Abstract
LocSigDB (http://genome.unmc.edu/LocSigDB/) is a manually curated database of experimental protein localization signals for eight distinct subcellular locations; primarily in a eukaryotic cell with brief coverage of bacterial proteins. Proteins must be localized at their appropriate subcellular compartment to perform their desired function. Mislocalization of proteins to unintended locations is a causative factor for many human diseases; therefore, collection of known sorting signals will help support many important areas of biomedical research. By performing an extensive literature study, we compiled a collection of 533 experimentally determined localization signals, along with the proteins that harbor such signals. Each signal in the LocSigDB is annotated with its localization, source, PubMed references and is linked to the proteins in UniProt database along with the organism information that contain the same amino acid pattern as the given signal. From LocSigDB webserver, users can download the whole database or browse/search for data using an intuitive query interface. To date, LocSigDB is the most comprehensive compendium of protein localization signals for eight distinct subcellular locations.
Database URL: http://genome.unmc.edu/LocSigDB/
Introduction
Proteins are synthesized in the cytoplasm and on ribosomes bound to the endoplasmic reticulum (ER), with a few proteins synthesized in mitochondria or chloroplasts (as in plant cells). However, the nucleus-encoded proteins are targeted to different subcellular locations to carry out defined functions. Aberrant mislocalization of proteins to unintended locations can interfere with numerous cellular processes, often leading to many diseases (1). Proteins are directed to their appropriate destinations by a process called protein targeting or protein sorting, which is primarily dependent upon the information contained in the targeted protein itself; known as ‘intrinsic signals’ or ‘address tags’ (2, 3). These signals are stretches of amino acid residues within a protein; they are present either at the N-terminus; as is the case in Golgi, the secretory pathway or mitochondrial proteins (4–6), or at the C-terminus; as in the case of peroxisomes and ER (7, 8). Signals at the N-terminus are usually referred to as signal peptides and they help direct proteins to cellular as well as extracellular locations. Generic structure of a signal peptide consists of a positively charged N-terminus followed by a long stretch of hydrophobic region at the core. The C-terminus amino acid composition varies depending upon the organelle the protein enters or is inserted into (in case of cellular membranes) or if the protein is secreted out of the cell. However, there are exceptions to these general rules: Golgi retention signals can be found at the C-terminus (9) and mitochondrial signal peptides can also be found at the internal positions (10) or at the C-terminus (11) of some proteins. Conversely, the nucleus has its own class of targeting signals (12, 13), which can be located anywhere on the peptide chain and usually contain basic, positively charged amino acids. The precise functioning of cells and tissues relies on the fidelity of protein targeting. Consequently, diseases like cancer and psoriasis and inflammatory conditions such as sepsis, rheumatoid arthritis and tissue rejection, can result from the malfunction of signalling pathways (14, 15). Furthermore, errors in these address tags can result in heritable diseases (16). As a result, the biomedical community will greatly benefit form a catalogue of experimentally identified protein subcellular localization signals as a way to manage and manipulate diseases by exploitation of these sorting signals (17–19).
Many computational methods have been developed for the prediction of protein subcellular localization (20); however, there was little or no emphasis on predicting the sorting signals. Some methods that predict the sorting signals limit their predictions to N-terminal signal peptides and their cleavage sites (21, 22). Likewise, few methods predict the internal nuclear localization signals (23, 24) and also C-terminal peroxisomal targeting signals (25), but again these methods are limited to single organelles. Also potential sorting signals are identified by LOCATE database based on PROSITE patterns and signal peptide prediction methods; however; it does not document any experimental confirmation of these signals (26). Similarly, databases that are dedicated to protein subcellular localization (26, 27) only house information on catalogs of proteins in different organelles and provide no information on the sorting signals. Among the existing localization signal databases (28, 29), NLSdb (28) contains manually curated experimentally validated nuclear localization signals but, it accounts for only one-third of the currently available experimental nuclear localization signals that are catalogued in our LocSigDB (refer to Database Statistics Section). As for SPdb (29), it is a database of experimentally and computationally predicted localization signals, but limited to only signal peptides. Moreover, both the databases have not been updated in the recent years. Also ELM (30), a database of eukaryotic linear motifs is an excellent resource on functional sites in proteins including targeting motifs. However, the number of ELMs classified as targeting motifs is relatively few. Another rich source of sorting signals information is UniProt (31), which offers information on experimental as well as predicted (potential, probable or similarity based) sorting signals; however, the criterion for a signal to be identified as being experimental was not as rigorous as reported by LocSigDB, where evidence for every entry is backed by at least one PubMed article. But, beginning of September 2014, UniProt has also begun to adopt the evidence ontology combined with source information and for the experimental evidence codes; this source information is most often in the form of a PubMed ID. This has resulted in fewer sorting signals than before being classified as ‘experimental’. Other than the protein localization signal databases, a few interesting review articles have also been published (14, 32, 33) which catalog the experimentally validated localization signals, although the proportion of signals reported is much smaller than LocSigDB and typically the reviews are focussed either on a single organelle or on a protein of interest. Therefore, there is a need for developing an up-to-date and comprehensive database of experimentally known protein localization signals for all the major subcellular locations of a cell.
Over the past decade, we have developed a variety of tools for predicting protein subcellular localization (34–38). This has motivated us to develop the LocSigDB web server, which we believe will fill in the current gap. LocSigDB is a unique, extensive and substantially large database when compared to any existing localization signal databases. LocSigDB comprises sorting signal information for 533 distinct experimentally validated signals, along with the proteins that harbor them for eight distinct subcellular locations. We believe that LocSigDB will act as a value-adding resource for the biomedical research community to learn about the localization signals that have already been identified, as well as help form a rationale to deduce new potential signals.
Aims of the database
LocSigDB is a database dedicated to protein subcellular localization signals and the proteins that harbor them, along with the research articles that have experimentally confirmed these signals. The main goal of this database is to collect and organize the information on localization signals and present it in a user-friendly and searchable format to facilitate easy retrieval of information. Our second goal is to ensure that the information in this database is up-to-date. In addition to monitoring the new literature, we also provide a signal submission form on our website for users to submit experimentally characterized sorting signals. We will review this information for authenticity before adding it to our database. We anticipate LocSigDB will serve as a comprehensive resource of protein targeting signals to the scientific community.
Database content
Collection of localization signals from literature
Over 1000 published articles related to protein-targeting signals were collected and reviewed to extract experimentally determined subcellular localization signals across eight major cellular compartments. Various keywords and their combinations were used in PubMed searches to retrieve appropriate literature; some examples are ‘Nuclear localization signal’, ‘NLS’, ‘Nuclear localization sequence’, ‘Mitochondrial targeting signal’, ‘Lysosome sorting signal’, and so forth. The returned literature was reviewed with a focus on the following criteria for selecting a signal as a valid sorting signal: (i) if a particular signal is able to target a non-resident protein to its own specific organelle (e.g. a signal is a valid nuclear localization signal if it can target a non-nuclear protein to nucleus), or (ii) by deleting or mutating some amino acids from the signal prevents import of the protein into its native subcellular location. Based on these criteria, only 518 articles were selected out of the 1000 articles that contained relevant information (some articles had information on more than one protein). Note that some researchers report a part of the protein sequence as a signal while others report rather a specific peptide sequence or a sequence pattern with a set of conserved residues or non-specific residues allowed in the patterns. Such signal patterns were translated into regular expressions in our database to facilitate pattern-based querying.
Database fields
Each entry in the LocSigDB annotates a localization signal with five descriptors: (i) Protein(s): The protein(s) in which the experimental localization signal was reported in the literature. (ii) Localization: The exclusive subcellular location where the protein containing the targeting signal is found. (iii) Reference(s): Published primary literature on the experimental localization signals. This field has URLs that cross-link to PubMed citations. (iv) Coordinate(s): The start and end positions of the signals found is a given sequence. This parameter is displayed only during search by ‘Protein ID' or ‘Protein Sequence'. (v) UniProt Accession(s): UniProt accessions of all proteins that contain the same amino acid pattern as that of a given signal. Preferably SwissProt accessions for the protein are presented but in few cases the signals could not be mapped onto SwissProt annotated proteins; there the TrEMBL accessions have been reported. This helps achieve complete integration with the UniProt. (vi) Organisms: The organism(s) where each signal is found and this information is retrieved from the UniProt as it corresponds to the UniProt accessions and the respective organism representing the same. But the organism field is unique such that each organism is reported only once although more than one protein sequences (accessions) from the same organism may contain the signal of interest.
Database statistics
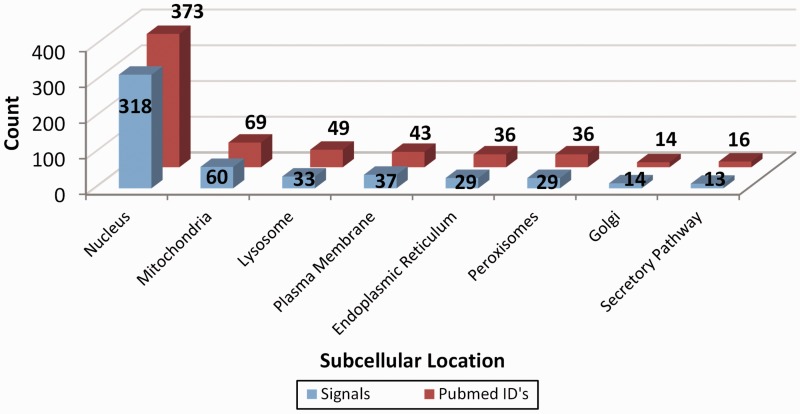
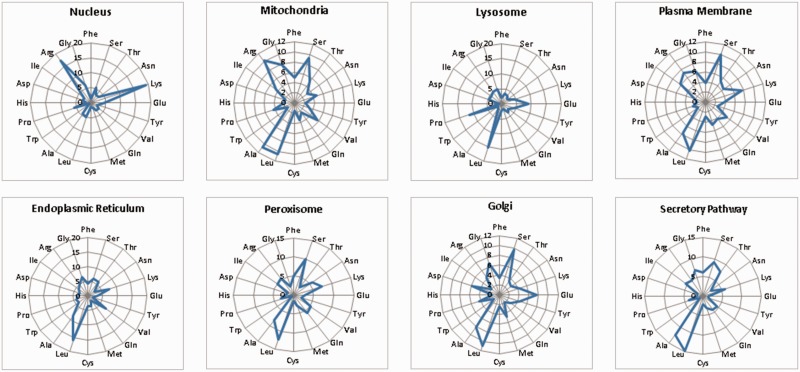
LocSigDB contains 533 experimentally validated localization signals. Figure 1 illustrates LocSigDB statistics with the number of localization signals and published articles for each distinct subcellular location. It can be observed from Figure 1, that the maximum number of studies have been done for inferring the localization signals for nucleus targeted proteins followed by mitochondria with a wide margin. Nuclear localization signals for 318 proteins accounted for a higher total number of signals than that of the remaining seven organelles combined. In comparison, the number of nuclear localization signals reported by NLSdb (28) accounts for only about one-third of those that are catalogued in LocSigDB. Organelle based radar plots were generated to look at the frequency distribution of each amino acid in the localization signals for each distinct subcellular location as shown in Figure 2. Here, the frequency of occurrence of an amino acid has been normalized by the number of experimentally verified localization signals as well as the average length of signals for each organelle to correlate the significance of certain amino acids in subcellular localization. In case of nuclear signals, it is evident that basic positively (39) charged amino acids, lysine (Lys) and arginine (Arg), significantly contribute in the nuclear localization of a protein. Similarly, mitochondrial signals have alternating hydrophobic and positively charged amino acids (40), which is evident from Figure 2; Mitochondrial signals are dominated by the frequent occurrence of hydrophobic amino acids leucine (Leu) and alanine (Ala), as well as positively charged amino acids like arginine (Arg) and small amino acids like glycine (Gly) and serine (Ser). Conversely, negatively charged amino acids like aspartic acid (Asp) and glutamic acid (Glu) are present in modest frequencies only in the localization signals of Golgi and ER. This observation is also consistent with the most common sequence patterns like KDEL and its related amino acid patterns (41). The most abundant amino acids: serine (Ser), leucine (Leu), alanine (Ala) and glycine (Gly), (42, 43) are prevalent in most of the localization signals except nuclear signals; while the least abundant amino acids: tryptophan (Trp), histidine (His) and cysteine (Cys) are present in lowest frequencies in the localization signals of all the eight subcellular locations.
Figure 1.
Representation of the database statistics showing the number of localization signals for distinct subcellular location along with the count of the research articles elucidating these signals. As clearly seen, most studies have been done on inferring the protein localization signals for nucleus followed by mitochondria and all the other six organelles with a wide margin.
Figure 2.
An overview of the frequency distribution of amino acids in the signal set for each of the eight subcellular organelles. As seen from the radar plots, there are clear differences in the frequency occurrence of amino acids for each distinct organelle. Nucleus is dominated by positively charged residues like lysines and arginines; whereas, mitochondrial signals have frequent occurrence of hydrophobic amino acids like: glycine, leucine and alanine as well as positively charged amino acids like arginine. Also, negatively charged amino acids like aspartic acid and glutamic acid are present only in the signals of organelles like Golgi and ER.
Database access and user interface
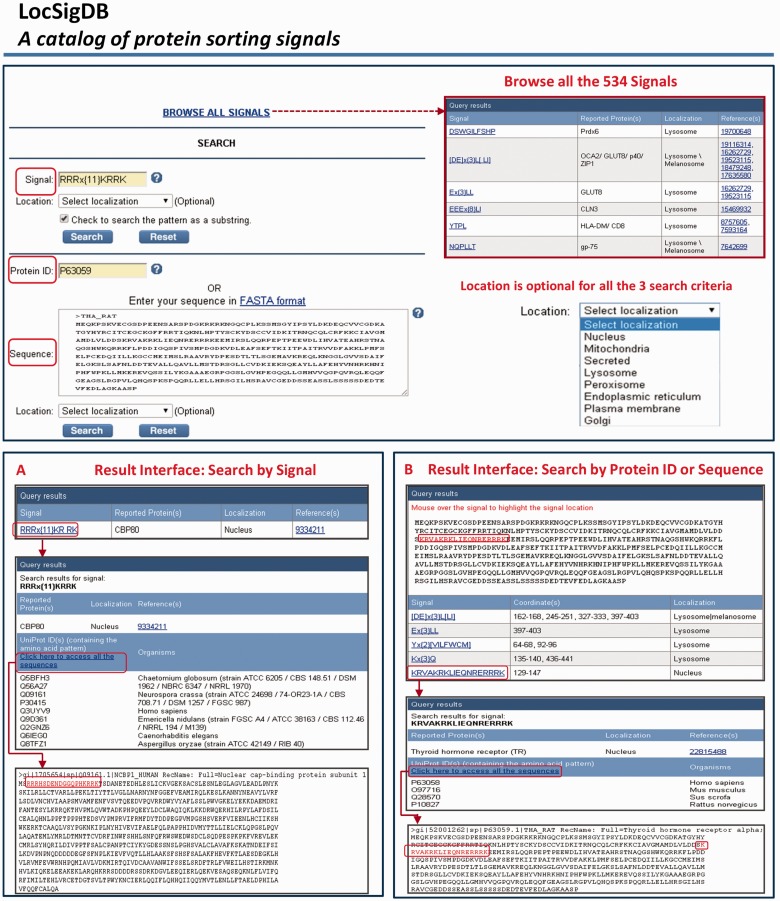
LocSigDB is freely accessible on the web at http://genome.unmc.edu/LocSigDB/. At this time, the database contains 533 experimentally identified protein localization signals, the proteins that harbour such signals and corresponding literature associated with each signal. LocSigDB provides three search functions to retrieve information pertaining to localization signals. Users can search the database by submitting (i) a defined localization signal or a motif pattern (using wild card characters), (ii) a protein identifier (NCBI’s RefSeq ID or UniProt’s protein accession or ID) or (iii) a protein sequence itself (in FASTA format). The web interface system is built using LAMP (Linux, Apache HTTP Server, MySQL and Perl) architecture. The data is stored in a MySQL database and is made available using a Perl/CGI backed user interface. When a user queries for a signal, the server searches for the signal in the LocSigDB and displays the results. The results page also contains links to further probe the information on signal motifs and PubMed citations. If the user wants to search using an NCBI protein identifier, the server automatically retrieves the sequence from the NCBI database using NCBI E-utilities and displays the results for the retrieved sequence. Similarly, a search can be made using UniProt/SwissProt IDs. In the third search option, a user can provide the full or a partial protein sequence in FASTA format. Certain sequence patterns (such as short repeats or common motifs) occur more frequently in protein sequences than others; thus, have a higher chance of randomly matching as substrings to a query pattern and generating false positives. Hence, we have made the substring search as ‘optional’ below the search window and the substring matches are displayed only if chosen by the user. Additionally, avoiding the use of wild character (*) or lax regex patterns in the queries will reduce the false positive matches. An overview of the query and result interface is shown in Figure 3, which demonstrates a step-by-step navigation through the database using relevant examples (given in the FAQs file) for the three query functions. The entire database can be downloaded as a tab-delimited, comma-delimited or as an excel file using the ‘Download’ link provided on the left panel. Users can browse the database contents and, highlight the special features, like signal highlights in the results page.
Figure 3.
An overview of the query and the search results interface in LocSigDB. LocSigDB provides three search functions to retrieve the localization signal information. (A) Query using a signal displays links to all the available descriptors for the signal in question; (B) Query by a protein ID or protein sequence (FASTA format) will retrieve corresponding sequence from the public database. The sequence is displayed in the results interface and the user can mouse over the corresponding signals identified in the protein, while highlighting the signal coordinates in red on the protein sequence. Each signal is in turn linked to the signal attribute window that displays the annotations for that signal.
Data submission
To encourage users to submit experimentally validated new localization signals/motifs, we have provided a ‘Submit signal’ link on the left panel. This link leads to a preformatted form for submission of signals on the LocSigDB webpage. We will review these signals and the literature for accuracy before including in our database. We intend to maintain and frequently update LocSigDB.
Conclusion and future direction
We present LocSigDB, a comprehensive database and web interface to search and explore localization signals for eight distinct subcellular locations in eukaryotic and bacterial cells. LocSigDB has been designed to help better understand the signal-dependent transport of various proteins; and as a result it is intended to help the development of therapies based on the interception of cellular signals in diseased cells (15, 44). A number of research projects in our laboratory are tied to protein subcellular localization; therefore, we are motivated to keep this database up-to-date. The website will be updated twice a year, as new data become available and is thus intended to be a long-term resource for the research community in this area. Often, mutations in the localization signal can alter the subcellular location of a protein; consequently, we also plan to add disease causing mutated signals to the database in our next update. While the crux of LocSigDB is the experimentally verified signals, it can also be used as an important resource to develop new methodologies for predicting localization signals/motifs, which in turn can help advance the field. Based on the provided information, our long-term goal is to develop and maintain LocSigDB as the most comprehensive resource for protein localization signals.
Acknowledgements
The authors thank the Bioinformatics and Systems Biology core facility at UNMC for providing infrastructure support for the database and webserver development and Megan Brown for proofreading the manuscript. The open-access publication costs for this article have been funded by 1R01GM086533-01A1 grant support to CG.
Funding
This research is fully supported by National Institutes of Health [1R01GM086533-01A1 to CG].
Conflict of interest. None declared.
References
- 1.Davis J.R., Kakar M., Lim C.S. (2007) Controlling protein compartmentalization to overcome disease. Pharm. Res., 24, 17–27. [DOI] [PubMed] [Google Scholar]
- 2.Blobel,G. and Dobberstein,B. (1975) Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol., 67, 835–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hegde R.S., Bernstein H.D. (2006) The surprising complexity of signal sequences. Trends Biochem. Sci. , 31, 563–571. [DOI] [PubMed] [Google Scholar]
- 4.Munro S. (1998) Localization of proteins to the Golgi apparatus. Trends Cell Biol. , 8, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifacino J.S., Traub L.M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem., 72, 395–447. [DOI] [PubMed] [Google Scholar]
- 6.von Heijne G., Steppuhn J., Herrmann R.G. (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem., 180, 535–545. [DOI] [PubMed] [Google Scholar]
- 7.Brocard C., Hartig A. (2006) Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim . Biophys. Acta., 1763, 1565–1573. [DOI] [PubMed] [Google Scholar]
- 8.Gomord V., Wee E., Faye L. (1999) Protein retention and localization in the endoplasmic reticulum and the golgi apparatus. Biochimie, 81, 607–618. [DOI] [PubMed] [Google Scholar]
- 9.Gao C., Yu C.K.Y., Qu S., et al. (2012) The Golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and Golgi retention signals at its C terminus. Plant Cell, 24, 2086–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fölsch H., Guiard B., Neupert W., et al. (1996) Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J., 15, 479–487. [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C.M. (1999) The DNA Helicase, Hmi1p, Is transported into mitochondria by a C-terminal cleavable targeting signal. J. Biol. Chem., 274, 20937–20942. [DOI] [PubMed] [Google Scholar]
- 12.Kosugi S., Hasebe M., Tomita M., et al. (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins. Proc. Natl. Acad. Sci. USA, 106, 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange A., McLane L.M., Mills R.E., et al. (2010) Expanding the definition of the classical bipartite nuclear localization signal. Traffic, 11, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLane L.M., Corbett A.H. (2009) Nuclear localization signals and human disease. IUBMB Life, 61, 697–706. [DOI] [PubMed] [Google Scholar]
- 15.Chahine M.N., Pierce G.N. (2009) Therapeutic targeting of nuclear protein import in pathological cell conditions. Pharmacol. Rev., 61, 358–372. [DOI] [PubMed] [Google Scholar]
- 16.Király O., Boulling A., Witt H., et al. (2007) Signal peptide variants that impair secretion of pancreatic secretory trypsin inhibitor (SPINK1) cause autosomal dominant hereditary pancreatitis. Hum. Mutat., 28, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro-Fernández C., Maya-Núñez G., Conn P.M. (2005) Beyond the signal sequence: protein routing in health and disease. Endocr. Rev., 26, 479–503. [DOI] [PubMed] [Google Scholar]
- 18.Datta R., Waheed A., Shah G.N., et al. (2007) Signal sequence mutation in autosomal dominant form of hypoparathyroidism induces apoptosis that is corrected by a chemical chaperone. Proc. Natl. Acad. Sci. USA, 104, 19989–19994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stern B., Olsen L. (2007) Improving mammalian cell factories: The selection of signal peptide has a major impact on recombinant protein synthesis and secretion in mammalian cells. Trends Cell Mol. Biol., 2, 1–17 [Google Scholar]
- 20.Imai K., Nakai K. (2010) Prediction of subcellular location of proteins: where to proceed? Proteomics, 22, 3970–3983. [DOI] [PubMed] [Google Scholar]
- 21.Petersen T.N., Brunak S., von Heijne G., et al. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- 22.Bannai H., Tamada Y., Maruyama O., et al. (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics, 18, 298–305. [DOI] [PubMed] [Google Scholar]
- 23.Cokol M., Nair R., Rost B. (2000) Finding nuclear localization signals. EMBO Rep., 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brameier M., Krings A., Maccallum R.M. (2007) NucPred—predicting nuclear localization of proteins. Bioinformatics, 19, 1159–1160. [DOI] [PubMed] [Google Scholar]
- 25.Neuberger G., Maurer-Stroh S., Eisenhaber B., et al. (2003) Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol., 3, 581–592. [DOI] [PubMed] [Google Scholar]
- 26.Sprenger J., Lynn F.J., Karunaratne S., et al. (2008) LOCATE: a mammalian protein subcellular localization database. Nucleic Acids Res., 36, D230–D233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pierleoni A., Martelli P.L., Fariselli P., et al. (2007) eSLDB: eukaryotic subcellular localization database. Nucleic Acids Res., 35, D208–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair R., Carter P., Rost B. (2003) NLSdb: database of nuclear localization signals. Nucleic Acids Res., 31, 397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choo K.H., Tan T.W., Ranganathan S. (2005) SPdb—a signal peptide database. BMC Bioinf. , 6, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinkel H., Michael S., Weatheritt R.J., et al. (2012) ELM—the database of eukaryotic linear motifs. Nucleic Acids Res., 40, D242–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The UniProt Consortium. (2014) Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res., 42, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jans D.A., Chan C.K., Huebner S. (1998) Signals mediating nuclear targeting and their regulation: application in drug delivery. Med. Res. Rev., 18, 189–223. [DOI] [PubMed] [Google Scholar]
- 33.Lee B.J., Cansizoglu A.E., Süel K.E., et al. (2006) Rules for nuclear localization sequence recognition by Karyopherinβ2. Cell, 3, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guda C., Guda P., Fahy E., et al. (2004) MITOPRED: a web server for the prediction of mitochondrial proteins. Nucleic Acids Res., 32, W372–W374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guda C. (2006) pTARGET: a web server for predicting protein subcellular localization. Nucleic Acids Res., 34, W210–W213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King B., Guda C. (2007) ngLOC: an n-gram-based Bayesian method for estimating the subcellular proteomes of eukaryotes. Genome Biol, 8, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King B.R., Latham L., Guda C. (2009) Estimation of subcellular proteomes in bacterial species. Open Appl. Inf. J., 3, 1–11. [Google Scholar]
- 38.Guda C. (2010) Towards cataloguing the subcellular proteomes of eukaryotic organisms. Seq. Genome Anal. Methods Appl. iConcepts Press, 259–269. [Google Scholar]
- 39.Pouton C.W., Wagstaff K.M., Roth D.M., et al. (2007) Targeted delivery to the nucleus. Adv. Drug Deliv. Rev., 59, 698–717. [DOI] [PubMed] [Google Scholar]
- 40.Mossmann D., Meisinger C., Vögtle F.N. (2011) Processing of mitochondrial presequences. Biochim Biophys Acta., 1819, 1098–1106. [DOI] [PubMed] [Google Scholar]
- 41.Teasdale R.D., Jackson M.R. (1996) Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu. Rev. Cell Dev. Biol., 12, 27–54. [DOI] [PubMed] [Google Scholar]
- 42.Dyer K.F. (1971) The quiet revolution: a new synthesis of biological knowledge. J. Biol. Educ., 5, 15–24. [Google Scholar]
- 43.King J.L., Jukes T.H. (1969) Non-Darwinian evolution. Science, 164, 788–798. [DOI] [PubMed] [Google Scholar]
- 44.Levitzki A. (1997) Targeting signal transduction for disease therapy. Med. Oncol., 14, 83–89. [DOI] [PubMed] [Google Scholar]