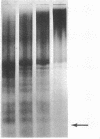
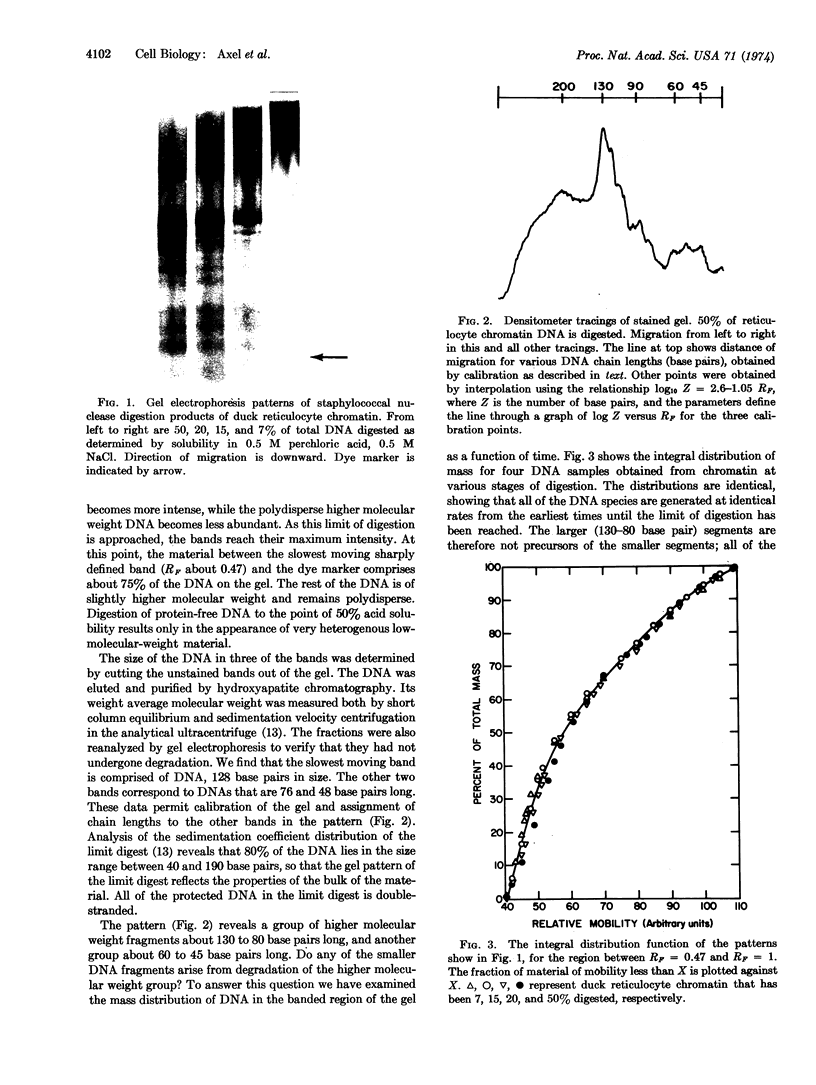
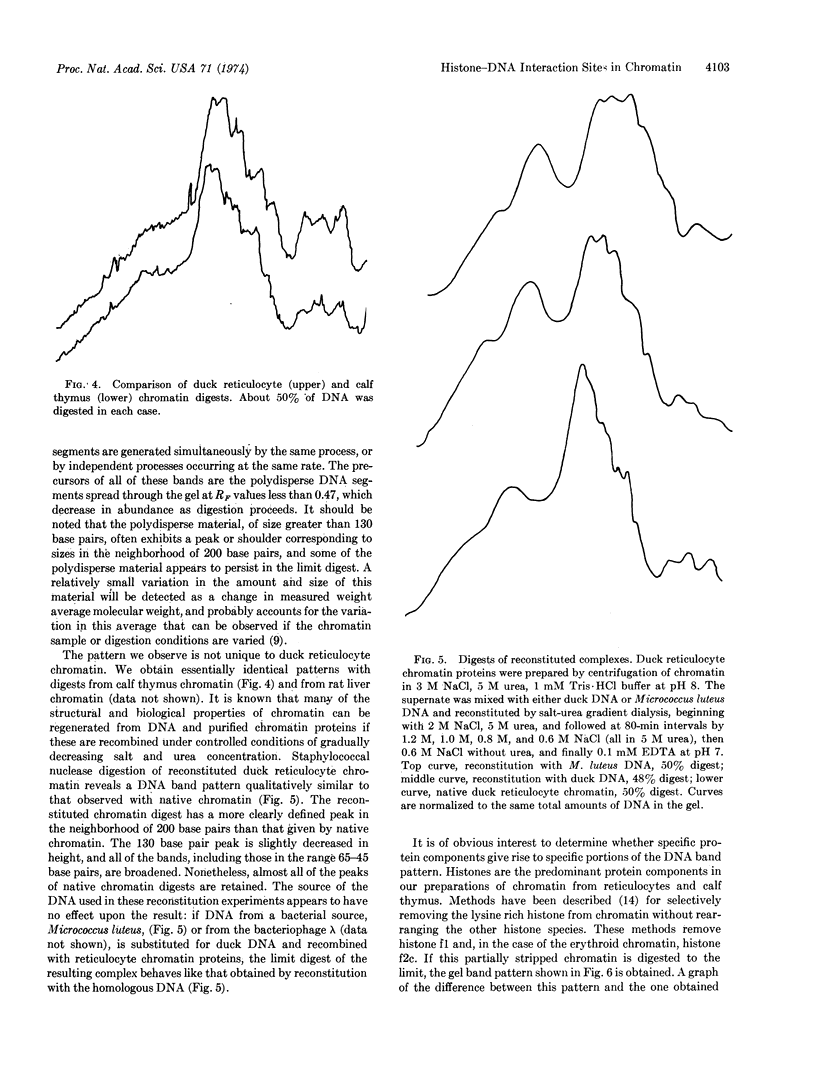
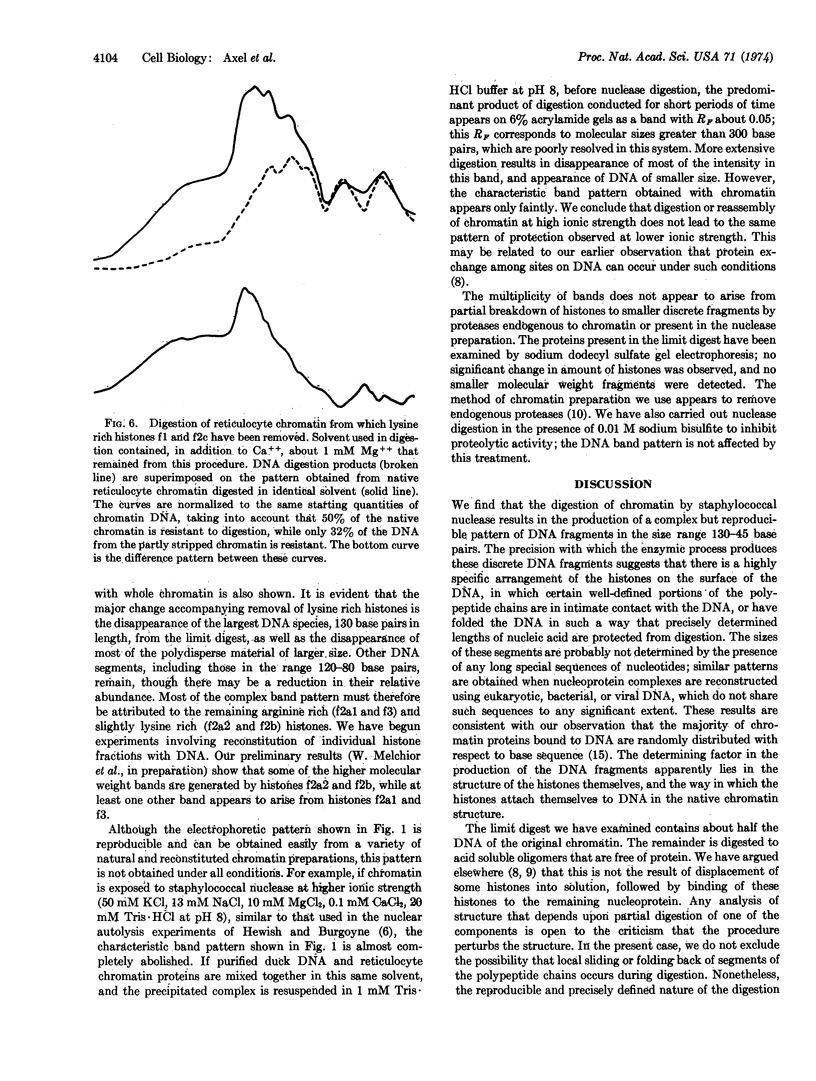
Abstract
Staphylococcal nuclease digestion of purified chromatin from duck reticulocytes or calf thymus results in the production of a series of double-stranded DNA fragments of discrete molecular size, ranging from about 130 to 45 base pairs, which can be detected by polyacrylamide gel electrophoresis. Similar patterns of protected DNA fragments are obtained from limit digests of chromatin “reconstituted” from purified DNA and chromatin proteins. The results obtained with reconstituted material do not depend upon the origin of the DNA, which may be derived from a bacterial, viral, or homologous source. The specificity of the protective mechanism, therefore, resides in the structure of the bound histones, and probably not in any special nucleotide sequences present in the DNA. Removal of lysine-rich histones from chromatin before digestion results principally in disappearance from the digest of a DNA fragment about 130 base pairs long. Our preliminary results suggest that other elements of the digest pattern can be assigned uniquely to the remaining histone components. These results indicate that the binding of histones to DNA in chromatin involves a limited number of specific and very well defined contacts between protein and nucleic acid, which arise from structural properties of the histones.
Keywords: nuclease, DNA-electrophoresis
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Cedar H., Felsenfeld G. Synthesis of globin ribonucleic acid from duck-reticulocyte chromatin in vitro. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2029–2032. doi: 10.1073/pnas.70.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Chemical probes of chromatin structure. Biochemistry. 1974 Aug 13;13(17):3622–3628. doi: 10.1021/bi00714a034. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- D'Anna J. A., Jr, Isenberg I. Interactions of histone LAK (f2a2) with histones KAS (f2b) and GRK (f2a1). Biochemistry. 1974 May 7;13(10):2098–2104. doi: 10.1021/bi00707a016. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Ilyin Y. V., Varshavsky A. Y., Mickelsaar U. N., Georgiev G. P. Studies on deoxyribonucleoprotein structure. Redistribution of proteins in mixtures of deoxyribonucleoproteins, DNA and RNA. Eur J Biochem. 1971 Sep 24;22(2):235–245. doi: 10.1111/j.1432-1033.1971.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Prunell A., Bernardi G. Fractionation of native and denatured deoxyribonucleic acid on agarose columns. J Biol Chem. 1973 May 25;248(10):3433–3440. [PubMed] [Google Scholar]
- Varshavsky A. J., Georgiev G. P. Clustered arrangement of histones F2al and F3 along DNA in chromosomal deoxyribonucleoproteins. Biochim Biophys Acta. 1972 Nov 9;281(4):669–674. doi: 10.1016/0005-2787(72)90166-9. [DOI] [PubMed] [Google Scholar]
- Williamson R. Properties of rapidly labelled deoxyribonucleic acid fragments isolated from the cytoplasm of primary cultures of embryonic mouse liver cells. J Mol Biol. 1970 Jul 14;51(1):157–168. doi: 10.1016/0022-2836(70)90277-9. [DOI] [PubMed] [Google Scholar]