Abstract
Age-related degenerative and malignant diseases represent major challenges for health care systems. Elucidation of the molecular mechanisms underlying carcinogenesis and age-associated pathologies is thus of growing biomedical relevance. We identified biallelic germline mutations in SPRTN (also called C1orf124 or DVC1)1–7 in three patients from two unrelated families. All three patients are affected by a new segmental progeroid syndrome characterized by genomic instability and susceptibility toward early onset hepatocellular carcinoma. SPRTN was recently proposed to have a function in translesional DNA synthesis and the prevention of mutagenesis1–7. Our in vivo and in vitro characterization of identified mutations has uncovered an essential role for SPRTN in the prevention of DNA replication stress during general DNA replication and in replication-related G2/M-checkpoint regulation. In addition to demonstrating the pathogenicity of identified SPRTN mutations, our findings provide a molecular explanation of how SPRTN dysfunction causes accelerated aging and susceptibility toward carcinoma.
Monogenic syndromes with highly penetrant tumor susceptibility and/or signs of premature aging affecting more than one tissue have been instrumental in identifying the genes and pathways involved in carcinogenesis and age-related diseases8,9. The latter are commonly defined as segmental progeroid syndromes10 and can be caused by germline mutations in genes encoding DNA repair proteins with concomitant cancer predisposition. Examples include WRN, the Werner helicase gene, in Werner syndrome or BLM, the Bloom helicase gene, in Bloom syndrome. In addition, mutations in nuclear lamina–associated genes, for example, LMNA (encoding lamin A/C) in Hutchinson-Gilford syndrome or BANF1 in Nestor-Guillermo progeria11,12, can result in segmental progeria. Although LMNA mutations are also found in a few atypical cases of Werner syndrome13, some patients with suspected Werner syndrome do not harbor mutations in any known progeria gene14.
Here we studied three patients from two unrelated families presenting with early onset hepatocellular carcinoma (HCC), genomic instability and progeroid features. Consanguineous family A (Fig. 1a) of Moroccan origin was referred to the International Registry of Werner Syndrome, and the clinical characteristics of the affected boy in the family, A-IV:1, have been described previously15. The patient had short stature, bilateral cataracts, premature hair graying and died of HCC at the age of 17 years. Family B is a nonconsanguineous Australian family of European ancestry (Fig. 1b). Both affected boys, B-II:1 and B-II:4, presented similar clinical features, including low body weight, micrognathia, triangular face, muscular atrophy, lipodystrophy, bilateral simian creases, delayed bone age and mild joint restrictions in the fingers and elbows. Although hepatitis A, B and C serologies and α-fetoprotein levels were normal in these two boys, both developed early onset HCC at age 16 and 14, respectively (Fig. 1c). B-II:1 died at age 18 years from complications of acute fulminant hepatic failure. The clinical characteristics of all three affected individuals are summarized and compared to those of known segmental progeroid syndromes in Table 1.
Figure 1.
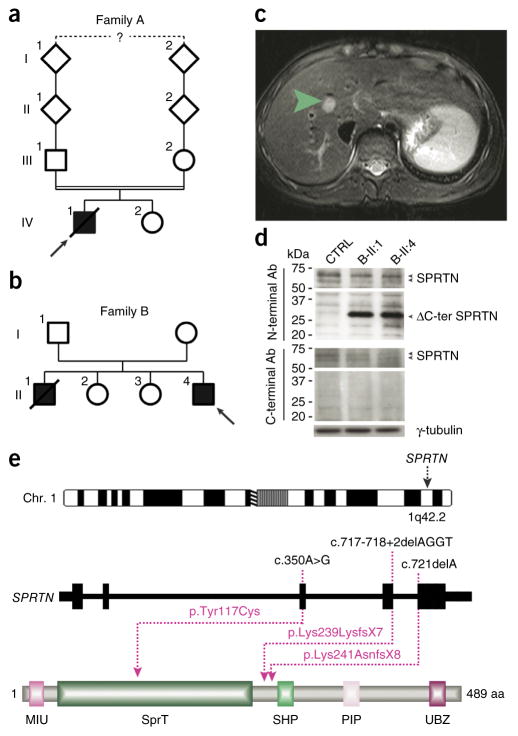
Identification of causative SPRTN mutations. (a,b) The pedigrees of families A and B. Filled and open symbols denote affected and healthy individuals, respectively; an arrow indicates the index patient, and diagonal lines indicate deceased status. The double line shows parental consanguinity, and the question mark denotes that the exact degree of consanguinity is unknown. (c) Axial view of magnetic resonance imaging of the liver of patient B-II:4. The green arrow indicates a 12 mm × 13 mm lesion mass with an absence of arterial phase enhancement within segment VIII of the liver that was subsequently shown to be a HCC. (d) Analysis of total cell extracts of patients’ LCLs with SPRTN antibodies (Ab) raised against the N- or C-terminal part of the protein. (e) Genomic localization and protein structure of SPRTN. The genomic structure is based on the longest ORF containing five coding exons (black rectangles). The positions of the identified mutations are shown at both the gene (top) and protein (bottom) levels. The protein diagram depicts the predicted functional domains of SPRTN. aa, amino acids.
Table 1.
Clinical and cellular findings in Werner syndrome, atypical Werner syndrome and patients described here
| Clinical findings | Werner syndrome | Atypical Werner syndrome | A-IV:1 | B-II:1 | B-II:4 |
|---|---|---|---|---|---|
| Short stature | + | + | + | − | − |
| Low body weight | + | + | + | + | + |
| Dermatological abnormalities | + | + | − | + | − |
| Cataracts | + | − | + | − | − |
| Sparse hair | + | + | − | − | − |
| Premature hair graying | + | + | + | − | − |
| Diabetes mellitus | + | + | − | − | − |
| Hypogonadism | + | − | − | − | − |
| Osteoporosis | + | − | − | − | − |
| General skeletal abnormalities | − | − | + | − | − |
| Atherosclerosis | + | + | − | − | − |
| Neoplasms | + | − | + | + | + |
| Hepatocellular carcinoma | Rare | − | + | + | + |
| Micrognathia | − | + | + | + | + |
| Lipodystrophy | + | + | ? | + | + |
| Muscular atrophy | + | + | ? | + | + |
| Attention deficit | − | − | − | + | + |
| Cellular findings | |||||
| Chromosomal instability | + | ? | + | + | + |
| Sensitivity to genotoxic agents | + | ? | + | + | + |
| Mutated gene | WRN | LMNA | SPRTN | SPRTN | SPRTN |
Atypical Werner syndrome includes cases with LMNA mutations. +, present in most patients; −, not present in most patients; ?, not known.
To identify the genetic cause of this putatively autosomal-recessive segmental progeroid disorder, we performed genome-wide linkage analysis (Supplementary Fig. 1) followed by exome sequencing of unrelated individuals A-IV:1 and B-II:4. Bioinformatic filtering identified SPRTN as the only gene with rare, biallelic mutations in the exomes of both individuals (Supplementary Tables 1 and 2). In A-IV:1, a 1-bp deletion at cDNA position 721 bp (c.721delA) was the only nonannotated sequence change with a severe impact on protein structure within the homozygous regions and is predicted to introduce a premature stop codon at amino acid 249 (p.Lys241AsnfsX8). B-II:4 was compound heterozygous for a c.350A>G missense alteration, resulting in the amino acid substitution p.Tyr117Cys, and a 4-bp deletion at cDNA position 717 bp (c.717_718+2delAGGT). At the cDNA level, this deletion predominantly caused intron inclusion, inducing a premature stop codon at amino acid 246 (p.Lys239LysfsX7). A very small fraction of cDNA demonstrated skipping of exon 4, resulting in a premature stop at position 161 bp (p.Val151IlefsX10) (Supplementary Fig. 2a–c). This finding was further supported by protein analysis, which identified a reduced amount of full-length protein and a new truncated protein (Fig. 1d). Sanger sequencing confirmed the mutations in all three patients (Supplementary Fig. 2d) and cosegregation with disease state in their families (Supplementary Table 3). None of these variants was present in dbSNP137 or the 1000 Genomes data. The substitution p.Tyr117Cys is located in a putative zinc metalloprotease SprT domain five amino acids upstream of Glu112, which was recently shown to be necessary for the regulation of error-prone translesional DNA synthesis (TLS)5. The identified truncating mutations (ΔC-ter SPRTN) lead to the loss of functionally important C terminal–located domains, including the ubiquitin-segregase p97 (VCP)16-interacting motif (SHP), the proliferating cell nuclear antigen interacting box (PIP) and the ubiquitin binding domain (UBZ4; Fig. 1e). The C-terminal part of SPRTN has an essential function at ultraviolet (UV)-induced stalled replication forks by the removal of DNA polymerase η in a p97-dependent manner after the completion of TLS2,3. Taken together, these genetic findings have already provided strong evidence for the pathogenicity of the identified mutations. The analysis of 48 additional patients with suspected Werner syndrome but without mutations in WRN or LMNA14 revealed no other SPRTN mutation, providing further evidence of extended locus heterogeneity for segmental progeroid syndromes.
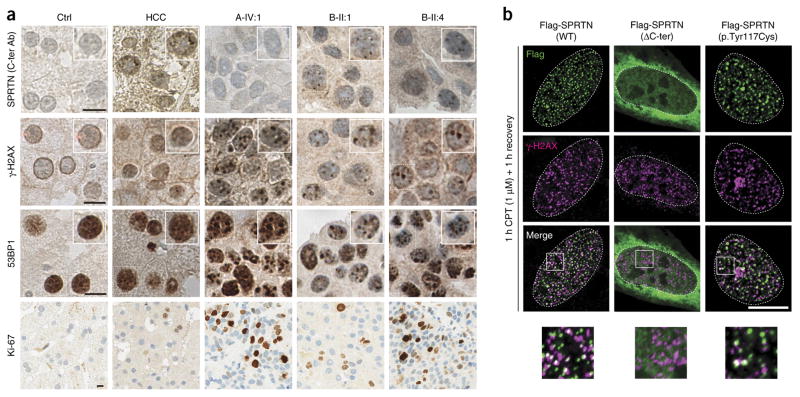
We next performed morphological and immunohistochemical analyses of patients’ liver tumor biopsies. Staining with a C-terminal SPRTN antibody (Fig. 2a) showed the absence of the C-terminal part of SPRTN in A-IV:1, thus confirming the truncation of the mutated protein in A-IV:1 in vivo. We observed focal accumulations of anti-SPRTN immunoreactive material in B-II:1 and B-II:4, as well as in idiopathic HCC. In vitro analysis of focal nuclear accumulation of ectopically expressed wild-type (WT) SPRTN and mutant SPRTN from patients additionally supported the in vivo finding and disclosed that WT and p.Tyr117Cys SPRTN are able to form nuclear foci, but ΔC-ter SPRTN is not (Fig. 2b). Analyses of cancer biomarkers revealed strong focal accumulations of both γ-H2AX (H2AFX) and 53BP1 (TP53BP1) (Fig. 2a). In addition, and opposite to what we observed in patient primary cell lines (Fig. 3a–c), Ki-67 (MKI67) staining in the biopsies of A-IV:1, B-II:1 and B-II:4 indicated a high proliferative index compared to healthy liver or idiopathic HCC (Fig. 2a). These data suggest a relatively aggressive neoplasm17,18.
Figure 2.
Severe DNA damage in hepatocellular carcinoma biopsies and focal nuclear accumulation of SPRTN. (a) Histological and immunohistochemical analyses of human liver biopsies from a healthy control (Ctrl), a patient with idiopathic, non–viral caused HCC and the HCC of patients with SPRTN mutations (A-IV:1, B-II:1 and B-II:4). The samples were stained with antibody raised against the C-terminal part of SPRTN (C-ter Ab) or with antibodies against γ-H2AX, 53BP1 or Ki-67. The insets in the top three rows are at 1.25× magnification. (b) U2OS cells were transiently transfected with Flag-tagged WT or mutant SPRTN and challenged with 1 μM of CPT to induce replication-related DSBs and thus mimic the DNA damage observed in patients’ livers. The images at the bottom of b are 3× magnified versions of the boxed areas in the merged images above. Scale bars, 10 μm (a,b).
Figure 3.
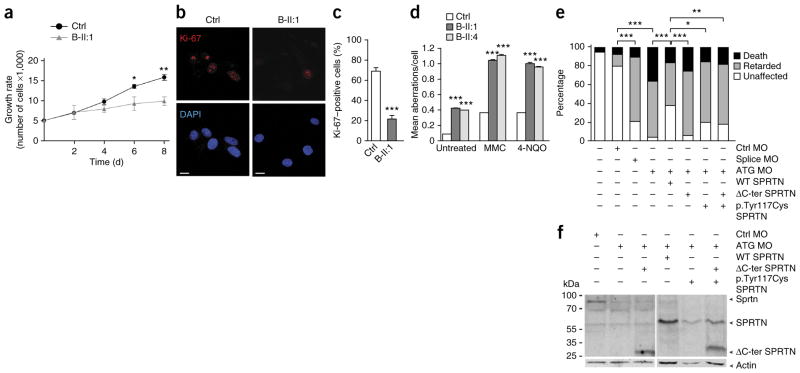
Genomic instability and cell proliferation defects. (a) A growth curve of control and patient primary skin fibroblasts. The data are from three independent experiments. (b) Primary skin fibroblasts stained with the proliferation marker Ki-67. Scale bars, 10 μm. DAPI, 4′,6-diamidino-2- phenylindole. (c) Quantification of the data in b. The data are from three independent experiments with greater than 100 cells scored per condition per experiment. (d) Chromosomal aberrations in patients’ LCLs under untreated conditions or the following genotoxic conditions: 40 ng/ml of MMC or 200 ng/ml of 4-NQO. The data are from three independent experiments with 33 analyzed metaphase cells per condition per experiment. The data in a, c and d were analyzed with unpaired t-test and are presented as the mean ± s.e.m. (e) Morpholino oligonucleotide (MO)-mediated gene downregulation of Sprtn in zebrafish at 10 hpf and a rescue experiment with human WT SPRTN or SPRTN harboring the two patient mutations. Ctrl MO denotes embryos injected with control MO, ATG MO denotes embryos injected with SPRTN MO targeting the start codon, and splice MO denotes embryos injected with a splice-site MO. The bar graph summarizes analyses in more than 100 embryos in three independent experiments showing the distribution of phenotypes observed after injections with 17.6 ng of MO and co-injections of 100 pg of SPRTN mRNA. P values in e were calculated using the χ2 test. *P < 0.01, **P < 0.001, ***P < 0.0001 (a,c–e). (f) Western blot analysis of the experiment shown in e. Sprtn, endogenous zebrafish protein; SPRTN, ectopically expressed human protein.
We next tested whether the cellular phenotypes described previously in A-IV:1, namely chromosomal instability with concomitant sensitivity toward genotoxic agents and severe proliferation defects15, were also present in primary cell lines from B-II:1 and B-II:4. Indeed, we found increased chromosomal instability in peripheral blood (Supplementary Fig. 3) and lymphoblastoid cell lines (LCLs) from the patients, which was enhanced after treatment with mitomycin C (MMC) and 4-nitroquinoline 1-oxide (4-NQO) (Fig. 3d). In patient fibroblasts, we observed multiple and variable aberrations that were clonal in nature (Supplementary Fig. 4), which is compatible with variegated translocation mosaicism, a phenomenon previously described in Werner syndrome cells19. Measurement of a proliferation index revealed a severe growth defect in cultured B-II:1 fibroblasts (Fig. 3a–c), thus confirming prior findings in cells from A-IV:1 (ref. 15). Further, short interfering RNA (siRNA)-mediated depletion of SPRTN in HEK293T and U2OS human cell lines also caused chromosomal instability and severe proliferation defects, respectively (Supplementary Figs. 5 and 6).
To assess the consequences of patient mutations in an in vivo complementation assay, we performed morpholino-mediated down-regulation of the SPRTN ortholog in zebrafish (LOC101886162, called here sprtn). sprtn silencing, verified by a GFP reporter assay (Supplementary Fig. 7a), led to a substantial increase of γ-H2AX foci (Supplementary Fig. 7b,c), indicating an evolutionarily conserved functional role of SPRTN in the DNA damage response. When injecting higher doses of morpholino, embryos displayed phenotypically normal development until the shield stage at 6 h post fertilization (hpf), a stage when maternal gene products are degraded20. At 10 hpf, however, embryos exhibited either early mortality or were delayed in development up to 4 h. This growth retardation phenotype is compatible with the proliferation defects observed in patients’ fibroblasts and the relative growth deficits observed in patients. This phenotype was partially rescued by co-injection of WT human SPRTN mRNA but not by equimolar amounts of mRNA from SPRTN harboring the identified mutations (Fig. 3e,f), thus further confirming their pathogenicity.
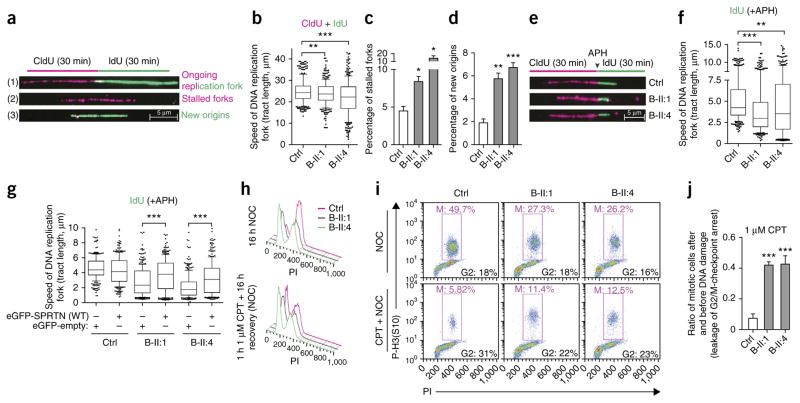
The lack of sun sensitivity and the presence of severe chromosomal breakage, proliferation defects and early onset HCC in patients indicated a more complex role of SPRTN in the maintenance of genome stability than solely TLS, as was proposed recently1–7. Defects in DNA replication have been proposed to be a major cause of the variegated translocation mosaicism and genomic instability that consequently lead to aging and cancer21–23. To test for a role of SPRTN in DNA replication, we analyzed the progression of replication forks directly by DNA fiber assay (Fig. 4a), the clearest method to unambiguously characterize the DNA replication machinery24. The speed of DNA replication was significantly but only mildly affected in patients’ LCLs under unchallenged conditions (Fig. 4b), but increased levels of stalled forks (Fig. 4c) and newly fired origins (Fig. 4d) indicated DNA replication stress as the cause of DNA damage in these patients25. When we treated LCLs with a low dose of aphidicolin (APH), mimicking the physiological barriers the DNA replication machinery approaches during DNA synthesis26, patients’ cells showed a typical signature of DNA replication stress, namely shorter replication forks than in control cells (Fig. 4e,f). This observation is comparable to cellular findings in Werner and Bloom syndromes27,28. Moreover, B-II:1 fibroblasts showed markedly increased numbers of double-strand breaks (DSBs) in S-phase cells (Supplementary Fig. 8). To further confirm that mutations in SPRTN are the cause of the DNA replication defect, we transfected patients’ cells with WT SPRTN, which almost completely corrected the replication defect (Fig. 4g and Supplementary Fig. 9a) and restored cellular proliferation (Supplementary Fig. 9b,c). siRNA-mediated SPRTN depletion severely affected the progression of DNA replication and induced an increased number of stalled forks, newly fired origins and formation of DSBs in the S phase of the cell cycle (Supplementary Fig. 10), providing further evidence of the role of SPRTN in general DNA replication. Notably, depletion of DNA polymerase η in patients’ cells or in SPRTN-depleted U2OS cells did not complement the DNA replication phenotype (Supplementary Fig. 11), suggesting that DNA polymerase η is not the main substrate, as reported previously2,3.
Figure 4.
DNA replication stress and leakage of the G2/M cell cycle checkpoint as the origin of genomic instability. (a) Schematic representation of a single DNA replication fork analysis by DNA fiber assay and its outcome ((1)–(3)). (b–d) DNA replication forks, as described in a, were analyzed in LCLs and quantified for velocity (b), percentage of stalled forks (c) and percentage of newly fired replication origins (d). n = 3; 100 CldU (5-chloro-2′-deoxyuridine) and 100 IdU (5-iodo-2′-deoxyuridine) symmetrical tracts were individually quantified, counted and are presented as total tract length (b), or 400 forks were randomly scored per condition per experiments (c,d). (e) Representative replication forks analyzed in LCLs when the incorporation of IdU was in the presence of a low dose of APH (0.1 μM). (f) The speed of replication forks under mild genotoxic conditions (IdU only). n = 3; 100 DNA fibers analyzed per experiment per condition. (g) LCLs were transfected with WT SPRTN, and the speed of replication fork was measured under mild genotoxic conditions, as in e. n = 2; 100 DNA fibers analyzed per experiment per condition. (h) Cell cycle profile of LCLs. NOC, nocodazole. PI, propidium iodide. (i) Mitotic (red triangle) LCLs without and with CPT treatment analyzed in the presence of NOC. (j) Quantification of i; n = 3. Data in b, f and g are shown as the median (bar) with the 25th–75th percentile range (box) and the 10th–90th percentile range (whiskers). Data in c, d and j are shown as the mean ± s.e.m. Data in b–d, f, g and j were analyzed with unpaired two-tailed t-test. *P < 0.0), **P < 0.001, ***P < 0.0001.
To evaluate how replication-related DNA damage is transferred to mitosis and may thus contribute to chromosomal instability, we measured the ability of patients’ cells to activate the G2/M checkpoint, the main guardian of genome stability after DNA replication stress29. We exposed patients’ LCLs to different genotoxic agents and measured the arrival of cells to mitosis by flow cytometry (Fig. 4h–j and Supplementary Figs. 12 and 13). We observed a severe G2/M-checkpoint defect in cells from B-II:1 and B-II:4 after treatment with genotoxic agents that interfere with DNA replication, such as camptothecin (CPT) (Fig. 4h–j), a topoisomerase I inhibitor that causes replication-related DSBs, or UV radiation (Supplementary Fig. 13). Notably, the G2/M checkpoint was completely functional when we created random and non–replication related DSBs using ionizing radiation (Supplementary Fig. 13). The hypersensitivity of patient cells to replication-related genotoxic agents but not to ionizing radiation (Supplementary Fig. 14) correlates with G2/M leakage.
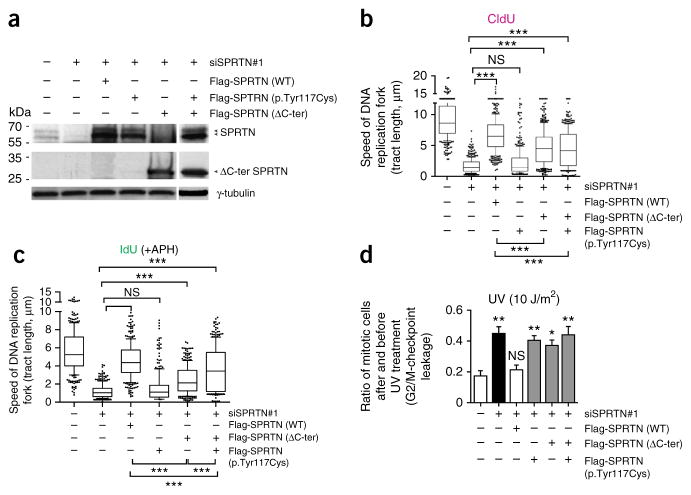
To further assess the role of patients’ mutations in DNA replication and G2/M-checkpoint regulation, we tested their function in U2OS cells that we depleted of endogenous SPRTN using siRNA (Fig. 5 and Supplementary Fig. 15). Ectopic expression of WT SPRTN restored DNA replication and G2/M-checkpoint defects. The expression of ΔC-ter SPRTN was also able to restore the progression of the DNA replication fork but to a much lesser extent then WT SPRTN, suggesting that this mutant is defective in proper DNA synthesis (Fig. 5b). The difference in efficacy was even more pronounced when we exposed cells to mild replication stress (APH treatment; Fig. 5c), further supporting the function of the C-terminal part of SPRTN in TLS. Notably, the cells ectopically expressing p.Tyr117Cys SPRTN were completely unable to restore DNA replication fork progression, suggesting the essential role of the SprT domain in general DNA replication. We obtained similar results in HEK293 cells (data not shown). In contrast to DNA replication, cells expressing either of the patient mutations or coexpressing both mutations were equally defective in activation of the G2/M checkpoint after exposure to UV radiation or CPT treatment (Fig. 5d and data not shown), indicating the communal function of the SprT domain and the C-terminal part of SPRTN in the regulation of the G2/M-checkpoint response. Taken together, these data demonstrate that SPRTN dysfunction leads to sustained DNA replication stress and consequent replication-related DNA damage, especially DSBs, which are transferred to the next cell generation because of a leakage of the G2/M checkpoint and, consequently, lead to cancer or aging.
Figure 5.
Characterization of patients’ mutations in DNA replication and G2/M-checkpoint regulation. (a) Western blot analysis of U2OS cells depleted of endogenous SPRTN by siRNA (siSPRTN#1) and simultaneously expressing siRNA-resistant WT SPRTN, ΔC-ter SPRTN or p.Tyr117Cys SPRTN or coexpressing of ΔC-ter SPRTN and p.Tyr117Cys SPRTN. (b,c) U2OS cells, as in a, labeled with CldU for 30 min (unchallenged conditions; b) or with IdU and treated with APH for 30 min (mild genotoxic conditions; c) were analyzed by DNA fiber assay. 1 μm of DNA tract length corresponds to 2.6 kb of newly synthesized DNA32. n = 3; more than 100 DNA fibers analyzed per experiment and per condition. (d) U2OS cells, as in a, were analyzed for the efficacy of the G2/M checkpoint after treatment with UV radiation, as described in Figure 4. The graph summarizes three independent experiments. The data in b and c are presented as the median (bar) with the 25th–75th percentile range (box) and the 10th–90th percentile range (whiskers). Data in d are shown as a bar graph with the mean ± s.e.m. Data in b–d were analyzed with unpaired two-tailed t-test. *P < 0.05, **P < 0.001, ***P < 0.0001. NS, no significant difference between the groups.
Studying the genetic and cellular basis of monogenic segmental progeroid and tumor susceptibility syndromes has been a meaningful approach in unraveling the molecular mechanisms and pathways involved in the regulation of cancer development and common diseases of the elderly8. HCC, although rarely occurring before the age of 40 years30, is the fifth most common malignancy and the third leading cause of cancer-related death worldwide31. Although major risk factors for HCC are well known, including hepatitis B and C infection and alcohol abuse, the molecular pathogenesis of HCC remains largely elusive. All three patients presented here—in addition to showing signs of accelerated aging in selected tissues—developed early onset HCC, identifying SPRTN as a monogenic and apparently highly penetrant susceptibility gene for HCC. Consequently, our findings suggest SPRTN as the subject of future studies of hepatocarcinogenesis and therapy.
URLs
The International Registry of Werner Syndrome, http://www.wernersyndrome.org/registry/registry.html.
ONLINE METHODS
Ethical approval and study procedures
The International Registry of Werner Syndrome has been recruiting patients suspected of having Werner syndrome since 1988. Studies of family A and B were approved by the University of Washington Institutional Review Board and the Human Research Ethics Committee of the Royal Children’s Hospital, Melbourne, Australia, respectively. DNA samples from whole blood were isolated by standard procedures after written informed consent of participating individuals.
Linkage analysis in family A
Linkage analysis was performed using Genome-Wide Human SNP Array 6.0. (Affymetrix, Inc., Santa Clara, CA, USA). Data handling, evaluation and statistical analyses have been described in detail before33.
Linkage analysis in family B
All individuals in pedigree B were genotyped using Illumina Human610-Quad BeadChips (USA) at the Australian Genome Research Facility, Melbourne. Genotypes were called using the GenCall algorithm implemented in Illumina’s BeadStudio package. The LINKDATAGEN script34 was used to select a subset of 11,913 SNP markers for analysis. These markers were chosen to be in approximate linkage equilibrium (spaced at least 0.15 cM apart) and to have high heterozygosity in the HapMap population of Utah residents with ancestry from northern and western Europe (CEU). Parametric linkage analysis was performed by MERLIN35 under a fully penetrant recessive inheritance model with a 0% phenocopy rate and a disease allele frequency of 0.00001. Allele frequencies from CEU were used.
Estimation of inbreeding coefficients in family B
FEstim36 was used to estimate genomic inbreeding coefficients for the genotyped members of family B. A subset of 11,374 high-heterozygosity markers in approximate linkage equilibrium were selected for analysis using LINKDATAGEN. FEstim was run in an independent model with starting values F = 0.05 and A = 0.05. All inbreeding coefficients were estimated to be 0.000, indicating no evidence of inbreeding.
Candidate gene sequencing
For candidate gene analysis, we designed intronic primers to PCR amplify coding exons and the respective exon-intron boundaries by using genomic DNA of the affected individual. Primer pairs for the amplification of the five SPRTN coding exons and their approximately 50 bp of flanking intronic sequences (RefSeq accession NM_032018.4) are available on request. PCR products were sequenced on an ABI 3730 DNA Analyzer with BigDye chemistry v3.1 (Applied Biosystems). Sequence traces were assembled, aligned and analyzed with SeqMan software (DNASTAR Lasergene). Cosegregation of the mutations in families A and B was tested by sequencing the PCR product of exon 5, or of exons 3 and 4, respectively, amplified from genomic DNA of all participating family members.
Exome sequencing in A-IV:1
We sequenced the exome of the proband on two lanes of an Illumina GAIIx Sequencer using a single-read 150-bp protocol after enrichment of exonic and splice-site sequences with the Agilent SureSelect Human All Exon 50 Mb kit. We mapped >194 million reads to the hg19 human reference genome. Approximately 89% of target sequences were covered at least 10-fold and 83% were covered at least 30-fold, with a mean coverage of about 112×. Data analysis of filter-passed reads was performed with the in-house pipeline V1.3 using BWA-short in combination with SAMtools pileup 0.1.7 for the detection of SNPs and short insertions and deletions (indels). In-house–developed scripts were applied to detect protein changes, affected splice sites and overlaps to known variations, with filtering against dbSNP build 137, the 1000 Genomes Project data build February 2012 and our in-house database of exome variants (with data from >200 exomes of individuals affected by different disorders). We focused our analysis on rare missense, nonsense, frameshift and splice-site mutations. The criteria for a variation to be taken into account were as follows: >6 reads, phred scaled quality score >15, population allele frequency <1%, <10 times seen in our in-house database and >15% of the reads supporting the allele.
Exome sequencing in B-II:4
The exome of the surviving affected male B-II:4 was sequenced by Axeq Technologies (Korea) on three lanes of an Illumina HiSeq sequencer using a paired-end 100-bp read protocol after exome capture with the Illumina TruSeq capture array. Novoalign (V2.08.03) alignment reads mapped 57,905,140 reads uniquely to the hg19 reference genome. Presumed PCR and optical duplicates were removed using Picard 1.65, and local realignment was performed using RealignerTargetCreator and IndelRealigner walkers from the Genome Analysis Toolkit (GATK)37,38. Greater than 80% of target sequences were covered at least tenfold, and the median coverage of targeted bases was 40×. Single-nucleotide variants and small indel variants were detected and genotyped using the UnifiedGenotyper walker from GATK version v2.3-3-g4706074. Variants were annotated using ANNOVAR39 against the RefSeq gene annotation; dbSNP build 137; 69 genomes from Complete Genomics40; 1,092 genomes from the 1000 Genomes project, February 2012 release; 6,503 European and African American ancestry exomes from the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP6500, https://esp.gs.washington.edu/drupal/); and an in-house database of 131 exomes of various ethnicities. Read-backed phasing was performed using HapCUT41. Variants located within the 33 linkage peaks were extracted and initially filtered for quality (phred scaled quality ≥13) and rarity (alternate allele frequency of ≤1% in the 1000 Genomes and NHLBI ESP6500 data sets and ≤4% in the Complete Genomics 69 and in-house data sets). We focused our analysis on rare homozygous or compound heterozygous variants predicted to affect protein sequence or splicing. Pairs of heterozygous variants located in the same gene that were inferred to be in cis phase by HapCUT41 were eliminated.
RT-PCR analysis
RT-PCR sequencing was done as described previously42,43. To distinguish between two SPRTN isoforms, we performed two RT-PCR reactions. Primer sequences used for reaction A to amplify the canonical 489-aa isoform 1 of SPRTN spanning exons 2 through 5 were SPRTN-2F and SPRTN-5R, with an expected WT size of 1,205 bp. Primer sequences used for reaction B to amplify the 250-aa isoform 2 of SPRTN spanning exons 2 through 4 were SPRTN-2F and intronic SPRTN-4r, with an expected WT size of 640 bp.
DNA fiber assay
The DNA fiber assay was performed as described previously44. Briefly, asynchronous LCL, U2OS or HEK293 cells were labeled with 30 μM of CldU for 30 min, washed three times with warm PBS and then labeled with 250 μM of IdU for an additional 30 min. The reaction was terminated by treating the cells with ice-cold PBS. Cells were lysed, and DNA fibers were spread onto glass slides, fixed with methanol and acetic acid, denatured with HCl, blocked with 2% BSA and stained with anti-rat and anti-mouse 5-bromo-2′-deoxyuridine (BrdU) that specifically recognize either CldU (Sigma, C6891) or IdU (Sigma, 17125). Anti-rat Cy3 (dilution 1:300, Jackson ImmunoResearch, 712-116-153) and anti-mouse Alexa-488 (dilution 1:300, Molecular Probes, A11001) were used as the respective secondary antibodies. Microscopy was done using a Leica DMRB microscope with a DFC360FX camera. The lengths of the CldU- and IdU-labeled tracts were measured by ImageJ software. Statistical analysis was done by GraphPad Prism software using unpaired t-test. For the DNA fiber assay under genotoxic stress, the second nucleotide (IdU) was incubated in the presence of 0.1 μM APH.
Flow cytometry and G2/M-checkpoint assay
These analyses were performed as described previously45. In brief, cells were harvested, washed with PBS and subsequently fixed in 3.6% formaldehyde at room temperature for 15 min. After washing, the cells were permeabilized and blocked with 1% FBS and 0.1% saponin in PBS for 30 min. For 5-ethynyl-2′-deoxyuridine (EdU) analysis, cells were incubated with 10 μM EdU before harvesting. EdU was detected with a Click-iT EdU Cell Proliferation Assay kit (Invitrogen, C10632). Alexa 647 ( 3458)– or Alexa 488 (9708)–conjugated anti–phosphorylated histone H3 (Ser10) (mitotic marker) was used (dilution 1:100, Cell Signaling). Mitotic index was determined as the ratio between the numbers of mitotic cells in the presence of nocodazole (400 nM for 16 h) after UV treatment compared to those in untreated cells. DNA content was analyzed by DAPI. Cells were analyzed on a Beckman Coulter CyAn ADP Analyzer. A minimum of 10,000 events were counted.
Growth assay
50,000 primary skin fibroblast or U2OS cells were seeded at day 0. Every 24 h, cells were washed, trypsinized, resuspended in 1 ml medium (DMEM, Sigma, D6429) and counted (TC10 automated cell counter, Bio-Rad).
Histology
Human liver biopsy specimens were obtained from University Hospital Zürich, Academic Medical Centre Amsterdam and Royal Children’s Hospital Parkville. Biopsy specimens were registered in respective biobanks and kept anonymous. The study protocol was in accordance with the ethical guidelines of the Helsinki declaration. Liver samples were prepared from paraffin blocks according to standard histological protocols and hemalauneosin stained, or immunohistochemical staining was performed using Leica Bond automated staining system.
Immunofluorescence studies
Cells were grown on glass coverslips, fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100, blocked with 5% BSA in PBS and immunostained with the respective antibodies. Images of immunostained cells were taken with an epifluorescent microscope (Olympus BX51) and acquired with a charge-coupled device (CCD) camera (Orca AG), a Zeiss LSM 510 META laser scanning microscope or an SP2 Leica confocal microscope.
Antibodies
The following antibodies were used in this study: anti-SPRTN (rabbit, polyclonal) raised against the N-terminal part (1–240 aa) of SPRTN (dilution 1:1,000, home made); anti-SPRTN raised against the C-terminal part of SPRTN (dilution 1:1,000, Atlas, HPA 025073); anti–DNA polymerase η (dilution 1:1,000, Abcam, Ab17725); anti–phosphorylated γ-H2AX (Ser139) (dilution 1:300, Millipore, 05-636); anti-zebrafish γ-H2AX (dilution 1:1,000, gift of J. Amatruda); anti-rabbit 53BP1 (dilution 1:200, Santa Cruz, sc-22760); anti–Ki-67 (dilution 1:200, Millipore, MAB 4190); anti-rat BrdU (dilution 1:500, Abcam, 6326); anti-mouse BrdU (dilution 1:100, Becton Dickinson, 347850); anti-mouse Alexa Flour 488 (dilution 1:300, Invitrogen, A21202), anti-mouse Alexa Flour 594 (dilution 1:300, Invitrogen, A11020); anti-mouse horseradish peroxidase (HRP) (dilution 1:10,000, Sigma, A2304); and anti-rabbit HRP (1:10,000, Sigma, A0545).
Cell lines
Primary skin fibroblasts, Epstein-Barr virus (EBV)-transformed LCLs, U2OS and HEK293T cells were used in this study. For stable transfected GFP-SPRTN (WT) or GFP–empty vector control or patient LCLs, cells were transfected by electroporation as indicated below and selected in a medium containing G418/Geneticin (Gibco, 10131-027).
DNA primers and siRNA sequences
The DNA primers and siRNA sequences used are listed in Supplementary Tables 4 and 5.
Plasmids
The I.M.A.G.E. full-length SPRTN cDNA clone (IRATp970E1156D, ImaGenes) was cloned into pFlag–CMV-1 (Sigma), peGFP-C1 (Clontech) or pcDNA3.1 (Invitrogen). Mutants were cloned using PCR amplification and restriction enzyme digestion and recombination. Site-directed mutagenesis was performed by PCR to introduce the desired mutations. The correctness of the DNA sequence was verified by sequencing.
Plasmid transfection
U2OS cells were transiently transfected with Lipofectamine 2000 (Life Technology), and primary skin fibroblasts and LCLs were transfected by electroporation (Amaxa Nucleofactor Technology, Lonza) following the manufacturer’s instructions.
siRNA transfection
siRNA depletion experiments in mammalian cells were conducted using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer’s instructions. A final concentration of 20 nM siRNA oligonucleotides was used. siRNA-transfected cells were analyzed 48 or 72 h after transfections.
Chromosome analysis
For chromosome analysis, primary skin fibroblasts, LCLs and siRNA-transfected HEK293T cells were incubated with 40 ng/ml MMC or 200 ng/ml 4-NQO or left untreated for an additional 24 h. Metaphase spreads and G banding were prepared using standard procedures, analyzed using an Axio imaging 2 microscope (Zeiss, Jena, Germany) and captured using Ikaros software (Metasystems, Altlussheim, Germany). 100 metaphase spreads were scored for chromosomal aberrations in three independent experiments. Based on 100 cells (untreated or treated with MMC or 4-NQO), the frequencies of aberrant cells were compared between LCLs from a healthy individual (AG1010) and LCLs of both affected individuals or between SPRTN siRNA treatment and a nonspecific siRNA using Fisher’s exact test, unless otherwise stated. The numbers of breaks per cell were contrasted by two-sample Poisson tests, where P values were approximated using the χ2 distribution with 1 degree of freedom.
Cloning of LOC101886162, the SPRTN ortholog in zebrafish, called here sprtn
The zebrafish sprtn ortholog was cloned using a reciprocal, tblastn protein query, BLAST (Basic Local Alignment Search Tool), which identified a partial sequence of a single zebrafish sprtn ortholog. Using 3′ RACE PCRs (first choice RLM RACE, Ambion, Life Technologies), the C-terminal part and the 3′ UTR of SPRTN were obtained. The complete ORF of SPRTN was subsequently amplified from the cDNA of eight somite-stage zebrafish and cloned into the pCS2+ vector.
Zebrafish maintenance and manipulation
Zebrafish were kept under controlled water and temperature conditions in a 14-h light and 10-h dark cycle. Fertilized eggs were allowed to develop at 28.5 °C up to the required stages and analyzed and processed as indicated. All husbandry procedures and experiments were approved by the ethics committee and research commission of the University of Ulm, Germany. For knockdown experiments, fertilized eggs were injected with RNA antisense MO or capped RNA transcribed with the mMessage mMachine Kit (Ambion) starting from linearized plasmids. Injections were carried out at the one- to two-cell stage with an Eppendorf Femtojet Microinjector (Germany). An antisense MO targeting the start codon of SPRTN and a splice-site MO were used to generate loss-of-function zebrafish. Injections were controlled against those with a five-base mismatch control (Ctrl) MO. 1.5–17.6 ng of antisense MO against zebrafish SPRTN or Ctrl MO were injected. The coding sequence of human SPRTN was amplified from pFlag–CMV-1–SPRTN and cloned into the pCRII-TOPO vector (Invitrogen, Darmstadt, Germany) and subsequently subcloned into vector pCS2+ by the use of BamHI and XhoI restriction sites followed by T4 ligation. The clinical mutations were introduced using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Waldbronn, Germany) and mutagenesis primers. Staining with antibody against phosphorylated zebrafish γ-H2AX (a gift from J. Amatruda) followed a standard protocol as described before46.
GFP reporter assay
To verify the efficacy and specificity of MO-induced knockdown, capped mRNA of GFP fused to the whole ORF of SPRTN and parts of the 5′ UTR was injected alone or along with MO or CtrlL MO into zebrafish eggs at the one-cell stage. At 24 hpf, embryos were assayed for GFP fluorescence on a Keyence BZ8000K fluorescent microscope.
Supplementary Material
Acknowledgments
We are thankful to the family members for participation, G. Gillies for assistance with patient samples, J. Schäfer for zebrafish care and Z. Garajova for technical assistance. We thank A. Ray Chaudhuri for initial help with the DNA fiber assay. We thank F. Böhm, Y. Böge and A. Weber from the University of Zurich and L. Campo and K. Myers from the University of Oxford for providing healthy and HCC human liver biopsies and performing histological and immunohistochemical staining. The zebrafish γ-H2AX antibody was a kind gift of J. Amatruda (University of Texas Southwestern). This work was supported by grants from Deutsche Forschungsgemeinschaft, the Cluster of Excellence ‘Macromolecular Complexes’ of Goethe University Frankfurt (EXC115), the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz program Ubiquitin Networks of the State of Hesse, Germany and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013)/European Research Council grant agreement number 250241-LineUb to I.D., the European Commission (Marie Curie Reintegration Grant 268333 to M.P.), the Deutsche Stiftung für Herzforschung (M.P.), the Medical Research Council (MC_PC_12001/1) and the Swiss National Science Foundation (31003A_141197) to K.R., grants from the US National Institutes of Health (NIH) National Cancer Institute (R24CA78088 and R24AG042328) to G.M.M., the NIH National Institute on Aging (R21AG033313) to J. Oshima, the Ellison Medical Foundation to J. Oshima, the German Research Foundation (DFG) in the framework of the Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases to C.K., an EMBO long-term fellowship to J.L.-M., a grant from the Croatian Ministry of Science, Education and Sport (216-0000000-3348) and a City of Split grant to J.T. and I.D. K.R.S. is supported by a PhD scholarship funded by the Pratt Foundation. M.B. is supported by an Australian Research Council Future Fellowship (FT100100764). P.J.L. is supported by a National Health and Medical Research Council (NHMRC) Career Development Fellowship (APP1032364). This work was made possible through Victorian State Government Operational Infrastructure Support and the Australian Government NHMRC Independent Research Institutes Infrastructure Support Scheme.
Footnotes
Accession codes. NCBI reference sequences: UniGene: NM_032018, protein: NP_114407.3. Microarray probes: NM_032018.4. UniProt: Q9H040. GenBank: AL512744. SPRTN mutations: NM_032018.5 (c.721delA, ss1387933564, rs527236212; c.350A>G, ss1387933565, rs527236213; c.717_718+2delAGGT, ss1387933572, rs587593493).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
D.L., B.V., S.H., P.J.L., I.M.-T., J.L.-M., M.P., J.C.H.S., K.R.S., J. Oehler, K.P., A.N., F.N., R.J.L., M.B.D., G.B., S.v.A., J.H., M.D., R. Fertig, M.D.B., K.H., H.T., J.A., G.N., P.N. and M.B. performed the experiments and did data analysis. E.C., R. Freire, J. Oshima, G.M.M. and C.M.A. contributed materials and reagents used in the study. D.L., K.R. and C.K. wrote the manuscript. J.T., D.J.A., I.D., K.R. and C.K. led and coordinated the entire project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Centore RC, Yazinski SA, Tse A, Zou L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol Cell. 2012;46:625–635. doi: 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis EJ, et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol. 2012;19:1093–1100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- 3.Mosbech A, et al. DVC1 (C1orf124) is a DNA damage–targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat Struct Mol Biol. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- 4.Juhasz S, et al. Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance. Nucleic Acids Res. 2012;40:10795–10808. doi: 10.1093/nar/gks850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MS, et al. Regulation of error-prone translesion synthesis by Spartan/C1orf124. Nucleic Acids Res. 2013;41:1661–1668. doi: 10.1093/nar/gks1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machida Y, Kim MS, Machida YJ. Spartan/C1orf124 is important to prevent UV-induced mutagenesis. Cell Cycle. 2012;11:3395–3402. doi: 10.4161/cc.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosal G, Leung JW, Nair BC, Fong KW, Chen J. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 is a regulator of translesion synthesis. J Biol Chem. 2012;287:34225–34233. doi: 10.1074/jbc.M112.400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burtner CR, Kennedy BK. Progeria syndromes and ageing: what is the connection? Nat Rev Mol Cell Biol. 2010;11:567–578. doi: 10.1038/nrm2944. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher O, Houlston RS. Architecture of inherited susceptibility to common cancer. Nat Rev Cancer. 2010;10:353–361. doi: 10.1038/nrc2840. [DOI] [PubMed] [Google Scholar]
- 10.Martin GM. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig Artic Ser. 1978;14:5–39. [PubMed] [Google Scholar]
- 11.Navarro CL, Cau P, Levy N. Molecular bases of progeroid syndromes. Hum Mol Genet. 2006;15:R151–R161. doi: 10.1093/hmg/ddl214. [DOI] [PubMed] [Google Scholar]
- 12.Puente XS, et al. Exome sequencing and functional analysis identifies BANF1 mutation as the cause of a hereditary progeroid syndrome. Am J Hum Genet. 2011;88:650–656. doi: 10.1016/j.ajhg.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, et al. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- 14.Oshima J, Hisama FM. Search and insights into novel genetic alterations leading to classical and atypical Werner syndrome. Gerontology. 2014;60:239–246. doi: 10.1159/000356030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruijs MW, et al. Atypical progeroid syndrome: an unknown helicase gene defect? Am J Med Genet A. 2003;116A:295–299. doi: 10.1002/ajmg.a.10730. [DOI] [PubMed] [Google Scholar]
- 16.Vaz B, Halder S, Ramadan K. Role of p97/VCP (Cdc48) in genome stability. Front Genet. 2013;4:60. doi: 10.3389/fgene.2013.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King KL, et al. Ki-67 expression as a prognostic marker in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 1998;13:273–279. doi: 10.1111/j.1440-1746.1998.01555.x. [DOI] [PubMed] [Google Scholar]
- 18.Nowsheen S, Aziz K, Panayiotidis MI, Georgakilas AG. Molecular markers for cancer prognosis and treatment: have we struck gold? Cancer Lett. 2012;327:142–152. doi: 10.1016/j.canlet.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Hoehn H, et al. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15:282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- 20.Aanes H, et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen JC, Loeb LA. The Werner syndrome gene: the molecular basis of RecQ helicase–deficiency diseases. Trends Genet. 2000;16:213–220. doi: 10.1016/s0168-9525(99)01970-8. [DOI] [PubMed] [Google Scholar]
- 22.Venkatesan RN, et al. Mutation at the polymerase active site of mouse DNA polymerase δ increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27:7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 24.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukas C, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 27.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 29.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 30.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 31.Caldwell S, Park SH. The epidemiology of hepatocellular cancer: from the perspectives of public health problem to tumor biology. J Gastroenterol. 2009;44 (suppl 19):96–101. doi: 10.1007/s00535-008-2258-6. [DOI] [PubMed] [Google Scholar]
- 32.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc Natl Acad Sci USA. 2008;105:20752–20757. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borck G, et al. Loss-of-function mutations of ILDR1 cause autosomal-recessive hearing impairment DFNB42. Am J Hum Genet. 2011;88:127–137. doi: 10.1016/j.ajhg.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahlo M, Bromhead CJ. Generating linkage mapping files from Affymetrix SNP chip data. Bioinformatics. 2009;25:1961–1962. doi: 10.1093/bioinformatics/btp313. [DOI] [PubMed] [Google Scholar]
- 35.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 36.Leutenegger AL, et al. Estimation of the inbreeding coefficient through use of genomic data. Am J Hum Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 41.Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24:i153–i159. doi: 10.1093/bioinformatics/btn298. [DOI] [PubMed] [Google Scholar]
- 42.Hisama FM, et al. Coronary artery disease in a Werner syndrome–like form of progeria characterized by low levels of progerin, a splice variant of lamin A. Am J Med Genet A. 2011;155A:3002–3006. doi: 10.1002/ajmg.a.34336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha B, et al. Ethnic-specific WRN mutations in South Asian Werner syndrome patients: potential founder effect in patients with Indian or Pakistani ancestry. Mol Genet Genomic Med. 2013;1:7–14. doi: 10.1002/mgg3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 46.Zhou W, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.