Summary
Spatial regression models have grown in popularity in response to rapid advances in GIS (Geographic Information Systems) technology that allows epidemiologists to incorporate geographically indexed data into their studies. However, it turns out that there are some subtle pitfalls in the use of these models. We show that presence of covariate measurement error can lead to significant sensitivity of parameter estimation to the choice of spatial correlation structure. We quantify the effect of measurement error on parameter estimates, and then suggest two different ways to produce consistent estimates. We evaluate the methods through a simulation study. These methods are then applied to data on Ischemic Heart Disease (IHD).
Keywords: Attenuation, Environmetal epidemiology, Geostatistics, Measurement Error, Mixed models, Random effects, SEIFA, Sensitivity, Spatial correlation, Spatial linear regression
1. INTRODUCTION
Advances in statistical methodology, together with the geographically referenced health databases, present an unique opportunity to investigate the environmental, social and behavioural factors underlying geographic variations (Elliott and Wartenberg, 2004). In health research, for example, social epidemiologists seek to assess the impact of socio-demographic characteristics of a community on the health of individuals living in that community (Elliot et al., 2000). Analysis of geo-coded data is complicated by correlations among observations located near each other. Regression analysis ignoring these spatial correlations leads to incorrect inference on the estimated regression coefficients by narrowing confidence intervals. Mixed effect models provide a convenient way of modelling spatial correlations by incorporating random effects with spatial correlation structure (Waller and Gotway, 2004). In this paper, we focus on how such models perform when covariates of interest are measured with error.
In the case study that motivates this paper, Australian researchers explored the relationship between the SEIFA index (an area-based measure of socio-economic status produced by the Australian Bureau of Statistics) and acute hospitalization for Ischemic Heart Disease (IHD) for approximately 600 postcodes in NSW, Australia (Burden et al., 2005; Guha et al., 2009). Regression models suggest a strong association between SEIFA and IHD, even after adjusting for factors such as age, gender, population density and other factors that might influence the outcome. However, exploratory analysis reveals that the estimated coefficient of the SEIFA index from such models depends strongly on the assumed spatial correlation structure. Briefly, the estimated SEIFA coefficients are all significantly negative, confirming that IHD rates decrease as social advantage increases. However, the magnitude of the effect varies by more than a factor of 2, depending on whether or not a spatial correlation adjustment is made. Similar sensitivity to assumed spatial correlation structure can be seen in analysis of the well-known Scottish Lip Cancer data (Breslow and Clayton, 1993; Clayton et al., 1993). In another spatial epidemiological study Molitor et al. (2007) fit a model for the effect of NO2 exposure on lung function. They considered a series of models included one based on a conditional autoregressive (CAR) model. They observed that models with spatial structure give smaller effect estimates as compared to models without spatial structure. These results suggest that estimated coefficients from a spatial regression model can be highly sensitive to whether and how spatial variation is accommodated. In this paper, we show that such sensitivity is specially likely to occur when the covariate of interest has been measured with error.
Presence of measurement error in the covariate of interest arises in many epidemiological and socio behavioural studies. For example, in the study of geographical variation in bladder cancer rates, lung cancer risk might be included in the model as a proxy for smoking exposure (Clayton et al., 1993). In environmental epidemiology, individual air pollution exposures might be approximated by the distance from the polluted sites or by using the measures at a few monitoring sites (Carroll et al., 1997). Further examples include geographical studies relating cancer incidence and mortality to dietary intakes (Cook and Pocock, 1983; Prentice and Sheppard, 1990).
Many papers have appeared in the literature over the years on covariate measurement error in the context of independent data (Carroll et al., 2006; Fuller, 2009; Wansbeek and Meijer, 2000; Ruppert et al., 2009). In case of linear regression with independent data, it is well known that presence of exposure measurement error causes estimated regression coefficients attenuate toward the null. However, relatively few have addressed the effect of exposure measurement error in the context of correlated data with spatial structure. In epidemiological studies of association between air pollutants and health outcome, typically data are available from few monitoring sites. Therefore, the measured exposure used in the analysis might be different from the underlying true exposure.
Xia and Carlin (1998) presented a spatio-temporal analysis of spatially correlated data with errors in the covariates, in the context of disease mapping. The authors empirically studied several alternative measurement error models using a metropolis gibbs algorithm. Li et al. (2009) derived asymptotic bias expressions for estimated regression coefficients in the context of a spatial linear mixed model. They showed that the regression estimates obtained from naive use of an error prone covariates attenuates the estimated regression coefficient and variance component estimates are inflated. They proposed the use of a maximum likelihood approach based on the EM algorithm to adjust for measurement error under the assumed error structure. However, their simulation assumes that the measurement error variance is known and they did not assess the performance of their method in the case of misspecification. Their approach is also subject to a high computational burden and may lead to spurious result in the presence of outliers or model misspecification (Gryparis et al., 2009; Szpiro et al., 2011). Furthermore, Szpiro et al. (2011) argued that in the presence of spatial correlation, joint modelling becomes challenging as it is very difficult to separate out the spatial correlation between exposure and outcome.
In this paper, we explore the sensitivity of estimated regression coefficients in spatial regression models, showing that it arises in settings where the covariate of interest has been measured with error. We show that ignoring measurement error attenuates estimated regression coefficients and observe that estimates can be very sensitive to the choice of assumed correlation structure in the model formulation. We derive expressions for the bias when measurement error is ignored and present some technical derivations that characterize the bias as a function of the degree of measurement error as well as the degree of spatial correlation in the covariate of interest and in the residuals. We show that the bias due to attenuation depends on the spatial correlation structure. When there is no or the same degree of spatial correlation in both covariate or the measurement error the bias in spatial linear model reduces to the familier attenuation factors under OLS modelling of independent data, namely , where is the variance of the true covariate and is the variance of the measurement error.
Based on these expressions, we propose two different strategies for obtaining consistent estimates: (i) adjusting the estimates using an estimated attenuation factor; and (ii) using an appropriate transformation of the error prone covariate. We then evaluate the performance of these two approaches via simulations. These approaches do not require complex programming and can be implemented via readily available mixed model software. Moreover, we suggest ways to estimate measurement error variance from the data rather than assuming measurement error variance as a known quantity. Our simulation results show that bias correction methods using the estimate of the measurement error work reasonably well in obtaining consistent estimates. However, estimation of the measurement error variance requires additional data or assumptions related to the underlying measurement error process. In the case of spatial epidemiology, validation data are typically rare. Therefore we suggest employing a sensitivity analysis when dealing with measurement error problems in practice. We illustrate the methods using data on Ischemic Heart Disease (IHD) and conclude with some practical guidelines.
2. MODEL FORMULATION
Suppose that Xi represents the true covariate of interest for spatial location i, i = 1, …, n, and suppose that it is related to an outcome Yi according to a linear model:
| (1) |
where ∊ = (∊1, …, ∊n)T ~ N(0, ∑∊) and ∑∊ is a covariance matrix, for now kept arbitrary. Let Wi be the observed covariate for spatial location i, related to the true covariate according to a classical measurement error model:
where U = (U1, …, Un)T ~ N(0, ∑U). When X = (X1, …, Xn)T is also normally distributed (say with mean μX and covariance ∑X), straightforward algebra establishes that Y = (Y1, …, Yn)T and W = (W1, …, Wn)T have a multivariate normal distribution,
where 1 is an n × 1 vector of ones. Standard theory for the multivariate normal establishes that Y∣W is normally distributed with conditional mean
| (2) |
and conditional variance
where
| (3) |
For ease of discussion, assume that the variable X has been centered so that μX = 0. In direct analogy with standard measurement error settings, these results suggest that regression coefficients obtained by regressing the outcome (Y) on the observed, but error prone covariate (W) will lead to bias as well as inaccurate variance modelling. We proceed now to explore the nature of this bias under varying assumptions about the correlation structure for Y, X and the measurement error term, U.
3. ASYMPTOTIC BIAS ANALYSIS
Suppose we fit model (1), naively replacing X with the error prone version of the covariate W and assuming independence of the error terms in the model on Y. The ordinary least squares estimate of β is
| (4) |
where W* is the n × 2 matrix with elements of the first column all equal to 1 and second column corresponding to the n × 1 vector W. Under the true model and assuming μX = 0, it is straight forward to show that the limiting value of this estimate is
where ρols = trace (∑X)/trace (∑X + ∑U), see the Appendix.
Using basic properties of the trace function, this simple formula leads to a number of interesting observations. For example, suppose that both ∑X and U have constant diagonal elements and , respectively, then the bias factor can be written as . This is the standard measurement error result (see Carroll et al. 2006), namely that the estimated regression coefficient is biased towards the null by an attenuation factor that reflects the proportion of the variability in the observed covariate W, explained by the true covariate X. Note that there is no bias in the estimated intercept in this case since we have assumed that X has mean zero. It is interesting to note that the result holds regardless of the correlation structures on the error term, ∑∊.
In the next section, we consider the bias associated with fitting a generalized least squares model in the presence of covariate measurement error. We will see that in this case, the degree of bias also depends on the assumed error structure.
3.1. Generalized Least Squares
Suppose we obtain a generalized least squares (GLS) estimator of β, under that assumption that the error term ∊ has covariance matrix ∑a, with the subscript “a” denoting “assumed”. For fixed ∑a, the estimator is:
| (5) |
In the limit under the true model and following similar arguments as in the OLS case, this estimate converges in probability to
where .
As in the OLS case, this simple formula also yields a number of interesting observations with important practical implications. First of all, because we can write
| (6) |
it follows that there will always be an attenuation of the estimated regression coefficient towards the null.
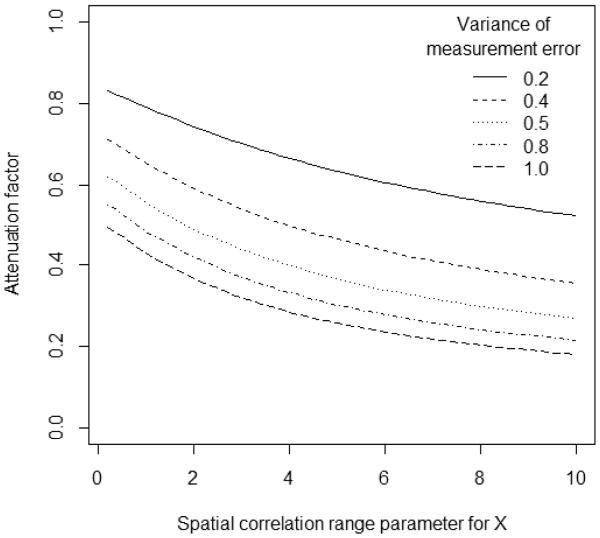
The following figure shows the attenuation factor associated with fitting generalized least squares, ρgls, under the assumption that X and ∊ each have unit variance and an exponential spatial covariance structure with the correlation between two observations a distance h units apart is given by Cor(h) = exp(−h/τ), where τ denote the range.
Each line in Figure 1, corresponds to a unique value of , the measurement error variance. The x-axis in the figure varies according to the value of the range parameter τx, which reflects the strength of the spatial correlation in the true covariate X. All calculations in the figure assume that there is zero spatial correlation in the measurement error term, U. Note that, as the range parameter goes to zero (τx → 0), the attenuation factor becomes identical to that which would be obtained if OLS were used instead of GLS. Of course, these results could change in the presence of other covariates in the model (see Zeger et al. 2000; Schwartz and Coull 2003).
Figure 1.
Attenuation factor associated with varying degree of measurement error.
From equation (6), it is clear that the two attenuation factors, ρgls and ρols, will equal the familiar attenuation factor under OLS modelling for independent data, namely , under a variety of circumstances, including:
and . That is, there is no spatial correlation in X or U and both random variables have homogenous variance.
When the degree of spatial correlation is the same for X and the measurement error, U. That is, and , where R is a spatial correlation matrix.
Note that these results holds regardless of the value of ∑a, the assumed correlation for the residuals in regression models. In practice, ρgls and ρols may differ depending on how well the assumed spatial correlation structure resembles the true process of the underlying covariance structure.
In the next section, we propose several approaches to adjust for measurement error in spatial regression settings.
4. BIAS CORRECTION
In the previous section, we have shown that presence of measurement error in covariates attenuates estimated regression coefficient to the null. A consistent estimate of the true regression coefficient can be obtained if we can estimate the various parameters that govern the measurement error process. This is possible if we have access to a validation data set without measurement error (Carroll et al., 2006). In the context of spatial epidemiology, however, validation data are rarely available. Therefore, we need additional assumptions to estimate the components of the attenuation factor. Without such assumptions or validation data, the measurement error and the true residual error variance are not identifiable in both case. In this paper we considered two different sets of assumptions that lead to the model identifiability. The first approach assumes that the true covariate X is smooth and that any observed nugget effect must be measurement error. The second assumes that measurement error variance is fixed and known over a feasible range. A sensitivity analysis is then carried out over the feasible range of known measurement error variance. Similar to Li et al. (2009), we assume that the underlying covariate process {Xi} defined in section 2 contains all the spatial correlation and that the measurement error is pure noise i.e., . Under this assumption, the attenuation factor, from equation (6) becomes
| (7) |
The OLS version can be obtained from the special case where .
We examine two different bias correction strategies to obtain a consistent estimate of the regression coefficient. The first approach deals with estimation of each of the components of ρgls and the second uses a linear transformation of the error prone covariate, W.
Both methods require knowledge of ∑X and or their estimated values. We estimated ∑X and by fitting the error prone covariate (W) in an intercept only model with an assumed spatial correlation structure. Under the assumption that measurement error is pure noise and ∑X, is a smooth spatial covariate with no nugget, the above model gives us a maximum likelihood estimate of the nugget effect in W, which corresponds to . Similarly, fitting Y on W with spatial correlation structure give us a maximum likelihood estimate of the underlying residual covariance structure, ∑∊. The first method additionally requires an estimate of ∑a.
4.1. Method I: Method of Moments
This method involves post analysis adjustment of the estimated regression coefficient using an estimate of the attenuation factor. Ignoring the measurement error and performing a likelihood analysis under the assumed covariance structure of Y∣X using W instead of X results in estimates denoted by or depending on whether ordinary or generalized least squares has been used. Let be the estimate of the corresponding slope from the above regression, where for the ease of exposition we leave off the superscript ‘ols’ or ‘gls’. We have shown that its limiting value is ρβ1. Denote its variance by . We then define an adjusted estimate, where is an estimate of the attenuation factor defined at equation (7) and where the estimated variance . An estimate of ρ is obtained by substituting and in equation (7).
4.2. Method II: Transformation method
Recall from equation (2) that E(Y∣W) = β01 + β1(I − Λ)μX + β1ΛW, where Λ = (∑X(∑X + ∑U)−1. This suggests the use of a linear transformation of W to achieve an appropriate linear regression model that can be fitted to yield a consistent estimate of β. Specifically, letting T = μX + Λ(W − μX), it follows that T ~ (μX, Λ∑X) and Cov(Y, T) = β1Λ∑X. Hence using the joint normality of W and Y, we have E(Y∣T) = β0 + β1T, and .
Define as the estimator of T, obtained by substituting in consistent estimates of μX and Λ. The outcome Y can then be regressed on , with an assumed spatial correlation structure, via a linear mixed model to obtain a consistent estimate of β1 and corresponding standard error.
5. SIMULATIONS
We conducted a simulation study to evaluate the finite sample properties of two methods proposed in the previous section to adjust for measurement error. We simulated 100 sample locations randomly within a d × d rectangular grid, where d is taken to have a value of either 40 or 80. Specifically, the ith random sample location si was generated by simulating two coordinates (e.g., latitude and longitude) from a Uniform[0, d] distribution. Given the set of si’s, the unobserved true covariate X was generated with mean 0 and covariance matrix ∑X, where ∑X was assumed to have an exponential correlation structure with unit variance with unit variance. This implies that the correlation between two observations distance h units apart is (1 − ηx) * exp(−h/τx), where τx is the range parameter and ηx characterizes the so called nugget effect. We considered three different range parameters (τx = 1, 5, 10) resulting in minimal, moderate and high correlation among the values of X’s with a nugget effect of ηx = 0.1.
The observed error-prone versions, W, of the true covariate were generated by adding Gaussian noise with variance to X. Outcome data, Y, were then generated according to equation (1), the slope and intercept parameter are taken as (β0, β1)T = (1, 2)T and the error variances were generated using a similar exponential correlation structure as ∑X, but with different range parameters. We also add a random Gaussian noise to the residual error variance (nugget effect).The variance parameter and the nugget for the residual error was taken as 0.5 and 0.1, respectively.
To generate simulated data with exponential spatial correlation and also in model fitting, we used the nlme package (Pinheiro et al., 2013). To extract the covariate matrices from the object of lme fit we used the mgcv package (Wood, 2006) in R (R Core Team, 2013).
To study the performance of our proposed methods under various degree of correlations within the rectangular grid and for various values of the measurement error variance, we simulated data based on various combinations of measurement error variances and 0.5). To simplify our presentation, only the results with measurement error variance in an 80 × 80 grid scales are illustrated. In general, the results obtained by varying the measurement error and/or size of the grid are similar.
Table 1 shows the average of the estimated regression coefficients, empirical standard errors and average of the estimated standard errors under 9 different combinations of spatial correlation in the covariate X and in the error for the model Y given X, based on 1000 simulations. The first column of table 1 specifies the combination of range parameters (τX, τ∊) used in that particular simulation. The 2nd and 3rd columns shows the estimated regression parameters under ordinary least squares based on using the true covariate X and the error prone covariates W, respectively (see equation 4). The 4th column shows the results from fitting a linear mixed model (using the ‘lme’ function in R) with assumed exponential correlation structure, but without adjusting for measurement error. The 5th and 6th columns present the bias corrected estimates of the regression parameter β1 using Method I and Method II, respectively. The next two columns of the table represent the results from method I and method II when true measurement error variances were used instead of estimated values. That is, results in column 5 use the estimated measurement error and column 7 uses the true value of the measurement error variance under method I. Similarly, columns 6 and 8 represent the results obtained using method II based on the estimated and true measurement error variances, respectively. The last column of the table shows results from method II when all the components of Λ were calculated using true values (i.e., values used in data generation).
Table 1.
Simulation results using different combinations of range parameters. Reported numbers are averaged over 1000 simulations with 100 observations per simulation with measurement error variance 0.2.
| Bias corrected lme using | ||||||||
|---|---|---|---|---|---|---|---|---|
| Range* | OLS | lme | estimated | true | true Λ | |||
| (τX, τ∊) | using X | using W | using W | Method1 | Method2 | Method1 | Method2 | Method2 |
| Estimated coefficient | ||||||||
|
| ||||||||
| (1,1) | 1.999 | 1.689 | 1.683 | 1.867 | 1.838 | 2.039 | 2.000 | 1.995 |
| (1,5) | 2.000 | 1.692 | 1.682 | 1.886 | 1.844 | 2.048 | 2.001 | 1.997 |
| (1,10) | 2.001 | 1.691 | 1.681 | 1.889 | 1.849 | 2.038 | 2.001 | 1.995 |
| (5,1) | 1.999 | 1.682 | 1.665 | 2.075 | 1.987 | 2.039 | 1.990 | 1.995 |
| (5,5) | 2.002 | 1.687 | 1.641 | 2.106 | 1.998 | 2.051 | 1.988 | 1.998 |
| (5,10) | 2.004 | 1.687 | 1.630 | 2.106 | 2.000 | 2.039 | 1.986 | 1.997 |
| (10,1) | 2.000 | 1.666 | 1.638 | 2.113 | 2.013 | 2.040 | 1.990 | 1.996 |
| (10,5) | 2.002 | 1.668 | 1.584 | 2.151 | 2.028 | 2.050 | 1.984 | 1.996 |
| (10,10) | 2.005 | 1.670 | 1.562 | 2.173 | 2.048 | 2.037 | 1.982 | 1.997 |
|
| ||||||||
| Empirical Standard error | ||||||||
|
| ||||||||
| (1,1) | 0.075 | 0.097 | 0.099 | 0.473 | 0.321 | 0.138 | 0.126 | 0.115 |
| (1,5) | 0.077 | 0.099 | 0.099 | 0.614 | 0.331 | 0.147 | 0.127 | 0.115 |
| (1,10) | 0.077 | 0.099 | 0.095 | 0.676 | 0.349 | 0.142 | 0.123 | 0.112 |
| (5,1) | 0.079 | 0.104 | 0.107 | 0.583 | 0.409 | 0.145 | 0.131 | 0.120 |
| (5,5) | 0.091 | 0.110 | 0.113 | 0.753 | 0.508 | 0.178 | 0.139 | 0.123 |
| (5,10) | 0.098 | 0.116 | 0.115 | 0.768 | 0.512 | 0.172 | 0.143 | 0.127 |
| (10,1) | 0.083 | 0.114 | 0.121 | 0.494 | 0.334 | 0.176 | 0.139 | 0.125 |
| (10,5) | 0.102 | 0.126 | 0.142 | 0.616 | 0.418 | 0.247 | 0.159 | 0.137 |
| (10,10) | 0.117 | 0.134 | 0.145 | 0.692 | 0.469 | 0.210 | 0.165 | 0.142 |
|
| ||||||||
| Average of estimated standard errors | ||||||||
|
| ||||||||
| (1,1) | 0.075 | 0.100 | 0.099 | 0.110 | 0.109 | 0.120 | 0.119 | 0.118 |
| (1,5) | 0.074 | 0.100 | 0.096 | 0.108 | 0.107 | 0.118 | 0.115 | 0.114 |
| (1,10) | 0.073 | 0.099 | 0.093 | 0.105 | 0.104 | 0.113 | 0.112 | 0.110 |
| (5,1) | 0.076 | 0.101 | 0.101 | 0.127 | 0.124 | 0.125 | 0.120 | 0.120 |
| (5,5) | 0.075 | 0.100 | 0.101 | 0.130 | 0.126 | 0.126 | 0.121 | 0.121 |
| (5,10) | 0.073 | 0.099 | 0.098 | 0.127 | 0.124 | 0.123 | 0.119 | 0.119 |
| (10,1) | 0.078 | 0.103 | 0.104 | 0.135 | 0.129 | 0.131 | 0.124 | 0.123 |
| (10,5) | 0.077 | 0.103 | 0.106 | 0.145 | 0.137 | 0.138 | 0.129 | 0.129 |
| (10,10) | 0.075 | 0.102 | 0.104 | 0.146 | 0.139 | 0.137 | 0.128 | 0.128 |
Range*- (τX, τ∊)values of the range parameter following exponential correlation in X and the error term in the model on Y respectively.
The simulation results confimed that the degree of bias for linear mixed model with error prone covariate varies with the strength of the spatial correlation structure of covariate as well as residuals. However, our proposed bias correction methods perform well interms of providing consistent estimates of the regression coefficient. Both methods under-estimate the true regression coefficient when measurement error is estimated and there is very low correlation in X. This makes sense because the nugget effect in X is non-identifiable in that setting. This is because the assumption that the true covariate X is smooth is no longer valid, hence estimates are not reliable in such situations.
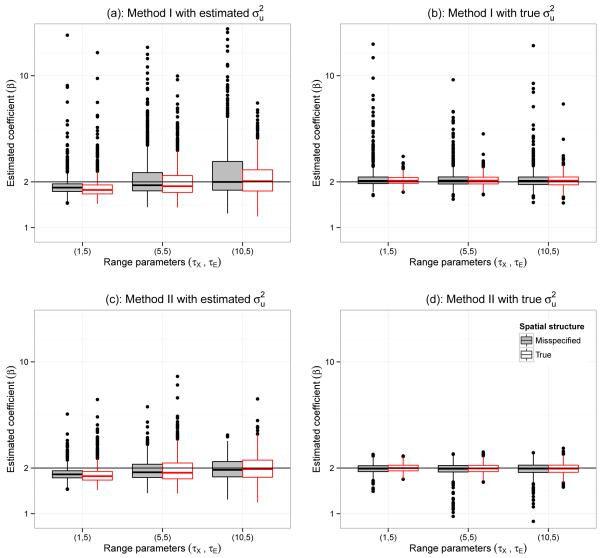
To assess the sensitivity of the true spatial correlation structure on parameter estimation, we run a simulation with misspecified spatial correlation structure. In this simulation we generated data using an exponential covariance structure, but fitted under the assumption of a Gaussian covariance structure. Figure 2 shows the distribution of estimated coefficients when estimated and true values of are used with Method 1 (a-b) and Method 2 (c-d), under different range parameters combined with true and misspecified covariate structure. For each combination of range parameters, the first boxplot (from left) represents results from the misspecified covariance structure. The results obtained for the other combination of range parameters (not shown in this figure) are similar.
Figure 2.
Distribution of estimated coefficient when estimated and true value of used with Method 1 (a-b) and Method 2 (c-d) under different range parameters combinations with true and misspecified covariate structure.
Our simulation results illustrate that proposed methods are quite robust in case of misspecification of underlying covariance structure. However the accuracy of the methods depends largely on the value of . Therefore, a close estimate of to the true value is more important than having a good estimates of underlying covariance structure. Hence, recommend a sensitivity analysis be used in practice.
To evaluate the performance of the proposed method under small samples, we also conducted a simulation with a sample size of 50. In this case, the estimates obtained from method I are slightly upwardly biased with higher standard errors. However method II adjusts for bias quite well and provides a reliable estimates of standard errors. In general, considering all the simulation senarios, the transformation method (Method II) outperforms the method of moments (Method I) in terms of standard errors.
We also run another set of simulations to ascertain whether the spatial configuration of the point locations is an important feature in determining the effectiveness of bias correction. We generated data based on a cubic function with Gaussian noise in a 80×80 grid rather than uniformly distributed within the grid. Our simulation results show that the spatial configuration affects the estimates of measurement error variance. However, our methods are quite robust in case of misspecification of underlying spatial configuation, hence adjust bias very well if true measurement error variances are known.
6. ANALYSIS OF ISCHEMIC HEART DISEASE DATA
Data on Ischemic Heart Disease (IHD) were collected from all hospitals in New South Wales (NSW), Australia between July 1, 1994 and June 30, 2002. A detailed description of the data has been given elsewhere (Burden et al., 2005). Briefly, patients who were admitted to the hospitals via the emergency room and discharged with a diagnosis of IHD were considered as acute IHD cases. Data also includes patient age, gender and geographic location reported via postcode of residence. Due to confidentiality issues our analysis is not based on actual data, but a simulated version that has very similar structure. Data from 579 postcodes were included in the analysis. IHD event data were linked with the Census data which contains age and gender-specific population counts. SEIFA (Socio-Economic Indexes For Areas) scores and centroid co-ordinates (latitude and longitude) for each postcode were obtained from Australian Bureau of Statistics (ABS). Since temporal patterns were not our main concern in this study, we averaged the 8 year SEIFA scores and aggregated values of the population size and number of IHD admitted cases for each postcode. We then calculated age-sex adjusted standardized incidence ratios (SIR) by dividing the observed number of IHD cases by the age-sex adjusted expected IHD cases (Breslow and Day, 1994).
Li et al. (2009) analysed square root transformed standard mortality ratios (SMR) to make them more normal distributed. However, we found that untransformed SIR values more closely approximated the normally distribution and hence we did not transform. We fit model (1) assuming an exponential correlation structure for data observed for each postcode,with distance based on lattitude and longitude of each postcode centroid. As Burden et al. (2005) noted, the principal component analysis that was used to derive the SEIFA score only accounts for about 30 percent of the total variation of the component used. Therefore, it is likely that the SEIFA score is subject to substantial measurement error. We standardized the SEIFA scores to have a mean of zero.
The results of our analysis are given in Table 2.
Table 2.
Analysis of Ischemic Heart Disease Data in NSW, Australia under different specification of measurement error
| Methods | Estimates for SEIFA | |
|---|---|---|
| Ignoring measurement error | ||
| Ordinary Least Squares | −0.062 | 0.014 |
| LME with spatial correlation | −0.141 | 0.015 |
| Accounting for measurement error bias | ||
| Method 1 | −0.377 | 0.041 |
| Method 2 | −0.278 | 0.015 |
The naive analysis ignoring spatial correlation, suggests a significant protective effect associated with higher SEIFA values (, SE=0.014). Analysis via a linear mixed model accounting for spatial correlation also suggests that the effect is very strong ( with SE=0.015). However, the magnitude of the effect is much larger.
We applied our bias correction methods on the result obtained from the linear mixed model. The linear mixed model of SEIFA based on a intercept only model with assumed exponential spatial correlation suggests the estimate of measurement error variance as 0.28. Both methods suggest a strongly significant effect of SEIFA ( and −0.278 with SE=0.041 and 0.015 respectively). A large difference in the estimated standard error for Method I and Method 2 is observed. As the standard error for method I is given by , where is the estimated attenuation factors, the estimated standard error for method I doesn’t account for the variability of the attenuation factor. In practice, a bootstrap procedure can be used to calculate the standard error for method I. We implement a block bootstrap procedure by leaving one block at each iteration. Blocks are automatically selected using the cluster separation method “clara” (Kaufman and Rousseeuw, 2005) in R (R Core Team, 2013). Specifically, this method selects k representative objects in the data set, where k is the number of clusters. The remaining objects are then assigned to the nearest representative object to form a cluster. The representative objects are selected in such a way that the average distance of the representative objects to all other objects in the same cluster is minimized. Our results show that the difference in estimated standard error reduces with large number of blocks while estimated standard error for method 2 remain unchanged (result not shown).
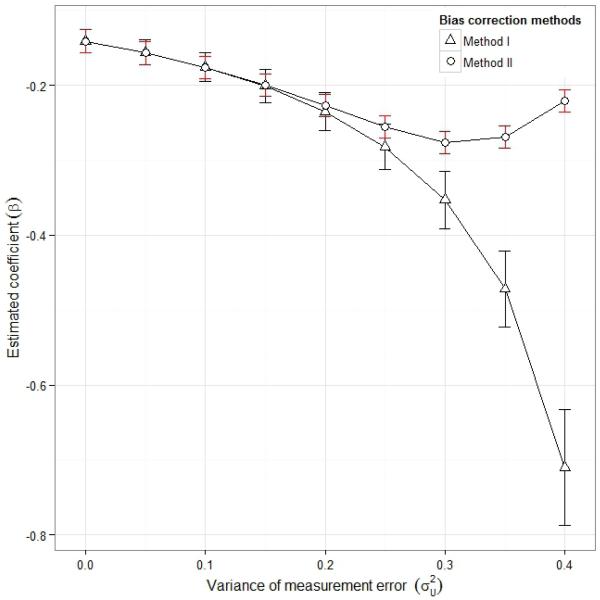
Since we do not have a validation dataset and thus cannot test the assumption underlying the bias correction methods, we conduct a sensitivity analysis to help in the interpretation of our results. We conducted sensitivity analysis varying measurement error variance, from 0.0 (naive) to 0.40. The result of the sensitivity analysis is presented in figure 3.
Figure 3.
Sensitivity analysis for IHD data. The assumed measurement error variance varied between 0 (naive) and 0.40
As measurement error variance, increases, the estimates obtained by method of moments also decreases. The estimates obtained using transformation methods also decreases until when the assumed measurement error variance is less than the estimated measurement error variance and then increases. We note that the transformation method appears to give stable results over the range of .
7. DISCUSSION
In this paper, we develop a framework to understand the nature of the bias of estimated regression coefficients when covariates are measured with error in spatial modelling settings. Both our empirical simulation results and asymptotic bias calculations suggest that ignoring measurement error and conducting naive analysis attenuates the estimated regression coefficient towards the null hypothesis of no effect. Our results extend classical measurement error theory in that the attenuation depends on the degree of spatial correlation in both X and the assumed random error from the regression model. We proposed two different strategies to obtain consistent estimates of the regression coefficients. Our strategies include 1) posthoc adjustment of estimated regression coefficients based on an estimated attenuation factor and 2) a linear transformation of the error prone covariates that can then be analysed to yield consistent results. We have shown that the bias correction methods both perform well in obtaining consistent estimates. We have applied our proposed approaches to the analysis of Ischemic Heart Disease data.
Li et al. (2009), report similar results for the asymptotic bias associated with regression analysis involving covariate measurement error. They also provide a modelling technique based on an EM algorithm. However, their approach can be difficult to apply, especially in situations involving large data sets. Our proposed approaches do not require sophisticated programming and can be handled by readily available packages such as lme in R.
Moreover, Li et al. (2009) use true values of the measurement error variance in all of their simulations and showed that EM algorithm performs quite well even in the case of small sample sizes. However, they did not evaluate the performance of their approach when measurement error variances are estimated. We showed that when the measurement error variance is known, both of our proposed methods provide reliable estimate of the true regression coefficient, even if the degree of bias varies with the strength of the spatial correlation structure. As expected, the performance of our methods reduces substantially when the measurement error variance is estimated.
Approaches to measurement error adjustment require either additional assumptions or additional data to quantify the magnitude of measurement error. Our theory suggests that, in the absence of measurement error, the estimates and corresponding standard errors obtained by method 1, method 2 and linear mixed model with spatial correlation would be identical. However, in the presence of measurement error the estimates obtained using these methods would be different. This is also true for the estimated standard errors. The estimated standard error of the regression coefficient, , corresponding to method 1 depends on assumed measurement error variance. Hence, the estimated standard errors along with corresponding confidence intervals obtained using method I ignore the variability of the assumed or estimated measurement error variance. In practice, a bootstrap procedure can be used to obtain appropriate standard errors for method I.
One observation from our simulation is that the use of empirical measurement error variance estimates from the data leads to under estimation of the regression coefficient when there is minimal spatial correlation in the covariates X. This might be due to the large grid scale compare to small correlation in the neighbourhood that makes model close to non-identifiable. However, reducing grid size to 40 × 40 improves the estimates. Similar results based on the grid size also reported previously by Bell and Grunwald (2004).
Furthermore, use of estimated measurement error variance resulted in different estimate of the average of estimated standard error and empirical error. However, when true measurement error variance is used, the average of the estimated standard error and empirical standard error became very similar. This indicates that knowledge of true measurement error variance is crucial not only in obtaining a consistent estimate of the regression coefficient but also in obtaining valid inference. A sensitivity analysis would be helpful in this regards.
Indeed, the assumption of smooth spatial surface for X that is all the nugget estimates is due to measurement error might not be reasonable in practice. We run a simulation adding additional nugget effect to the covariate, X. All our simulations suggest that the proposed bias correction methods works reasonably well when there is some information about the measurement error variance is available. Moreover, estimation of measurement error variance depends on the spatial configurations. Therefore, knowledge of measument error variance is important. In practice, if measurement error is unknown, the best way is to run a sensitivity analysis within the reasonable range of measurement error variance.
Our heart disease data example shows that there is substantial increase in the magnitude of the significant protective effect when adjusted for measurement error. This is consistent with the results from systematic review that the neighbourhood socio-economic effect on health is consistent across studies (Pickett and Pearl, 2001). However, cautions should be taken to infer as these results might be susceptible to ecological bias or ecological fallacy.
The ecological bias resulted from the disconnection between the level of analysis and level of inference. In the group level data analysis, ecological bias is susceptible if there is an additional effect due to the overall exposure level on the group above its effect on individuals (Sheppard, 2003). In case of aggregate group level analysis Prentice and Sheppard (1995) showed that using group level covariates in the analysis reduces the effects of measurement error in covariates. However, Greenland (2001) and Jackson et al. (2006) noted that ecological covariates are subject to non-random survey errors and may not be addressed by aggregation of group level analysis of covariates. Moreover, in many research area group level data were only source of data available for analysis. A classic example is air pollution studies where individual measurement on exposure are rarely collected for the cost and feasibility (Sheppard et al., 2012). Often air pollution data are measured with error, therefore, it is of natural interest to obtain the bias corrected estimates of the effect.
In our simulation we have considered only a single covariate measured with error in a spatial linear mixed model with Gaussian error. It would be of interest to explore the effect of covariate measurement error in the presence of multiple covariates and also omitted covariates. Future work can also be done on extending our formulation to the spatial generalized linear mixed model with non-Gaussian outcomes. However, such explorations are beyond the scope of this present paper.
In light of the increasing popularity of multi-level models that include both individual and area-specific covariates, it is important that practitioners be aware of the importance, not only of careful modelling of the mean function, but also of accounting for the measurement error and appropriate spatial structure of their data.
APPENDIX.
The ordinary least squares estimate of β is
| (8) |
with W* defined in the text. Under the true model, Y = X*β + ∊, we have
Now under certain regularity conditions (Zheng and Zhu, 2012) and by the weak law of large numbers, and . It follows that, . Since W* and X* have first column equals to 1 corresponding to intercept of the model and assuming μX = 0, it follows,
where ρols = trace (∑X)/trace (∑X + ∑U).
LIST OF CHANGES
REFERENCES
- Bell ML, Grunwald GK. Mixed models for the analysis of replicated spatial point patterns. Biostatistics. 2004;5(4):633–648. doi: 10.1093/biostatistics/kxh014. [DOI] [PubMed] [Google Scholar]
- Breslow N, Day N. Statistical methods in cancer research. International Agency for Research on Cancer; 1994. [Google Scholar]
- Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. Journal of the American Statistical Association. 1993;88(421):9–25. [Google Scholar]
- Burden S, Guha S, Morgan G, Ryan L, Sparks R, Young L. Spatio-temporal analysis of acute admissions for ischemic heart disease in nsw, australia. Environmental and Ecological Statistics. 2005;12(4):427–448. [Google Scholar]
- Carroll R, Chen R, George E, Li T, Newton H, Schmiediche H, Wang N. Ozone exposure and population density in harris county, texas. Journal of the American Statistical Association. 1997;92(438):392–404. [Google Scholar]
- Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement error in nonlinear models: a modern perspective. Chapman and Hall/CRC; 2006. [Google Scholar]
- Clark I. Statistics or geostatistics? sampling error or nugget effect?. Fourth World Conference on Sampling & Blending; The Southern African Institute of Mining and Metallurgy; 2009. pp. 13–18. [Google Scholar]
- Clayton DG, Bernardinelli L, Montomoli C. Spatial correlation in ecological analysis. International Journal of Epidemiology. 1993;22(6):1193–1202. doi: 10.1093/ije/22.6.1193. [DOI] [PubMed] [Google Scholar]
- Cook DG, Pocock SJ. Multiple regression in geographical mortality studies, with allowance for spatially correlated errors. Biometrics. 1983:361–371. [PubMed] [Google Scholar]
- Elliot P, Wakefield J, Best N, Briggs D. Spatial epidemiology: methods and applications. Oxford University Press; 2000. [Google Scholar]
- Elliott P, Wartenberg D. Spatial epidemiology: current approaches and future challenges. Environmental health perspectives. 2004;112(9):998. doi: 10.1289/ehp.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller W. Measurement Error Models. Wiley Series in Probability and Statistics, Wiley; 2009. [Google Scholar]
- Greenland S. Ecologic versus individual-level sources of bias in ecologic estimates of contextual health effects. International Journal of Epidemiology. 2001;30(6):1343–1350. doi: 10.1093/ije/30.6.1343. [DOI] [PubMed] [Google Scholar]
- Gryparis A, Paciorek CJ, Zeka A, Schwartz J, Coull BA. Measurement error caused by spatial misalignment in environmental epidemiology. Biostatistics. 2009;10(2):258–274. doi: 10.1093/biostatistics/kxn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Ryan L, Morara M. Gauss–seidel estimation of generalized linear mixed models with application to poisson modeling of spatially varying disease rates. Journal of Computational and Graphical Statistics. 2009;18(4):818–837. [Google Scholar]
- Jackson C, Best N, Richardson S. Improving ecological inference using individual-level data. Statistics in Medicine. 2006;25(12):2136–2159. doi: 10.1002/sim.2370. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw P. Finding Groups in Data: An Introduction to Cluster Analysis. Wiley Series in Probability and Statistics, Wiley; 2005. [Google Scholar]
- Li Y, Tang H, Lin X. Spatial linear mixed models with covariate measurement errors. Statistica Sinica. 2009;19(3):1077. [PMC free article] [PubMed] [Google Scholar]
- Molitor J, Jerrett M, Chang CC, Molitor NT, Gauderman J, Berhane K, McConnell R, Lurmann F, Wu J, Winer A, et al. Assessing uncertainty in spatial exposure models for air pollution health effects assessment. Environmental Health Perspectives. 2007;115(8):1147. doi: 10.1289/ehp.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett KE, Pearl M. Multilevel analyses of neighbourhood socioeconomic context and health outcomes: a critical review. Journal of epidemiology and community health. 2001;55(2):111–122. doi: 10.1136/jech.55.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team nlme: Linear and Nonlinear Mixed Effects Models. 2013 R package version 3.1-109. [Google Scholar]
- Prentice RL, Sheppard L. Dietary fat and cancer: consistency of the epidemiologic data, and disease prevention that may follow from a practical reduction in fat consumption. Cancer Causes & Control. 1990;1(1):81–97. doi: 10.1007/BF00053187. [DOI] [PubMed] [Google Scholar]
- Prentice RL, Sheppard L. Aggregate data studies of disease risk factors. Biometrika. 1995;82(1):113–125. [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- Ribeiro PJ, Diggle PJ. geoR: a package for geostatistical analysis. R-NEWS. 2001;1(2):14–18. iSSN 1609-3631. [Google Scholar]
- Ruppert D, Wand M, Carroll RJ. Semiparametric regression during 2003–2007. Electronic Journal of Statistics. 2009;3:1193. doi: 10.1214/09-EJS525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J, Coull BA. Control for confounding in the presence of measurement error in hierarchical models. Biostatistics. 2003;4(4):539–553. doi: 10.1093/biostatistics/4.4.539. [DOI] [PubMed] [Google Scholar]
- Sheppard L. Insights on bias and information in group-level studies. Biostatistics. 2003;4(2):265–278. doi: 10.1093/biostatistics/4.2.265. [DOI] [PubMed] [Google Scholar]
- Sheppard L, Burnett RT, Szpiro AA, Kim SY, Jerrett M, Pope CA, III, Brunekreef B. Confounding and exposure measurement error in air pollution epidemiology. Air Quality, Atmosphere & Health. 2012;5(2):203–216. doi: 10.1007/s11869-011-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiro AA, Sheppard L, Lumley T. Efficient measurement error correction with spatially misaligned data. Biostatistics. 2011;12(4):610–623. doi: 10.1093/biostatistics/kxq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller LA, Gotway CA. Applied spatial statistics for public health data. ume 368. John Wiley & Sons; 2004. [Google Scholar]
- Wansbeek TJ, Meijer E. Measurement error and latent variables in econometrics. ume 37. Elsevier Amsterdam; 2000. [Google Scholar]
- Wood S. Generalized additive models: an introduction with R. Chapman and Hall/CRC; 2006. [Google Scholar]
- Xia H, Carlin BP. Spatio-temporal models with errors in covariates: mapping ohio lung cancer mortality. Statistics in Medicine. 1998;17(18):2025–2043. doi: 10.1002/(sici)1097-0258(19980930)17:18<2025::aid-sim865>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Zeger SL, Thomas D, Dominici F, Samet JM, Schwartz J, Dockery D, Cohen A. Exposure measurement error in time-series studies of air pollution: concepts and consequences. Environmental health perspectives. 2000;108(5):419. doi: 10.1289/ehp.00108419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhu J. On the asymptotics of maximum likelihood estimation for spatial linear models on a lattice. Sankhya A. 2012;74(1):29–56. [Google Scholar]