Abstract
The Parkinson's disease (PD) associated gene PINK1 encodes a protein kinase that mediates the phosphorylation of multiple proteins involved in mitochondrial homeostasis. The broader downstream signaling events mediated by PINK1 kinase activity have not been well documented. We combine quantitative phosphoproteomic strategies with siRNA mediated PINK1 knock down in mammalian cells to identify alterations of phosphorylation events downstream of PINK1. Although down-regulation of PINK1 has no major effect on the proteome expression in these cells, phosphorylation of over one hundred proteins was reduced reflecting basal levels of phosphorylation signaling events downstream of PINK1. Motif analysis of the residues flanking the phosphorylation sites indicates proline-directed kinase specificity. Surprisingly, we found that the downstream signaling nodes included many transcription factors, as well as nuclear proteins involved in DNA and RNA metabolism. Thus, PINK1 dependent phosphorylation signaling may regulate nuclear activities.
Keywords: PINK1, mass spectrometry, phosphorylation, quantification, SILAC
Introduction
Loss-of-function mutations of PINK1, the gene encoding PTEN-induced putative kinase 1 (PINK1), cause autosomal recessive early-onset Parkinson’s disease.1 PINK1 is known primarily as a mitochondrial serine/threonine kinase, acting to promote the survival of neurons and muscle cells through a mitochondrial quality control mechanism.2,3,4 Upon sustained mitochondrial membrane depolarization or other factors that compromise mitochondria, PINK1 accumulated on the mitochondrial outer membrane and autophosphorylated leading to the recruitment of Parkin to the mitochondria. 5 Activated PINK1 kinase phosphorylates Parkin and promotes Parkin-mediated ubiquitination of mitochondrial outer membrane proteins including TRAP1, Mitofusin, Miro and VDAC.6,7,8,9 These processes lead to the degradation of ubiquitinated proteins and mitophagy, preventing further damage to cells. The kinase activity of PINK1 appears to be essential since the most frequent familial PD-associated PINK1 mutations locate at the kinase domain and these mutations disrupt PINK1 kinase function. 10,11 Furthermore, autophospohrylation upon mitochondrial membrane dissipation has been demonstrated to be the first step to activate PINK1 for subsequent Parkin recruitment. 5 Although this model links PINK1 function to mitochondria and provides a mechanism for mitochondria-mediated neurodegeneration, the complete picture of signaling downstream of PINK1 is still lacking. In the kinome tree PINK1 appears to be a solitary family member, 12 the lack of specificity in many kinases prompted us to look for proteins other than mitochnodrial components that could be phosphorylated after PINK1 activation. Furthermore, identifying PINK1 substrates and downstream phosphorylation targets in a systematic manner is crucial to understand the physiological and pathological functions of PINK1 in a broader sense.
Large-scale, mass spectrometry-based quantitative proteomics provides a systematic means to identify global protein alterations of the biological system upon perturbations.13,14 Among a number of quantitative approaches coupled with mass spectrometry, stable-isotope labeling in cell culture (SILAC) has been widely used to profile proteome and phosphoproteome changes in the cell.15,16 As a sensitive means to respond to intrinsic and environmental cues, protein phosphorylation is generally dynamic and of low stoichiometry, making it difficult to identify phosphorylation events in large scale without enrichment of phosphorylated species. With the advance of new biological materials, a large variety of enrichment strategies have been developed to improve phosphoproteomic coverage. Strong-cation exchange (SCX) followed either by immobilized metal ion chromatography (IMAC)17 or by TiO2 affinity capture,18, 19 hydrophilic interaction chromatography (HILIC) followed by IMAC,20 and affinity capture using certain transitional-metal compounds,21 all show dramatic improvements in phosphopeptide coverage. More straightforward methods include enrichment through chemical derivatization,22 or simply a single step IMAC23 or TiO2 affinity capture.24 Each of these methods has their own preferences for phosphopeptides with unique properties including charge and hydrophobicity, and also differs in requirement for the amount of starting materials. We chose a combination of HILIC-IMAC for tandem enrichment because it offers a balance between reproducibility and compatibility with small sample sizes at 1 mg level.
We applied the SILAC method in mammalian cells to quantify the phosphorylation events after siRNA mediated PINK1 knocking down. We enriched phosphopeptides for mass spectrometry analysis using a combined HILIC-IMAC approach. Among 4500 quantified phosphorylation events, 166 showed a statistically significant decrease in phosphorylation, and 47 showed an increase. We found that there was enrichment in nuclear proteins among these phosphorylated proteins. Sequence motif analysis of the phosphorylation sites identified a proline-directed serine/threonine kinase recognition preference, and sequence alignment of PINK1 with several other serine/threonine kinases indicated a distinct substrate interaction sequence. Taken together, our data represent the first large-scale analysis of phosphoproteome after PINK1 depletion.
Results
The proteome is not changed in steady-state cells after PINK1 knock down
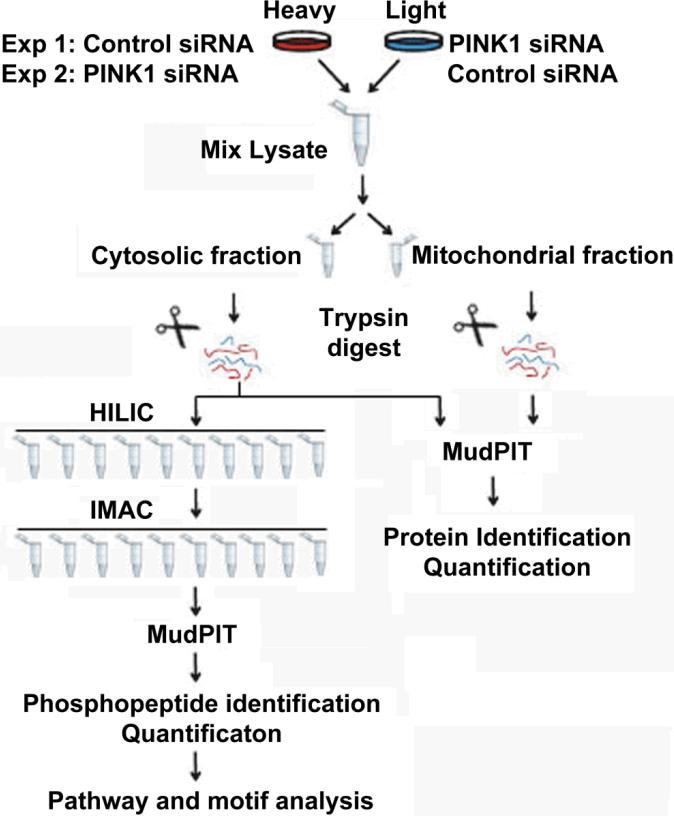
We hypothesize that the phosphorylation state of proteins downstream of PINK1 could be affected by knocking down the expression of PINK1, and designed a strategy depicted in Figure 1 to identify these changes. We label HEK293 cells with either regular medium (hence “light” cells) or “heavy” medium containing 13C15N enriched arginine and lysine (hence “heavy” cells). Small interference RNA with scrambled sequence (non-targeting siRNA) as a control or with specific sequence targeting human PINK1 (PINK1 siRNA) was applied to either heavy or light cells, respectively. In replicate experiments, application of the control or PINK1 siRNA to either heavy or light cells was swapped so that the quantitative changes in protein phosphorylation would be inverse to each other between these switching experiments. Homogenates from the two cell populations were mixed and half of the mixture were enriched for mitochondria followed by analysis using multi-dimensional protein identification technology (MudPIT).25 While the remaining mixture were digested with trypsin, fractionated by hydrophilic interaction chromatography (HILIC),20 and phosphopeptides were further enriched from each fraction using immobilized metal ion affinity chromatography (IMAC).17 Finally, the phosphopeptide-enriched fractions were pooled and analyzed by MudPIT to identify and quantify the phosphoproteome. Five percent of the digested lysates were also analyzed by MudPIT to quantify the proteome of the cell lysates. The ratio between the light and the heavy version of each peptide and phosphopeptide was calculated, which represents the quantitative differences of proteins or phosphorylation sites.26
Fig. 1.
Quantitative phosphoproteomic strategies to analyze the effect of PINK1 knocking down.
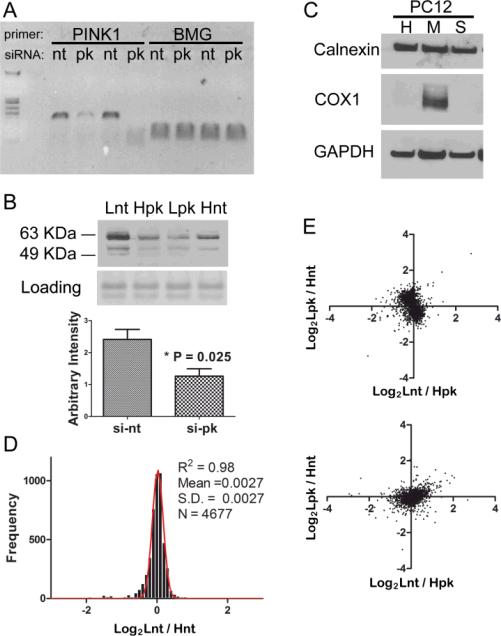
Forty-eight hours post transfection, effective knocking down of PINK1 mRNA was detected by RT-PCR in Figure 2A, comparing non-target siRNA (nt) to that of PINK1 siRNA (pk) transfection using human β2-microglobulin (BMG) as the loading control. Two sets of PINK1 siRNAs were used with slightly different efficiency of knocking down. PINK1 protein level was also reduced by half as determined by Western blot (Figure 2B). Because mounting evidence has converged on the notion that PINK1 primarily functions as a mitochondrial kinase,27,8 we also prepared mitochondrial fractions using a standard protocol. Enrichment of mitochondrial protein COX1 was evident, while there was no enrichment for the endoplasmic reticulum protein calnexin and marginal enrichment for the cytosolic protein GAPDH (Figure 2C).
Fig. 2.
The proteome was not altered after knocking down PINK1. (a) RT-PCR shows effective knocking down of PINK1 mRNA (pk) comparing to non-target control (nt), with β2-microglobulin (BMG) mRNA serving as a loading control. The specific amplicon of 116 bp is shown in the gel.(b) Western blot analysis shows that endogenous PINK1 protein is greatly reduced after introducing siRNA against PINK1. Both full length and processed PINK1 at an apparent molecular weight between 50-64 KDa are detected. Lnt: light-isotope labeled cells were treated with non-target siRNA, Hpk: heavy-isotope labeled cells were treated with PINK1 siRNA, Lpk and Hnt represent the labeling strategy in the swapped experiment. (c) Western blot analysis shows the enrichment of mitochondrial (M) components compared with whole cell homogenate (H) and soluble cell fraction (S). (d) The same batch of PC12 cells were grown in either light or heavy SILAC media, equal amount of total cellular protein were mixed and analyzed by the MudPIT procedure and thousands of proteins were quantified. The distributions of protein ratios between light and heavy cells follow normal distribution with very small deviation. (e) Two biological experiments with swap labeling show light versus heavy protein ratios representing PC12 cells transfected with either PINK1 siRNA or control siRNA . Both the cytosolic fraction (upper panel) and the mitochondria fraction (lower panel) were plotted.
To assess whether heavy isotope labeling has any effect on protein expression, we analyzed the mixture from an equal amount of light or heavy labeled rat pheochromocytoma cells (PC12). Histogram of log transformed ratios of 4677 non-redundant proteins fit well to a Gaussian distribution, with the measurement of goodness of fit (R2) equal to 0.98 (Figure 2D). This indicates that heavy isotope labeled PC12 cells express proteome at almost identical levels to that of the light cells. We also plotted protein expression ratios between the two swap-labeled controls and PINK1 deficient PC12 cells. Overall, there were no obvious shift of data points from the origin after PINK1 knocking down, as shown by the two correlation plot in Figure 2E (whole cell lysate) and Figure 2F (mitochondria fraction). Similar results were found in human embryonic kidney (HEK293) cells. The correlation plot of the quantitative results for whole cell lysates from the two swap labeling experiments (Figure 3A) showed a distribution centered on the origin without obvious changes, whereas only a few proteins had consistent alteration in the mitochondrial components (Figure 3B). These data suggest that knocking down PINK1 in either PC12 or HEK293 cells has no ostensible effect on proteome expression under normal conditions.
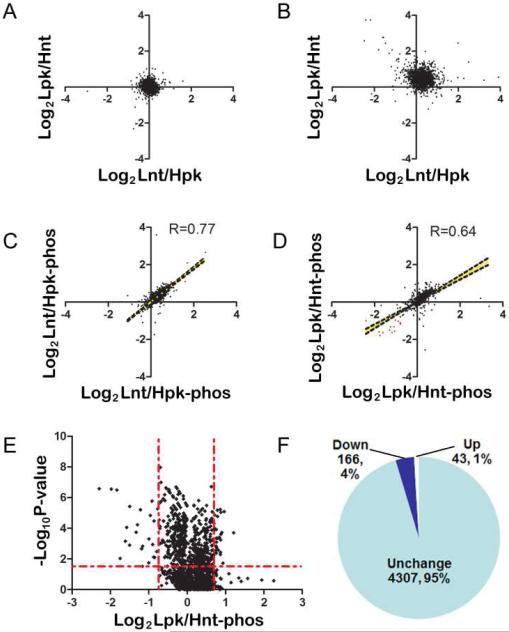
Fig. 3.
Identification of phosphoproteome changes after knocking down PINK1. (a) and (b) Similar to Figure 2e except that HEK293 cells were used in the experiment. (c) and (d) Quantification of phosphopeptide from two instrument runs of two biological experiments shows good correlation. The representation for the experiment scheme was the same as used in Figure 2. (e) Volcano plot of the phosphopeptide ratios shows a trend of reduced phosphorylation after knocking down PINK1. (f) Schematic representation of the quantitative phosphorylation results.
Identification of phosphoproteome changes after PINK1 down-regulation
HEK293 cells that showed effective knock down of PINK1 were used for SILAC experiments (Figure 2B right panel). To enrich for phosphopeptides, we fractionated the trypsin digested, 1:1 mixture of light and heavy cells using HILIC into 10 fractions. Phosphopeptides from each fraction were then captured using the IMAC approach. To ensure high quality identification of phosphopeptides, we used a 10-ppm precursor ion mass tolerance for all phosphorylated peptides and a false discovery rate at the peptide level lower than 1%. From two biological replicate experiments, we identified nearly five thousand phosphopeptides with phosphopeptide enrichment ratios ranged between 60% and 80%, consistent with previous reports. 28,19,23 From these phosphopeptides, we quantified forty five hundred unique ones (Supplemental Table 1), giving rise to over two thousand unique phosphoprotein groups.
Repetitive quantitative measurements of the enriched phosphopeptides were analyzed using Pearson's correlation algorithm. We found that the results were reproducible with a correlation coefficient of 0.77 and 0.64, respectively, and the P values for both experiments were statistically significant (P<0.0001, Figure 3, C and D). A total of 1658 out of the 4307 quantified phosphopeptides had replicate measurements, allowing us to calculate the probability values using T distribution and corrections using Benjamini-Hochberg method.29 A volcano plot of the p-values for the log-transformed phosphopeptide ratios between the PINK1 knockdown and control showed obvious down regulation of dozens of phosphopeptides although no change in the majority of the phosphopeptide ratios were observed (Figure 3E). Among the phosphopeptides with corrected P values less than 0.05, we found 166 with ratios less than 0.7 and 43 with ratios greater than 1.42. These results are summarized in Figure 3F.
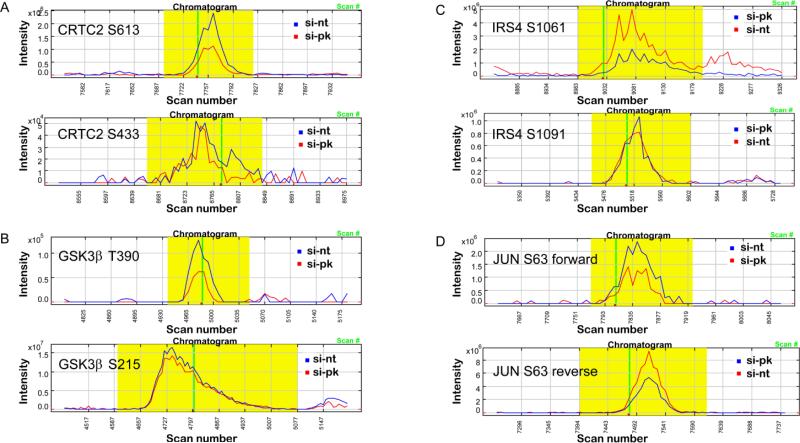
Several of the differentially phosphorylated proteins are shown in Figure 4. While phosphorylation of S613 was reduced by about two fold on CREB regulated transcription coactivator 2 (CRTC2), phosphorylation of S433 was unchanged (Figure 4A). An almost identical fold reduction in phosphorylation was found on T390 of glycogen synthase kinase 3β (GSK3β) and S1061 of insulin receptor substrate 4 (IRS4); while at the same time we found no change in phosphorylation on S215 of GSK3β and S1091 of IRS4 (Figure 4, B and C). Interestingly, we found a reciprocal reduction in phosphorylation on S63 of JUN transcription factor in the forward and reverse labeling experiments (Figure 4D). Taken together, these data indicate that although phosphorylation events of most proteins are not altered after reducing PINK1 protein level and presumably the kinase activity, there is an overall trend of reduction in phosphorylation in a subset of proteins.
Fig. 4.
Phosphorylation changes of several nuclear proteins. Reconstructed chromatograms of phosphopeptides showing phosphorylation sites from the following proteins: (a) CRTC2, (b) GSK3β, (c) IRS4, (d) JUN.
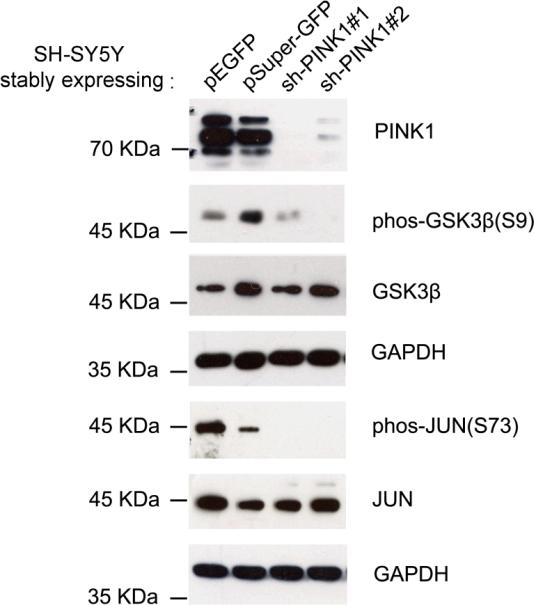
To validate phosphorylation changes detected by our proteomic approach, we created SHSY5Y cell lines stably expressing pEGFP and pSuper-GFP as controls, as well as the same cell lines expressing small hairpin RNAs (shRNA) against PINK1 with two different seed sequences. Western blot analysis showed that cells expressing both PINK1 shRNAs dramatically down regulated the expression of endogenous PINK1 protein (Figure 5). We used these cells to characterize phosphorylation levels of GSK3β and JUN. At steady state, while antibodies against unphosphorylated GSK3β and JUN detected no ostensible changes among all four cell lines, phosphorylation-specific antibodies against GSK3β at S9 and JUN at S73 showed dramatic reduction in phosphorylation in PINK1 deficient cells (Figure 5), consistent with our phosphoproteomic results.
Fig. 5.
Validation of protein phosphorylation changes. (a) SH-SY5Y cells stably expressing either pSuper-GFP vector or pEGFP vector were used as control cells to compare with SH-SY5Y cells expressing pSuper-GFP containing shRNA against PINK1 at two different exonic locations. Western blot analysis of the cell homogenates using antibodies against either non-phosphorylated or phosphorylated GSK3β and c-Jun. Stable cell lines expressing different plasmids are designated.
Bioinformatic analysis identifies nuclear proteins as potential downstream targets of PINK1
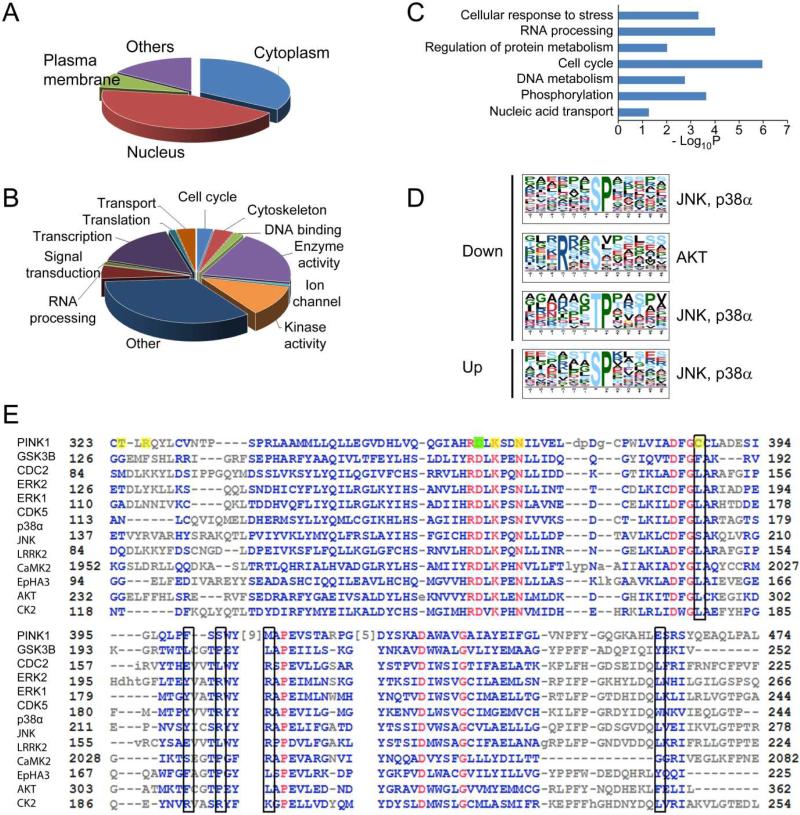
The 166 proteins with changed phosphorylation states were further analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID).30 Gene Ontology annotations indicated that about 7% of these proteins had plasma membrane localization, 34% were cytosolic proteins, and interestingly, 43% of these proteins had nuclear localization (Figure 6A). Functional annotation suggested that a large portion of these proteins had enzymatic activity (including kinases) as well as transcription activity. Proteins involved in RNA processing, transport, cell cycle and cytoskeletal organization compose a lower portion (Figure 6B). Fisher's exact test of the molecular pathways that these proteins can fit into indicates that pathways related to cell cycle, kinase signaling-mediated protein phosphorylation events, RNA processing, DNA metabolism, as well cellular response to stress were significantly enriched (Figure 6C).
Fig. 6.
Bioinformatics analysis of phosphorylation events. Gene Ontology analysis of changed phosphoproteins in two categories, (a) cellular localization and (b) biological function, indicates an enrichment of nuclear proteins, and proteins involved in DNA and RNA metabolism. (c) Pathway analysis indicates a significant over-representation of proteins in several pathways including cell cycle and cellular response to stress. (d) Motif analysis of the phosphopeptides with altered phosphorylation identified proline-directed kinase recognition motif, which was recognized by JNK, p38α, and Akt. (e) Alignment of PINK1 with that of many kinases both proximal and distal to PINK1 in the kinome tree, the sequence that covers substrate recognition residues is used for the alignment. The proton acceptor (D362) was labeled with a filled green box, and the rest of the substrate interacting residues including ATP binding sites were labeled with filled yellow boxes. The open black boxes indicates proposed +1 proline interacting residues.
Most kinases recognize substrate phosphorylation sites with well-defined amino acid sequence flanking the substrate residue.31 We extracted the amino acid sequence motifs using Motif-X, a pattern recognition algorithm32 that has been demonstrated valuable in previous phosphoproteomic studies.28 From the peptides that had reduced phosphorylation after PINK1 knock down, Motif-X extracted predominantly proline-directed motifs, which is recognized by kinases like JNK and p38α. 33 A basophilic serine phosphorylation site resembling the Akt recognition motif 33 was also found. For those phosphopeptides that had increased phosphorylation upon PINK1 knockdown, only the proline-directed motif resembling the JNK and p38α site was extracted (Figure 6D).
To better understand the substrate specificity and domain functions, we aligned PINK1 protein sequence with other kinases in the kinome tree with known substrate structure preferences including both nearest and distal neighbors (Figure 6E). The alignment showed an overall highly conserved catalytic domain between PINK1 and other kinases, especially residues surrounding the proton acceptors (D362, labeled with green box) (residues that extract protons from the hydroxyl group of serine, threonine or tyrosine) and ATP binding sites. However, more discrepancies were observed near the substrate binding region. In the alignment, known substrate interacting residues including ATP binding sites of PINK1 were labeled yellow. Beside the proton accepting site, other substrate interaction residues were not highly conserved between PINK1 and other kinases which suggests distinct substrate specificity. The black boxes in our alignment analysis shows proposed +1 proline interacting residues indicated from previous work on other proline-directed kinases including CDC2, ERK1 and CDK5. 34
Discussion
The causal link between the gene PINK1 and a subset of familial autosomal-recessive Parkinson's disease has been established for almost ten years. Although numerous studies on its cellular function followed and its kinase activity was well defined, identifying bona fide substrates of PINK1 kinase remained a daunting task. So far a majority of the studies converge on mitochondrial signaling downstream of PINK1 activation. PINK1 anchors to the mitochondria outer membrane upon stabilization and activation, with its kinase domain is shown to face the cytosol.35 It is therefore conceivable that cytosolic proteins could be phosphorylated and the signal event could be transduced to other cellular compartments. To test this hypothesis, we designed quantitative phosphoproteomic strategies to identify phosphorylation changes after transient knock down of PINK1 in different cell lines. We found that knocking down PINK1 in a relatively short period of time (2-4 days) has negligible influence on the overall proteome of either human or rodent cell lines under standard culture conditions. This is consistent with the general view that the major function of PINK1 is neuroprotection, which is activated primarily under cellular stress conditions. Among the changed phosphorylation events there was a trend of reduced phosphorylation after knocking down PINK1, suggesting that a basal level of PINK1 dependent phosphorylation and downstream signaling events is operating under steady-state conditions. A smaller fraction of phosphorylation events were found to be increased after PINK1 deficiency, possibly resulting from a negative feedback control mechanism of the kinase signaling network. Surprisingly, we found that the downstream signaling nodes included many transcription factors as well as nuclear proteins involved in DNA and RNA metabolism. Thus, PINK1 dependent phosphorylation signaling might influence nuclear activity. To our knowledge, this is the first large scale quantitative study of the phosphoproteome with loss of PINK1 function.
Up till now, designated features of PINK1 substrates are not fully elucidated. Our approach can only detect protein phosphorylation changes caused by PINK1 down regulation, while these proteins may or may not be the direct substrates of PINK1. Due to the lack of crystal structure of PINK1, it is still challenging to predict the structure characteristics of its substrates. Sequence alignment between PINK1 and other kinases showed an overall highly conserved catalytic domain surrounding the proton acceptors (D362) and ATP binding sites. In contrast, there are more discrepancies near the substrate binding region. It was previously assumed that substrates containing proline at +1 position of the phosphorylation site should not fit well into the catalytic pocket of most of serine/threonine kinases since most kinases are expecting to encounter a hydrogen-bond partner for the backbone substrate.34 However, more than a quarter of the sites identified in many large-scale phosphoproteomic studies as well as our results were proline-directed sites. It is hypothesized that proline-directed kinases can create a hydrophobic pocket to accommodate the proline ring.36 An arginine was predicted at the bottom of this hydrophobic pocket (corresponding to R170 of CDC2) to exclude the binding of other larger side chains and water molecules. Proline can be accommodated because it is the only natural secondary amine that does not need to dissociate the hydrogen bonded water molecule to come into this hydrophobic binding pocket.34 For PINK1, methionine at this position is also highly conserved between species.37 With a long tail and a hydrophobic bulky head, it is presumably guiding the proline residue into this hydrophobic pocket like arginine does. Regardless, obtaining the intact protein structure is greatly needed to fully understand its substrate selectivity.
The PINK1-Parkin pathway in regulating mitophagy probably plays an important role in the pathogenesis of familial PD that harboring PINK1 mutations.2,3,5,6,7 Whether this pathway operates in sporadic PD that comprise a much greater patient population remains to be determined. Our quantitative proteomic screen identified alterations in phosphorylation state from multiple kinase-mediated signal transduction pathways, some of these pathways may play a role in dopaminergic neuronal survival--potentially involved in the pathogenesis of sporadic PD. The reduced phosphorylation at serine/threonine sites in GSK3β and JUN found in the proteomic screen was substantiated by immunoblotting assay using phosphorylation site-specific antibody. Although the antibody we used recognized a different site (S9 in GSK3β and S73 in JUN) from the proteomic data, it was well known that phosphorylation of S9 in GSK3β is a priming event that facilitates subsequence phosphorylation of multiple sites,38 and that both S63 and S73 in JUN are phosphorylated upon activation.39 In support of our findings, Akundi et al. found that phosphorylation of Akt, GSK3β and ribosomal protein S6 were all reduced in PINK1 deficient mouse embryonic fibroblast cells in response to insulin-like growth factor-1 stimulation.40 We also found reduction in phosphorylation of multiple transcriptional regulators including JUN, CRTC2, NCOR1, NFATC4, etc. While the contribution of each signaling node to the pathogenesis of PD requires further dissection in great detail, overall our findings suggest that PINK1 may play important roles in multiple growth factor-dependent signal transduction and these processes may be involved in dopaminergic neuronal survival under physiological and stress-induced conditions.
Among thousands of phosphoproteins detected, we did not identify mitochondrial proteins with dramatic changes in phosphorylation. This is consistent with the idea that PINK1 is recruited to the mitochondrial outer membrane upon mitochondria depolarization or other stress conditions that compromise mitochondria, and execute its protective kinase functions. Under physiological condition the basal kinase activity of PINK1 is not known to act on mitochondrial substrates. Several recent studies have identified PINK1 kinase activity beyond mitochondria. For example, Murata et al. found that over expression of PINK1 can enhance phosphorylation of Akt via activation of mTORC2, a signal pathway associated with increased cell motility.41 Berthier et al. showed that PINK1 interacted with and phosphorylated the polycomb protein EED/WAIT1 thus sequestrated the repressor complex from the nuclei, resulting in changes in histone H3K27 trimethylation state and gene transcription.42 It was proposed that reactive oxygen species (ROS)-mediated mitochondria-to-nucleus signaling was a key to aging and radical-related diseases including Parkinson's disease.43 Mutations in PINK1 and other neural protective genes including Parkin and DJ-1 compromise neuron's detoxification ability against ROS-mediated mitochondrial damages over the patient's life span, leading to accumulative death of neurons. Further studies on the precise nuclear signaling events regulated by PINK1 under various physiological and pathological conditions are needed to expand our understanding of PINK1 function and its role in PD.
Methods
Oligos and siRNAs and antibodies
The primer sequences used in the RT-PCR and real-time PCR to quantify PINK1 mRNA are: CTACCGCTTCTTCCGCCAGT (forward) and AGCCCGAAGGCCAGAAAGAC (reverse). The SMARTpool siRNAs against human PINK1 and the non-target siRNA was purchased from Dharmacon, the sequences are: 1. GAAAUCCGACAACAUCCUU 2. GAGCAUCGGCCUGCAGUUG 3. GGAGCCAUCGCCUAUGAAA 4. GCAAAUGUGCUUCAUCUAA. The antibodies used for immunoblot analysis are listed below: rabbit against PINK1 (1:1000, Cayman Chemical, Ann Arbor, MI), rabbit against calnexin (1:1000, Abcam, Cambridge, MA), mouse antibodies against COX1 (1:1000) and GAPDH (1:1000) (Cell Signaling Technology, Danvers, MA).
Cell culture and sample preparation
Rat PC12 cell and human HEK293 cells were cultured in 10 cm culture plates using Dubelco’s minimal essential medium (DMEM) without lysine and arginine, and supplemented with 10% dialysed fetal bovine serum and 100 units/ml of penicillin together with 100 µg/ml of streptomycin. Either heavy isotope (12C15N)-enriched arginine and lysine (Spectra Isotopes, Cambridge, MA), or light isotope (12C14N)-enriched arginine and lysine (Sigma, St. Louis, MO) were added to the medium to make a complete SILAC medium. The cells were grown in either light or heavy medium for seven passages to ensure complete labeling of the heavy isotope (MCP 2002 Mann). In two biological replicate experiments, either heavy cell or light cells were transfected with PINK1 siRNA, while the respective light or heavy cells were transfected with non-targeting scrambled siRNA, using DharmaFect transfection reagents. Seventy two hours post transfection, the cells were collected with HEPES-buffered sucrose (10 mM HEPES, pH 7.4, 0.32 M sucrose, protease inhibitor cocktails (Roche, Mannheim, Germany), 2 mM NaF, 1 mM Na3VO4), light and heavy cells were mixed with equal number of cells and then homogenized. The lysates were centrifuged at 700 X g for 10 minutes to remove the cell debri. The resulting supernatant was designated cytosolic fraction and used for further analysis.
For mitochondrial preparation, cells were collected with the same HEPES-buffered sucrose solution and mixed with equal number of cells. A differential centrifugation method was used based on previously published protocol.44
Stable cell lines were created using the neuroblastoma SH-SY5Y cells based on a commercially available pSUPER RNAi system (OligoEngine, Seattle, WA). The two sequences targeting PINK1 were GGCAATTTTTACCCAGAAA (shPINK1#1) and GAAATCCGACAACATCCTT (shPINK1#2). The oligos containing the PINK1 target sequence of both sense and antisense strands were synthesized, annealed, and cloned into the pSUPER-GFP vector. Validated shPINK1-pSUPER-GFP clones were transfected into SH-SY5Y cells using Lipofectamine 2000 (Invitrogen, Calsbad, CA). The cells were maintained in DMEM medium supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, 100 μg/ml of streptomycin and selected with 600 μg/ml of G418. Single clones were picked and cultured separately and pooled clones were also collected. All resulting stable cells express GFP after the cell lines were created.
Detection of mRNA and protein levels after PINK1 knocking down
Real-time PCR analysis was performed using the ABI PRISMs 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) on the reverse transcribed total RNA preparations from the transfected cells. Primers were designed to direct against the exonic regions of PINK1 using Invitrogen's Primer Designer and their specificity for binding to the desired sequences was verified against the NCBI database. The amplicon size of 116 bp was used to visualize PINK1 mRNA levels.
For immunoblot analysis, 30 μg of protein from either whole cell lysate or mitochondrial preparation was used. Western blot analyses were performed on samples from at least duplicate experiments. All antibodies were obtained from commercial sources. The blots were scanned and band intensities were analyzed using AlphaEaseFC (Alpha Innotech, San Leandro, CA). The intensity values were background-subtracted, and arbitrary intensity values were obtained by dividing the intensity value of the loading control. Statistical analysis was performed on the normalized intensity values using unpaired Student-t test assuming two tailed distribution. A P value of greater than 0.05 was considered as statistically significant.
Phosphopeptide enrichment combining hydrophilic interaction chromatography (HILIC) and immobilized metal ion affinity chromatography (IMAC)
A previously published procedure was used to prepare cellular fractions for phosphopeptide enrichment. 45 Two milligrams of protein samples from the cytosolic fraction of the mixed light-and heavy-isotope encoded cell lysate were precipitated with cold acetone at -20°C overnight. The precipitates were centrifuged at 14,000 x g for 15 minutes at 4°C. The pellets were solubilized and reduced with 100 mM Tris-HCl/8 M urea/5 mM DTT 100 μl, cysteine alkylated with 10 mM iodoacetamide, and digested with 20 μg of trypsin at 37°C overnight. The digestion was terminated by adding TFA to 0.4%. The resulting peptide solution was desalted with SepPak cartridge (Waters, #WAT054955, 100 mg of C18 beads for 2.5 mg digest) according to the manufacturer's instructions, and lyophilized for HILIC separation. The HILIC separation was on a 13071TSKgel Amide-80 column (5 um, 4.6x250 mm) from TOSOH Biosciences based on a previously described method 46. The gradient was started with 80% of HILIC buffer B (98% acetonitrile with 0.1% TFA, buffer A composed of 98% water and 0.1% TFA) running at 0.5 ml/min. The separation gradient is as follows: the constant flow is set at 0.5 ml/min, and followed by 5 minutes 80% B, 40 minutes 80-60% B, 5 minutes 60-10% B, 5 minutes 10% B, and 5 minutes 80% B. Fractions were collected every 5 minutes from the start of the gradient until 55 minutes and a total of 11 fractions were collected. All fractions were snap-frozen in liquid nitrogen and lyophilized. Each fraction was dissolved in 400 µl of IMAC sample buffer (250 mM acetic acid/40% acetonitrile, pH 2.5-3.0), and phosphopeptides were enriched with PHOS-Select Iron Affinity Gel slurry (Sigma, St. Louis) based on a published method 17 using a pull-down approach instead of the StageTip approach describe in the reference, and eluted with 400 mM ammonium hydroxide.
Analysis of phosphopeptides by multi-dimensional protein identification technology (MudPIT) and linear iontrap-orbitrap mass spectrometer
For each combined phosphopeptide sample, two 10-step MudPIT 25 experiments were performed to maximize the coverage. Peptides were pressure-loaded onto a 100 μm i.d. fused silica capillary column packed with strong cation exchanger (SCX, Whatman, Clifton, NJ) and a C18 material (Phenomenex, Ventura, CA), with the SCX end fritted with immobilized Kasil 1624 (PQ Corperation, Valley forge, PA). After desalting, an analytical column of 100-μm i.d. capillary packed with another C18 material was attached to the SCX end with a ZDV union, and the column was placed in line with a HPLC pump as a nanospray ionization source, and interfaced with an LTQ-Orbitrap mass analyzer. The buffer solutions were: 5% acetonitrile/0.1% formic acid (buffer A); 80% acetonitrile/0.1 % formic acid (buffer B), and 500 mM ammonium acetate/5 % acetonitrile/0.1 % formic acid (buffer C). The first step consisted of a 100 min gradient from 0-100 % buffer B. Each of the remaining steps are composed of a 5 min of step-increased salt buffer (buffer C), a 10 min gradient from 0-15% buffer B, and a 130 min gradient from 15-45% buffer B, followed by a 20 min gradient increase to 100% buffer B, and a reverse of gradient to 100% buffer A. As peptides were eluted from the microcapillary column they were electrosprayed directly into the LTQ-Orbitrap (Thermo, San Jose, CA) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan with 60,000 resolution at 400 m/z by Orbitrap (400-1400 m/z) followed by five data-dependent MS/MS scan by LTQ were repeated continuously throughout each step of the multidimensional separation.
Identification, quantification of phosphopeptides and phosphoproteins
The MS/MS spectra were searched with the ProLucid algorithm47 against an IPI rat or human database (Version 3.48, released at 2008/09/01: ftp://ftp.ebi.ac.uk/pub/databases/IPI/) that was concatenated to a decoy database in which the sequence for each entry in the original database was reversed.48 To identify phosphopeptides, the search parameters included a differential modification on serine, threonine and tyrosine residues of 79.9663 amu, indicating the addition of phosphorous group(s) on those residues. The database search results were assembled and filtered using the DTASelect program 49.
The assembled database matching file was used to obtain quantitative ratios between the light and heavy version of each peptide using the software Census.50 The ratio between the light and the heavy version of the peptides is the surrogate of the protein expression ratio. And the ratio between the light and the heavy version of the phosphopeptide represented the difference in the phophorylation level at the identified site after knocking down PINK1.
Statistical and bioinformatics analysis
Two batches of the experiments were performed to generate biological replicates. To assess the reproducibility of mass spectrometry measurement, two MudPIT runs were performed in the first batch of samples. To test the significance of changed phosphorylation events, Student's T-test was used when the phosphopeptides were quantified in both biological replicates and multiple hypothesis testing was performed using Benjamini-Hochburg method.
To extract common sequence similarities among the changed phosphorylation sites, Motif-X algorithm32 was applied to extract amino acid sequence patterns flanking the phosphorylation sites. The identified phosphopeptides were first evaluated using a binomial probability based algorithm (A-score) 51 to obtain confidence levels of the site localization. Only highly confidently localized phosphorylation sites (P<0.05) were used for motif analysis. To be considered as motifs, we allow the occurrences of the same pattern to be at least 20 times, and a significance value of less than 0.000001. The IPI human protein database was used as the background reference database.
Gene Ontology and pathway analysis was performed using the online bioinformatics tool Database for Annotation, Visualization and Integrated Discovery (DAVID).30 Phosphoproteins with changed phosphorylation sites were loaded into the DAVID knowledgebase. As a control, we randomly generated the same number of phosphoproteins from the list of proteins with no change in phosphorylation, and mapped into the signal pathways in Ingenuity with identical parameters.
Supplementary Material
Acknowledgements
The authors would like to acknowledge financial support from the US National Institutes of Health grant 5R01 MH067880-02 to JRY, the New investigator fund from Shanghai Institute Biochemistry and Cell Biology, and the National “985” Project grant from ECNU to LL. We would also like to thank Drs. Zhiqi Xiong, Zilong Qiu and Xiaotao Li for generously providing backbone plasmids to clone shRNAs as well as antibodies. Fianly, we thank Dr. Xiaotao Li for critically reading the manuscript and Sung Kyu Park for assistance in phosphorylation site localization.
Footnotes
Conflict of Interest
The authors declare no conflict of interest for this study.
References
- 1.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304(5674):1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 2.Cookson MR, Dauer W, Dawson T, Fon EA, Guo M, Shen J. The roles of kinases in familial Parkinson's disease. J Neurosci. 2007;27(44):11865–8. doi: 10.1523/JNEUROSCI.3695-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105(32):11364–9. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105(5):1638–43. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okatsu K, Oka T, Iguchi M, Imamura K, Kosako H, Tani N, Kimura M, Go E, Koyano F, Funayama M, Shiba-Fukushima K, Sato S, Shimizu H, Fukunaga Y, Taniguchi H, Komatsu M, Hattori N, Mihara K, Tanaka K, Matsuda N. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun. 2012;3:1016. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pridgeon JW, Olzmann JA, Chin LS, Li L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007;5(7):e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340(6131):471–5. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147(4):893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J, Batchu VV, Schubert M, Castellani RJ, Russell JW. A novel PGC-1alpha isoform in brain localizes to mitochondria and associates with PINK1 and VDAC. Biochem Biophys Res Commun. 2013;435(4):671–7. doi: 10.1016/j.bbrc.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, Chen F, Gu Y, Hasegawa H, Salehi-Rad S, Wang L, Rogaeva E, Fraser P, Robinson B, St George-Hyslop P, Tandon A. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem. 2005;280(40):34025–32. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 11.Song S, Jang S, Park J, Bang S, Choi S, Kwon KY, Zhuang X, Kim E, Chung J. Characterization of PINK1 (PTEN-induced putative kinase 1) mutations associated with Parkinson disease in mammalian cells and Drosophila. J Biol Chem. 2013;288(8):5660–72. doi: 10.1074/jbc.M112.430801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1(5):252–62. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 14.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;2005(267):pl2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 15.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 16.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7(12):952–8. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 17.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3(10):1630–8. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinkse MW, Mohammed S, Gouw JW, van Breukelen B, Vos HR, Heck AJ. Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J Proteome Res. 2008;7(2):687–97. doi: 10.1021/pr700605z. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 20.McNulty DE, Annan RS. Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol Cell Proteomics. 2008;7(5):971–80. doi: 10.1074/mcp.M700543-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Kweon HK, Hakansson K. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal Chem. 2006;78(6):1743–9. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 22.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat Methods. 2005;2(8):591–8. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 23.Liao L, McClatchy DB, Park SK, Xu T, Lu B, Yates JR., 3rd Quantitative analysis of brain nuclear phosphoproteins identifies developmentally regulated phosphorylation events. J Proteome Res. 2008;7(11):4743–55. doi: 10.1021/pr8003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundby A, Andersen MN, Steffensen AB, Horn H, Kelstrup CD, Francavilla C, Jensen LJ, Schmitt N, Thomsen MB, Olsen JV. In vivo phosphoproteomics analysis reveals the cardiac targets of beta-adrenergic receptor signaling. Sci Signal. 2013;6(278):rs11. doi: 10.1126/scisignal.2003506. [DOI] [PubMed] [Google Scholar]
- 25.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 26.MacCoss MJ, Wu CC, Liu H, Sadygov R, Yates JR., 3rd A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal Chem. 2003;75(24):6912–21. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, Kitada T, Kim JM, Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377(3):975–80. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 28.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 30.Huang DH, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Manning BD, Cantley LC. Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE. 2002;2002(162):pe49. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23(11):1391–8. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 33.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, DeMaggio AJ, Hoekstra MF, Blenis J, Hunter T, Cantley LC. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16(11):6486–93. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105(33):12022–7. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubersax JA, Ferrell JE., Jr. Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8(7):530–41. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 37.Woodroof HI, Pogson JH, Begley M, Cantley LC, Deak M, Campbell DG, van Aalten DM, Whitworth AJ, Alessi DR, Muqit MM. Discovery of catalytically active orthologues of the Parkinson's disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 2011;1(3):110012. doi: 10.1098/rsob.110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frame S, Cohen P, Biondi RM. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001;7(6):1321–7. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 39.Cho YY, Tang F, Yao K, Lu C, Zhu F, Zheng D, Pugliese A, Bode AM, Dong Z. Cyclin-dependent kinase-3-mediated c-Jun phosphorylation at Ser63 and Ser73 enhances cell transformation. Cancer Res. 2009;69(1):272–81. doi: 10.1158/0008-5472.CAN-08-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akundi RS, Zhi L, Bueler H. PINK1 enhances insulin-like growth factor-1-dependent Akt signaling and protection against apoptosis. Neurobiol Dis. 2012;45(1):469–78. doi: 10.1016/j.nbd.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murata H, Sakaguchi M, Jin Y, Sakaguchi Y, Futami J, Yamada H, Kataoka K, Huh NH. A new cytosolic pathway from a Parkinson disease-associated kinase, BRPK/PINK1: activation of AKT via mTORC2. J Biol Chem. 2011;286(9):7182–9. doi: 10.1074/jbc.M110.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berthier A, Jimenez-Sainz J, Pulido R. PINK1 regulates histone H3 trimethylation and gene expression by interaction with the polycomb protein EED/WAIT1. Proc Natl Acad Sci U S A. 2013;110(36):14729–34. doi: 10.1073/pnas.1216844110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Storz P. Reactive oxygen species-mediated mitochondria-to-nucleus signaling: a key to aging and radical-caused diseases. Sci STKE. 2006;2006(332):re3. doi: 10.1126/stke.3322006re3. [DOI] [PubMed] [Google Scholar]
- 44.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured filroblasts. Nature Protocols. 2007;2(2):9. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 45.Liao L, Sando RC, Farnum JB, Vanderklish PW, Maximov A, Yates JR. 15N-labeled brain enables quantification of proteome and phosphoproteome in cultured primary neurons. J Proteome Res. 2012;11(2):1341–53. doi: 10.1021/pr200987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNulty DE, Annan RS. Hydrophilic interaction chromatography for fractionation and enrichment of the phosphoproteome. Methods Mol Biol. 2009;527:93–105. x. doi: 10.1007/978-1-60327-834-8_8. [DOI] [PubMed] [Google Scholar]
- 47.Xu TV, J. D., Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell. Proteomics. 2006;5(S174) [Google Scholar]
- 48.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 49.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SK, Venable JD, Xu T, Yates JR., 3rd A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5(4):319–22. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–92. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.