (Right). (M) Young SGs exist in three dynamic pools; old SGs unless localized within MGBs do not display highly dynamic component. Depolymerization of MTs immobilizes young as well as old SGs. Depolymerization of FA results in the increased highly dynamic fraction on the expense of the immobile fraction among young SGs and increased restricted fraction among old SGs. MGBs move on MTs. Results are derived from the same dataset as in Fig. 4B.
Because Lifeact-mCherry+ objects were more frequent among old than among young SGs, we evaluated how their omission affected our analysis. Contrary to our original conclusion (Fig. 1 D and G), bona fide young SGs (i.e., Ins-SNAPOG+, Lifeact-mCherry− objects) were more mobile than their older counterparts. This was documented by the presence of all three dynamic components among bona fide young SGs (Fig. 7 C and D). In contrast, bona fide old SGs were either restricted or nearly immobile, with predominance of the latter. Accordingly, the collective mean speed of bona fide young SGs was significantly higher than that of old bona fide SGs (Fig. 7F).
Finally, we analyzed the impact of MTs or FA depolymerization on the mobility of bona fide young and old SGs. Consistent with previous results (Fig. 3 A and B), in nocodozole-treated cells bona fide young SGs were either restricted or nearly immobile (Fig. 7 G and H) and their collective mean speed was decreased (Fig. 7K). Likewise, FA depolymerization reduced the D of highly dynamic bona fide young SGs to ∼50% (Fig. 7G), whereas it increased their frequency by approximately twofold at the expense of the nearly immobile component (Fig. 7H). As previously assessed (Fig. 3E), latrunculin A treatment increased the collective mean speed of bona fide young SGs (Fig. 7K). In the case of bona fide old SGs, MT depolymerization decreased their D (Fig. 7I) and speed (Fig. 7L) such that only nearly immobile SGs were present (Fig. 7J). FA depolymerization, instead, did not affect the D of bona fide old SGs (Fig. 7I) but increased the frequency of the restricted component (Fig. 7J) and their collective mean speed (Fig. 7L). These data are consistent with bona fide SGs of all ages being restricted in their motion by FA, but progressively losing with time their competence for MT-mediated transport (Fig. 7M).
Discussion
In the present study we show that age affects the mobility of cortical insulin SGs and their relationship with MTs and FA (Fig. 7M). Our initial analyses indicated that different motion modes contribute to the collective mobility of insulin SGs, with highly dynamic and processively moving old (28- to 30-h-old) SGs being more mobile than their young (3- to 5-h-old) counterparts. We then uncovered that FA, unlike MTs, plays opposite roles on the dynamics of SGs depending on their age, with old SGs being more frequently associated with FA and endosomal–lysosomal markers and localized within MGBs. These findings led us to demonstrate that bona fide old SGs, contrary to our initial assessment, are less mobile than young SGs and identify FA as a marker of aged SGs within MGBs/lysosomes. Importantly, glucose stimulation increased the mobility of young, but not old, SGs (Fig. 2) while increasing the fraction of FA+ old SGs.
Recent studies have unraveled the significant contribute of highly mobile, so-called restless newcomer SGs to both phases of glucose-stimulated insulin secretion (15–19). Independent, long-standing evidence indicates the preferential release of newly synthesized insulin (24–26). However, accurate information on insulin SG mobility in relationship to their age is not yet available. Development of the Ins-SNAP reporter for marking age-distinct pools of SGs enabled us to formally verify the preferential exocytosis of newly generated SGs and to show that the mobility of insulin SGs decreases within a few hours after their biogenesis (31). In that instance, however, we neither investigated how glucose stimulation affects the mobility of age-distinct SGs nor analyzed the different motion patterns contributing to the collective mobility of time-resolved SG pools and their dependence on MTs or FA. Such in-depth, quantitative analysis was carried out here and resulted in the identification of three dynamic components, termed highly dynamic, restricted, and nearly immobile, differing in their diffusion coefficients by one order of magnitude. These three motion modes were necessary and sufficient to account for the collective dynamics of young SGs, whereas old SGs were either restricted or nearly immobile.
Our results are in agreement with an earlier report of the three dynamic patterns, namely directed planar movement, random movement (otherwise defined as free planar diffusion), and restricted movement, that account for the motion of SGs in INS-1 cells (58). These motion modes most likely correspond to those classified here as highly dynamic, restricted, and nearly immobile. Here we extended the knowledge by providing information about the relative contribution of each dynamic component to the collective dynamics of age-distinct SG pools. Interestingly, the motion patterns identified for insulin SGs displayed diffusion coefficients and frequencies comparable to those reported for the regulated insertion of the TRPM8 ion channel into the PM of peripheral neurons (59) and for lytic granules in natural killer cells (60). These observations suggest that dynamic properties of regulated secretory vesicles are conserved among different cell types.
Nocodazole treatment affected both the highly dynamic and the restricted components (Figs. 3 B and D and 7 H and J), indicating their dependence on MTs. In contrast to young SGs, bona fide old SGs display no highly dynamic motion (Fig. 7 C and D), suggesting that they have lost their capability for long-range MT-mediated excursions. Disruption of FA allowed the transition of ∼15% of nearly immobile bona fide old SGs into the restricted pool and increased their collective mean speed (Fig. 7 J and L). Hence, old SGs may couple with MTs, but only for short-range excursions. These observations are robust, being supported by the concordance of statistical analyses on two independent large datasets, including movies of single labeled Ins-SNAPTMR-Star+ SGs or Ins-SNAPOG+, Lifeact-mCherry− SGs. Thus, we propose that SGs lose their capability to couple with MT-associated motor proteins over time, thereby decreasing their competence for MT-mediated transport (61). Notably, the motion of SGs could also reflect in part their binding to MTs undergoing remodeling (62), because MT stabilization by taxol reduced the D of highly dynamic young and old and restricted old SGs (Fig. 7 A and C). A higher density of microtubules upon taxol treatment could also increase the probability of SGs to switch direction, thereby decreasing their D.
Glucose stimulation, which induces FA remodeling (52, 53), recruited SGs to the highly dynamic component (Fig. 2C). Likewise, upon pharmacological disruption of FA, which also promotes insulin secretion (46–49), ∼50% of highly dynamic young SGs originated from SGs residing in the nearly immobile pool (Figs. 3B and 7H). These transitions correlated with an increased collective mean speed of young SGs (Figs. 2D, 3E, and 7K), whereas their D decreased to ∼50% (Figs. 2B, 3A, and 7I). Such apparently opposite findings (i.e., increased speed but decreased D) can be explained assuming that FA restricts not only the motion but also the likelihood of SGs to change direction by switching between MT tracks. In support of this interpretation is the observation that stabilization of FA with jasplakinolide increased the D of highly dynamic young SGs but decreased their frequency. Increased freedom of cortical SGs to explore the surrounding space upon actin depolymerization could in turn enhance their probability to land at sites competent for exocytosis and thereby undergo exocytosis. In view of these considerations it is plausible that greater mobility and competence for exocytosis of young SGs are tightly linked.
SGs that do not undergo exocytosis are eventually degraded intracellularly within MGBs (37, 56, 63). Our TIRFM data revealed the preferential association of old Ins-SNAPOG+ with FA+ vesicles that seemed morphologically distinct from bona fide SGs. In particular, Ins-SNAPOG+, FA+ objects were complex because they included approximately three Ins-SNAPOG+ cores, which moved synchronously along the same trajectory. Analyses by correlative light electron microscopy (CLEM) confirmed the greater occurrence of old Ins-SNAPTMR-Star within MGBs (Fig. 5E). Moreover, in INS-1 cells aged Ins-SNAP SGs were more frequently associated with endosomal–lysosomal markers (Fig. 6A), and a similar trend was observed in mouse primary beta cells (Fig. 6 B and C). Evidence that the combined amount of Ins-SNAPTMR-Star in cells and their media after 25 h in culture only accounted for ∼50% of the originally labeled protein further implies degradation of aged SGs.
Old Ins-SNAPOG+ SGs colocalized with the endosomal–lysosomal marker LAMP2 were also positive for FA (Fig. 6 D–F). The role of FA in disposal of aged SGs remains to be defined. As SGs age, their protein and/or lipid composition, such as phosphoinositides, may be altered, leading to the recruitment of actin nucleation promoting factors. Notably, glucose stimulation has been shown to activate N-WASP (53) and in our case tended to increase the occurrence of old FA+ SGs (Fig. 4C). To maintain homeostasis of insulin stores glucose stimulation may, on the one hand, induce insulin SG biogenesis and secretion of young SGs and, on the other hand, mark old SGs for degradation. At this time, however, we cannot yet discern whether FA coats individual SGs before their engulfment into MGBs/lysosomes or instead is peripherally associated with the membrane of the latter. In conclusion, selective marking of aged SGs with FA will provide a means to further dissect the molecular steps leading to their intracellular degradation.
In conclusion, our data show the tight correlation between aging of SGs and their mobility as well as the different relationship of young and old SGs with FA and localization of aged SGs to MGBs. They should enable a more accurate time–space-based modeling of insulin SG dynamics and exocytosis. According to these findings, it is also possible to envision how loss of glucose responsiveness could deplete beta cells of young, highly dynamic SGs competent for insulin release as well as impair the disposal of aged SGs and thus lead to deficient insulin secretion in type 2 diabetes.
Materials and Methods
Reagents.
TMR-Star, OG, and blocker were from NEB. Latrunculin A (used at 1 µM concentration for 30 min), jasplakinolide (used at 1µM concentration for 1 h), and OG–phalloidin were from Molecular Probes. Nocodazole (used at concentration 16.6 µM for 1 h) and taxol (used at 0.1 µM concentration for 1 h) were from Sigma-Aldrich.
DNA Constructs.
pEG-hIns-SNAP was described earlier (31). pLifeact-mCherry (54) and pEGFP-β-actin were gifts from E. Paluch, University College, London; pEGFP-RILP was from M. Zerial, MPI-CBG, Dresden; and pEGFP-α-tubulin was from G. Montagnac, Institute Curie, Paris.
Cell Culture.
The INS-1 cell line originally derived from a rat insulinoma (64, 65) was a gift from C. Wollheim, University of Geneva, Geneva. INS-1 cells used throughout this study were between passages 22 and 35. The cells were grown in standard INS-1 media [RPMI medium 1640 with l-glutamine (PAA Laboratories) supplemented with 20 mM Hepes, pH 7.4, 10% (vol/vol) heat-inactivated FBS (PAA Laboratories), 2 mM l-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 1 mM sodium pyruvate and 50 µM 2-mercaptoethanol, and 11 mM glucose] and incubated at 37 °C in a humidified 95% air, 5% CO2 atmosphere. The cells were trypsinized by incubation with 0.5 mg/mL trypsin-EDTA (PAA Laboratories) in Dulbecco’s PBS (PAA Laboratories) for 3 min at 37 °C. Trypsin was inactivated by addition of 10 mL of standard INS-1 medium. Cell culture was split 1:3 every 3–4 d and medium was changed 2 d after splitting.
Labeling of Age-Distinct Ins-SNAP+ SGs.
INS-1 cells transiently transfected with pEG-hIns-SNAP and grown in standard culture conditions for 4 d were incubated for 30 min with high-potassium buffer (15 mM Hepes, pH 7.4, 55 mM KCl, 70 mM NaCl, 24 mM NaHCO3, 1 mM MgCl.6H2O, 2 mM CaCl2.H2O, and 1 mg/mL ovalbumin fraction 5) to stimulate the release of predocked insulin SGs. Cells were then incubated for 30 min with high-potassium buffer containing 2 µM SNAP blocker (blocking buffer). After removal of the blocking buffer the cells were washed twice with 2 mL of PBS and incubated for 2 h in phenol red-free INS-1 cell medium. During this incubation the cells were washed twice, 30 and 60 min after the removal of the blocking buffer. Two hours after the removal of the blocking buffer cells were incubated for 30 min in phenol red-free INS-1 cell medium containing either 0.8 µM TMR-Star or 2 µM OG SNAP substrates (labeling media). Cells were immediately washed twice with PBS and again 30 and 60 min upon the removal of the labeling media and then incubated in phenol red-free INS-1 cell medium until used for imaging of insulin SGs with desired age. In some instances TMR-Star–labeled Ins-SNAP cells were placed in media without glucose (resting media) for 1 h and then imaged for 15 min while remaining in resting media or incubated in stimultion buffer (25 mM glucose, 5 mM KCl, 120 mM NaCl, 24 mM NaHCO3, 1 mM MgCl2, 2 mM CaCl2, 1 mg/mL ovalbumin, and 15 mM Hepes, pH 7.4).
Islet Isolation and Labeling.
Islets from SOFIA mice were isolated and dual-color SNAP-labeled as described (31) to visualize age-distinct Ins-SNAP SGs. After fixation at the desired time points with 4% (wt/vol) paraformaldehyde (PFA) in PBS for 30 min at 37 °C, islets were cryosectioned according to the Tokuyasu technique (66) with an EM UC6 ultramicrotome equipped with an EM FC6 cryodevice (Leica Microsystems) and 300-nm frozen sections were collected. Sections were then placed on glass coverslips, blocked with 1% BSA, and immunofluorescently stained for 1 h with rabbit anti-mouse-LAMP2 antibody (Abcam) diluted 1:50 in 1% BSA in PBS and secondary Alexa633-conjugated goat anti-rabbit IgG diluted 1:200 for 30 min at room temperature.
Immunocytochemistry.
Primary mouse monoclonal anti-α-tubulin (Sigma-Aldrich) and polyclonal rabbit anti-rat-LAMP2 (Invitrogen) antibodies were used at 1:1,000 and 1:50 dilutions, respectively. Alexa488-conjugated goat anti-mouse, Alexa568-conjugated goat anti-mouse, and Alexa633-conjugated goat anti-rabbit IgG (Molecular Probes) were used at 1:200 dilutions. Cells grown on coverslips were fixed with 4% PFA (20 min, 37 °C), permeabilized, and blocked with 0.2% saponin in goat-serum dilution buffer (GSDB) [16.5% (vol/vol) goat serum bovine, 20 mM sodium phosphate, pH 7.4, and 450 mM NaCl] for 1 h before overnight incubation at 4 °C with the primary antibodies diluted in GSDB. Next, cells were incubated with the Alexa-conjugated IgG or OG–phalloidin in GSDB for 45 min at room temperature. Coverslips were then mounted on glass slides with ProLong Gold antifade (Invitrogen).
Confocal Microscopy.
Samples were imaged with an LSM700 (Zeiss) equipped with λ = 488, 561- and 633-nm lasers, Plan-Apochromat 63×/1.4 N.A. oil DIC objective, BP505-530, LP575, MBS 405/488/555/639, and two photomultiplier tubes. Image processing was performed with the open source image software Fiji (67).
TIRFM.
Living cells seeded on 25-mm coverslips (ne = 1.5255; Warner Instruments) and SNAP-labeled were placed into a Bachhoffer chamber (Pekon) with an open metal holder (Okolab) heated to 37 °C. Imaging was performed with a Nikon TIRF system including a Nikon Eclipse Ti inverted microscope, motorized stage, and a Nikon 100×/1.49 N.A. Apo TIRF objective and controlled with the NIS Elements software. TMR-Star and mCherry were excited with 1% and 0.5% power of the λ = 561 laser (Coherent), whereas OG and EGFP were excited with 2% and 1% power of the λ = 488 nm laser, respectively. The fluorescence signals filtered through BP647/57 (TMR-Star, mCherry) and BP512/18 (OG, EGFP) filters were recorded with Andor iXon DU-897 cameras (pixel size 0.16 µm) connected with a TuCam adapter (Andor Technology), operated at EM gain 10 MHz at 14-bit, 2.4× conversion gain, multiplier 300, and speed of ∼33 fps.
Cryosectioning and Correlative Light Electron Microscopy of INS-1 Cells.
SNAP-labeled cells were fixed at the desired time points with 4% (wt/vol) PFA in PBS for 20 min at 37 °C, washed with PBS, and stored at 4 °C. Fixed cells were then sectioned according to the Tokuyasu technique (67) to collect 150-nm cryosections on finder grids (Maxtaform) for counterstaining with 5 µg/mL DAPI in PBS and imaging. Fluorescence images were acquired with a Delta Vision OMX structured illumination microscope (GE Healthcare) equipped with an Olympus UPlanSApo 100×/1.4 N.A. oil objective, 405-nm and 561-nm diode lasers, and sCMOS cameras (Andor Technology) to identify DAPI-stained nuclei and Ins-SNAPTMR-Star+ structures. The microscope was operated with the API software and the high-resolution reconstruction was performed with the OMX software. After fluorescence imaging, the sections were contrasted with 0.4% uranyl acetate and 1.8% (wt/vol) methylcellulose. Transmission electron microscopy (TEM) images were acquired with a Tecnai 12 Biotwin transmission electron microscope (FEI Company) operated at 120 kV equipped with a bottom-mount 2 × 2K F214 CCD camera (TVIPS). Fluorescent and corresponding TEM images were then aligned with the AMIRA software (VSG) before processing with the GIMP and Fiji (66) software.
Image Analysis.
TIRFM movies were analyzed using the Motion Tracking software (http://motiontracking.mpi-cbg.de) as described previously (68).
Frequency of Ins-SNAPOG+, Lifeact-mCherry+ Objects Within the Ins-SNAPOG+ Population.
Lifeact-mCherry was used to stain FA (Fig. S3), whereas young and old Ins-SNAP+ SGs were labeled with OG as described above. The frequency of Ins-SNAPOG+, Lifeact-mCherry+ objects was measured with Motion Tracking. To quantify the frequency of Ins-SNAPOG+, Lifeact-mCherry+ objects the number of tracks was evaluated, because the abundance of cytosolic Lifeact-mCherry signal could result in the detection of false Ins-SNAPOG+, Lifeact-mCherry+ objects owing to random encounters. Dual-color TIRFM of Ins-SNAPOG or Lifeact-mCherry was performed also in single transfected cells and the thresholds for the detection of double-positive fluorescent objects were chosen such that no Ins-SNAPOG+ objects were detected in Lifeact-mCherry single transfected cells, and vice versa.
Analysis of the Dynamic Components.
Diffusion coefficients and contribution of different motion modes to collective dynamics were calculated using the Motion Tracking analytical tool based on Bayesian probability theory (38). Briefly, given a set of M experimental curves each curve is a linear combination of K components with different and unknown contributions of individual components:
| [1] |
where is a -th component, is parameter of -th component, is a contribution of -th component into the -th curve, and is additional experimental noise. Independently fitting every single curve by maximizing the likelihood function allowed us to define two sets of parameters . In the case of normal noise the logarithm of the likelihood function is
| [2] |
in which are points where the experimental curve was measured and is the uncertainty of the -th measurement of the experimental curve. Individual fitting gives multiple sets of component parameters that were averaged using
| [3] |
to obtain one set of component parameters.
Fluorimetry.
Cells expressing Ins-SNAP were labeled by TMR-Star as described above. They were then lysed with lysis buffer [20 mM Tris⋅Cl (pH 8.0), 140 mM NaCl, 1 mM EDTA, 1 mM Triton X-100, and 1% protease inhibitors mixture P8340 (Sigma)] at desired time points to measure the signal of 3- to 5- and 28- to 30-h-old Ins-SNAPTMR-Star within cells and in the culture media. For background subtraction, cells not expressing Ins-SNAP were treated in the same way. Fluorimetry of the samples was performed with an EnVision plate reader (PerkinElmer). Fluorescence of intracellular 3- to 5-h-old Ins-SNAPTMR-Star was set to 100%.
Statistical Analysis.
Data are given as means and error bars are SEM. χ2-test and Student’s t test was calculated with Motion Tracking or Excel (Microsoft), respectively. Statistical significance is indicated either numerically or as *P < 0.05, **P < 0.01, and ***P < 0.005.
Supplementary Material
Acknowledgments
We thank C. Münster for isolation of mouse islets; M. Chernykh for assistance with Motion Tracking; S. Kretschmar, T. Kurth (Center for Regenerative Therapies Dresden), J. Meissner, and J.-M. Verbavatz (Max Planck Institute of Molecular Cell Biology and Genetics) for help with cryosectioning; the Zentrum für Informationsdienste und Hochleistungsrechnen at Technische Universität Dresden for providing resources on their Atlas PC cluster; S. Diez, E. Paluch, and members of the M.S. laboratory for fruitful discussions and suggestions; and K. Pfriem and D. Krüger for administrative assistance. This work was supported with funds from the Innovative Medicines Initiative Joint Undertaking under Grant Agreement 155005 (Improving beta-cell function and identification of diagnostic biomarkers for treatment monitoring in diabetes), resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations companies’ in-kind contribution. Additional funds were provided by the German Ministry for Education and Research to the German Center for Diabetes Research and the German Clinical Competence Network for Diabetes Mellitus (FKZ:01GI1102).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409542112/-/DCSupplemental.
References
- 1.De Camilli P, Jahn R. Pathways to regulated exocytosis in neurons. Annu Rev Physiol. 1990;52:625–645. doi: 10.1146/annurev.ph.52.030190.003205. [DOI] [PubMed] [Google Scholar]
- 2.Arvan P, Castle D. Sorting and storage during secretory granule biogenesis: looking backward and looking forward. Biochem J. 1998;332(Pt 3):593–610. doi: 10.1042/bj3320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes CJ, White MF. Molecular insights into insulin action and secretion. Eur J Clin Invest. 2002;32(Suppl 3):3–13. doi: 10.1046/j.1365-2362.32.s3.2.x. [DOI] [PubMed] [Google Scholar]
- 4.Cerasi E, Luft R. The plasma insulin response to glucose infusion in healthy subjects and in diabetes mellitus. Acta Endocrinol (Copenh) 1967;55(2):278–304. doi: 10.1530/acta.0.0550278. [DOI] [PubMed] [Google Scholar]
- 5.Curry DL, Bennett LL, Grodsky GM. Dynamics of insulin secretion by the perfused rat pancreas. Endocrinology. 1968;83(3):572–584. doi: 10.1210/endo-83-3-572. [DOI] [PubMed] [Google Scholar]
- 6.Porte D, Jr, Pupo AA. Insulin responses to glucose: Evidence for a two pool system in man. J Clin Invest. 1969;48(12):2309–2319. doi: 10.1172/JCI106197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rorsman P, et al. The cell physiology of biphasic insulin secretion. News Physiol Sci. 2000;15(2):72–77. doi: 10.1152/physiologyonline.2000.15.2.72. [DOI] [PubMed] [Google Scholar]
- 8.Henquin JC, Ishiyama N, Nenquin M, Ravier MA, Jonas JC. Signals and pools underlying biphasic insulin secretion. Diabetes. 2002;51(Suppl 1):S60–S67. doi: 10.2337/diabetes.51.2007.s60. [DOI] [PubMed] [Google Scholar]
- 9.Kahn SE, et al. Importance of early phase insulin secretion to intravenous glucose tolerance in subjects with type 2 diabetes mellitus. J Clin Endocrinol Metab. 2001;86(12):5824–5829. doi: 10.1210/jcem.86.12.8105. [DOI] [PubMed] [Google Scholar]
- 10.Fava E, et al. Novel standards in the measurement of rat insulin granules combining electron microscopy, high-content image analysis and in silico modelling. Diabetologia. 2012;55(4):1013–1023. doi: 10.1007/s00125-011-2438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barg S, Eliasson L, Renström E, Rorsman P. A subset of 50 secretory granules in close contact with L-type Ca2+ channels accounts for first-phase insulin secretion in mouse beta-cells. Diabetes. 2002;51(Suppl 1):S74–S82. doi: 10.2337/diabetes.51.2007.s74. [DOI] [PubMed] [Google Scholar]
- 12.Schulla V, et al. Impaired insulin secretion and glucose tolerance in beta cell-selective Ca(v)1.2 Ca2+ channel null mice. EMBO J. 2003;22(15):3844–3854. doi: 10.1093/emboj/cdg389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohara-Imaizumi M, et al. TIRF imaging of docking and fusion of single insulin granule motion in primary rat pancreatic beta-cells: Different behaviour of granule motion between normal and Goto-Kakizaki diabetic rat beta-cells. Biochem J. 2004;381(Pt 1):13–18. doi: 10.1042/BJ20040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohara-Imaizumi M, et al. Imaging analysis reveals mechanistic differences between first- and second-phase insulin exocytosis. J Cell Biol. 2007;177(4):695–705. doi: 10.1083/jcb.200608132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibasaki T, et al. Essential role of Epac2/Rap1 signaling in regulation of insulin granule dynamics by cAMP. Proc Natl Acad Sci USA. 2007;104(49):19333–19338. doi: 10.1073/pnas.0707054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuda T, et al. Rim2alpha determines docking and priming states in insulin granule exocytosis. Cell Metab. 2010;12(2):117–129. doi: 10.1016/j.cmet.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 17.Xie L, Zhu D, Gaisano HY. Role of mammalian homologue of Caenorhabditis elegans unc-13-1 (Munc13-1) in the recruitment of newcomer insulin granules in both first and second phases of glucose-stimulated insulin secretion in mouse islets. Diabetologia. 2012;55(10):2693–2702. doi: 10.1007/s00125-012-2640-z. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, et al. The Rab27a effector exophilin7 promotes fusion of secretory granules that have not been docked to the plasma membrane. Mol Biol Cell. 2013;24(3):319–330. doi: 10.1091/mbc.E12-04-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu D, et al. Syntaxin-3 regulates newcomer insulin granule exocytosis and compound fusion in pancreatic beta cells. Diabetologia. 2013;56(2):359–369. doi: 10.1007/s00125-012-2757-0. [DOI] [PubMed] [Google Scholar]
- 20.Gomi H, Mizutani S, Kasai K, Itohara S, Izumi T. Granuphilin molecularly docks insulin granules to the fusion machinery. J Cell Biol. 2005;171(1):99–109. doi: 10.1083/jcb.200505179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasai K, Fujita T, Gomi H, Izumi T. Docking is not a prerequisite but a temporal constraint for fusion of secretory granules. Traffic. 2008;9(7):1191–1203. doi: 10.1111/j.1600-0854.2008.00744.x. [DOI] [PubMed] [Google Scholar]
- 22.Degtyar VE, Allersma MW, Axelrod D, Holz RW. Increased motion and travel, rather than stable docking, characterize the last moments before secretory granule fusion. Proc Natl Acad Sci USA. 2007;104(40):15929–15934. doi: 10.1073/pnas.0705406104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seino S, Shibasaki T, Minami K. Dynamics of insulin secretion and the clinical implications for obesity and diabetes. J Clin Invest. 2011;121(6):2118–2125. doi: 10.1172/JCI45680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schatz H, Nierle C, Pfeiffer EF. (Pro-) insulin biosynthesis and release of newly synthesized (pro-) insulin from isolated islets of rat pancreas in the presence of amino acids and sulphonylureas. Eur J Clin Invest. 1975;5(6):477–485. doi: 10.1111/j.1365-2362.1975.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 25.Gold G, Gishizky ML, Grodsky GM. Evidence that glucose “marks” beta cells resulting in preferential release of newly synthesized insulin. Science. 1982;218(4567):56–58. doi: 10.1126/science.6181562. [DOI] [PubMed] [Google Scholar]
- 26.Halban PA. Differential rates of release of newly synthesized and of stored insulin from pancreatic islets. Endocrinology. 1982;110(4):1183–1188. doi: 10.1210/endo-110-4-1183. [DOI] [PubMed] [Google Scholar]
- 27.MacGregor RR, Hamilton JW, Cohn DV. The by-pass of tissue hormone stores during the secretion of newly synthesized parathyroid hormone. Endocrinology. 1975;97(1):178–188. doi: 10.1210/endo-97-1-178. [DOI] [PubMed] [Google Scholar]
- 28.Piercy M, Shin SH. Newly synthesized prolactin is preferentially secreted by the adenohypophysis in a primary cell culture system. Mol Cell Endocrinol. 1981;21(1):75–84. doi: 10.1016/0303-7207(81)90032-0. [DOI] [PubMed] [Google Scholar]
- 29.Duncan RR, et al. Functional and spatial segregation of secretory vesicle pools according to vesicle age. Nature. 2003;422(6928):176–180. doi: 10.1038/nature01389. [DOI] [PubMed] [Google Scholar]
- 30.Tsuboi T, Kitaguchi T, Karasawa S, Fukuda M, Miyawaki A. Age-dependent preferential dense-core vesicle exocytosis in neuroendocrine cells revealed by newly developed monomeric fluorescent timer protein. Mol Biol Cell. 2010;21(1):87–94. doi: 10.1091/mbc.E09-08-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanova A, et al. Age-dependent labeling and imaging of insulin secretory granules. Diabetes. 2013;62(11):3687–3696. doi: 10.2337/db12-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juillerat A, et al. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol. 2003;10(4):313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 33.Keppler A, et al. Labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase with small molecules in vivo and in vitro. Methods. 2004;32(4):437–444. doi: 10.1016/j.ymeth.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc Natl Acad Sci USA. 2004;101(27):9955–9959. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keppler A, Arrivoli C, Sironi L, Ellenberg J. Fluorophores for live cell imaging of AGT fusion proteins across the visible spectrum. Biotechniques. 2006;41(2):167–170, 172, 174–175. doi: 10.2144/000112216. [DOI] [PubMed] [Google Scholar]
- 36.Tirat A, Freuler F, Stettler T, Mayr LM, Leder L. Evaluation of two novel tag-based labelling technologies for site-specific modification of proteins. Int J Biol Macromol. 2006;39(1-3):66–76. doi: 10.1016/j.ijbiomac.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Halban PA, Wollheim CB. Intracellular degradation of insulin stores by rat pancreatic islets in vitro. An alternative pathway for homeostasis of pancreatic insulin content. J Biol Chem. 1980;255(13):6003–6006. [PubMed] [Google Scholar]
- 38.Sivia DS, Carlile CJ, Howells WS, König S. Bayesian analysis of quasielastic neutron scattering data. Physica B. 1992;182:341–348. [Google Scholar]
- 39.Hao M, et al. Regulation of two insulin granule populations within the reserve pool by distinct calcium sources. J Cell Sci. 2005;118(Pt 24):5873–5884. doi: 10.1242/jcs.02684. [DOI] [PubMed] [Google Scholar]
- 40.Lacy PE, Walker MM, Fink CJ. Perifusion of isolated rat islets in vitro. Participation of the microtubular system in the biphasic release of insulin. Diabetes. 1972;21(10):987–998. doi: 10.2337/diab.21.10.987. [DOI] [PubMed] [Google Scholar]
- 41.Pipeleers DG, Pipeleers-Marichal MA, Kipnis DM. Microtubule assembly and the intracellular transport of secretory granules in pancreatic islets. Science. 1976;191(4222):88–90. doi: 10.1126/science.1108194. [DOI] [PubMed] [Google Scholar]
- 42.Suprenant KA, Dentler WL. Association between endocrine pancreatic secretory granules and in-vitro-assembled microtubules is dependent upon microtubule-associated proteins. J Cell Biol. 1982;93(1):164–174. doi: 10.1083/jcb.93.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dentler WL, Suprenant KA. Isolation of microtubule-secretory granule complexes from the anglerfish endocrine pancreas. Ann N Y Acad Sci. 1986;466:813–831. doi: 10.1111/j.1749-6632.1986.tb38465.x. [DOI] [PubMed] [Google Scholar]
- 44.Varadi A, Ainscow EK, Allan VJ, Rutter GA. Involvement of conventional kinesin in glucose-stimulated secretory granule movements and exocytosis in clonal pancreatic beta-cells. J Cell Sci. 2002;115(Pt 21):4177–4189. doi: 10.1242/jcs.00083. [DOI] [PubMed] [Google Scholar]
- 45.Varadi A, Tsuboi T, Johnson-Cadwell LI, Allan VJ, Rutter GA. Kinesin I and cytoplasmic dynein orchestrate glucose-stimulated insulin-containing vesicle movements in clonal MIN6 beta-cells. Biochem Biophys Res Commun. 2003;311(2):272–282. doi: 10.1016/j.bbrc.2003.09.208. [DOI] [PubMed] [Google Scholar]
- 46.Malaisse WJ, Hager DL, Orci L. The stimulus-secretion coupling of glucose-induced insulin release. IX. The participation of the beta cell web. Diabetes. 1972;21(2, Suppl):594–604. doi: 10.2337/diab.21.2.s594. [DOI] [PubMed] [Google Scholar]
- 47.Orci L, Gabbay KH, Malaisse WJ. Pancreatic beta-cell web: Its possible role in insulin secretion. Science. 1972;175(4026):1128–1130. doi: 10.1126/science.175.4026.1128. [DOI] [PubMed] [Google Scholar]
- 48.van Obberghen E, et al. Dynamics of insulin release and microtubular-microfilamentous system. I. Effect of cytochalasin B. J Clin Invest. 1973;52(5):1041–1051. doi: 10.1172/JCI107269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Obberghen E, et al. Dynamics of insulin release and microtubular-microfilamentous system. VII. Do microfilaments provide the motive force for the translocation and extrusion of beta granules? Diabetes. 1975;24(10):892–901. doi: 10.2337/diab.24.10.892. [DOI] [PubMed] [Google Scholar]
- 50.Li G, et al. Effect of disruption of actin filaments by Clostridium botulinum C2 toxin on insulin secretion in HIT-T15 cells and pancreatic islets. Mol Biol Cell. 1994;5(11):1199–1213. doi: 10.1091/mbc.5.11.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varadi A, Tsuboi T, Rutter GA. Myosin Va transports dense core secretory vesicles in pancreatic MIN6 beta-cells. Mol Biol Cell. 2005;16(6):2670–2680. doi: 10.1091/mbc.E04-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomas A, Yermen B, Min L, Pessin JE, Halban PA. Regulation of pancreatic beta-cell insulin secretion by actin cytoskeleton remodelling: Role of gelsolin and cooperation with the MAPK signalling pathway. J Cell Sci. 2006;119(Pt 10):2156–2167. doi: 10.1242/jcs.02942. [DOI] [PubMed] [Google Scholar]
- 53.Uenishi E, et al. Actin dynamics regulated by the balance of neuronal Wiskott-Aldrich syndrome protein (N-WASP) and cofilin activities determines the biphasic response of glucose-induced insulin secretion. J Biol Chem. 2013;288(36):25851–25864. doi: 10.1074/jbc.M113.464420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riedl J, et al. Lifeact: A versatile marker to visualize F-actin. Nat Methods. 2008;5(7):605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halban PA. Structural domains and molecular lifestyles of insulin and its precursors in the pancreatic beta cell. Diabetologia. 1991;34(11):767–778. doi: 10.1007/BF00408349. [DOI] [PubMed] [Google Scholar]
- 56.Orci L, et al. Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. J Cell Biol. 1984;98(1):222–228. doi: 10.1083/jcb.98.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harashima S, Clark A, Christie MR, Notkins AL. The dense core transmembrane vesicle protein IA-2 is a regulator of vesicle number and insulin secretion. Proc Natl Acad Sci USA. 2005;102(24):8704–8709. doi: 10.1073/pnas.0408887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivarsson R, Obermüller S, Rutter GA, Galvanovskis J, Renström E. Temperature-sensitive random insulin granule diffusion is a prerequisite for recruiting granules for release. Traffic. 2004;5(10):750–762. doi: 10.1111/j.1600-0854.2004.00216.x. [DOI] [PubMed] [Google Scholar]
- 59.Veliz LA, et al. Near-membrane dynamics and capture of TRPM8 channels within transient confinement domains. PLoS ONE. 2010;5(10):e13290. doi: 10.1371/journal.pone.0013290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu D, Meckel T, Long EO. Distinct role of rab27a in granule movement at the plasma membrane and in the cytosol of NK cells. PLoS ONE. 2010;5(9):e12870. doi: 10.1371/journal.pone.0012870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gross SP, Vershinin M, Shubeita GT. Cargo transport: Two motors are sometimes better than one. Curr Biol. 2007;17(12):R478–R486. doi: 10.1016/j.cub.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 62.Blocker A, Griffiths G, Olivo J-C, Hyman AA, Severin FF. A role for microtubule dynamics in phagosome movement. J Cell Sci. 1998;111(Pt 3):303–312. doi: 10.1242/jcs.111.3.303. [DOI] [PubMed] [Google Scholar]
- 63.Halban PA, Mutkoski R, Dodson G, Orci L. Resistance of the insulin crystal to lysosomal proteases: Implications for pancreatic B-cell crinophagy. Diabetologia. 1987;30(5):348–353. doi: 10.1007/BF00299029. [DOI] [PubMed] [Google Scholar]
- 64.Chick WL, et al. A transplantable insulinoma in the rat. Proc Natl Acad Sci USA. 1977;74(2):628–632. doi: 10.1073/pnas.74.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asfari M, et al. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology. 1992;130(1):167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 66.Tokuyasu KT. Immunochemistry on ultrathin frozen sections. Histochem J. 1980;12(4):381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- 67.Schindelin J, et al. Fiji: An open source platform for biological image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122(5):735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.