Abstract
The three dimensional structure of the lysozyme from bacteriophage T4 has been determined from a 2.5 Å resolution electron density map. About 60% of the molecule is in a helical conformation and there is one region consisting of antiparallel β-structure. The polypeptide backbone folds into two distinct lobes linked in part by a long helix. In the region between the two lobes, there is a cleft which deepens into a hole or cavity, about 6-8 Å in diameter, extending from one side of the molecule to the other. This opening is closed off by side chains which extend to within 3-5 Å of each other. A number of mutant lysozymes in which residues in the vicinity of the opening are modified have markedly reduced catalytic activity, suggesting that this region of the molecule may be catalytically important. The three dimensional structure of T4 phage lysozyme is quite different from that of hen egg-white lysozyme although it is not clear at this time whether or not the mechanisms of catalysis of the respective enzymes are related.
Keywords: protein structure, x-ray diffraction, mutants
Full text
PDF
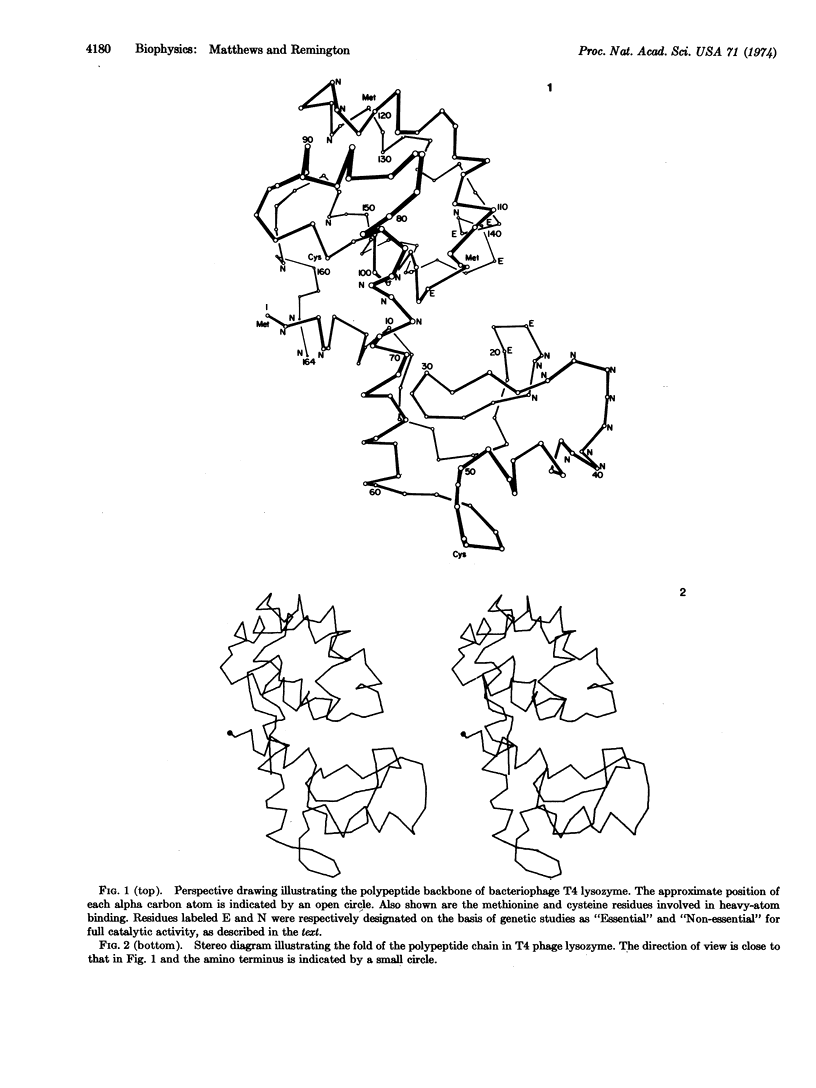


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake C. C., Johnson L. N., Mair G. A., North A. C., Phillips D. C., Sarma V. R. Crystallographic studies of the activity of hen egg-white lysozyme. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):378–388. doi: 10.1098/rspb.1967.0035. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Koenig D. F., Mair G. A., North A. C., Phillips D. C., Sarma V. R. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965 May 22;206(4986):757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- Blake C. C., Mair G. A., North A. C., Phillips D. C., Sarma V. R. On the conformation of the hen egg-white lysozyme molecule. Proc R Soc Lond B Biol Sci. 1967 Apr 18;167(1009):365–377. doi: 10.1098/rspb.1967.0034. [DOI] [PubMed] [Google Scholar]
- Blow D. M., Birktoft J. J., Hartley B. S. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969 Jan 25;221(5178):337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- Colman P. M., Jansonius J. N., Matthews B. W. The structure of thermolysin: an electron density map at 2-3 A resolution. J Mol Biol. 1972 Oct 14;70(3):701–724. doi: 10.1016/0022-2836(72)90569-4. [DOI] [PubMed] [Google Scholar]
- Dunnill P. Sequence similarities between hen egg-white and T4 phage lysozymes. Nature. 1967 Aug 5;215(5101):621–622. doi: 10.1038/215621a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Dahlquist F. W., Maynard A. Y. Letter: crystallographic data fro lysoxyme from bacteriophage T4. J Mol Biol. 1973 Aug 15;78(3):575–576. doi: 10.1016/0022-2836(73)90478-6. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Sigler P. B., Henderson R., Blow D. M. Three-dimensional structure of tosyl-alpha-chymotrypsin. Nature. 1967 May 13;214(5089):652–656. doi: 10.1038/214652a0. [DOI] [PubMed] [Google Scholar]
- Ocada Y., Amagase S., Tsugita A. Frameshift mutation in the lysozyme gene of bacteriophage T4: demonstration of the insertion of five bases, and a summary of in vivo codons and lysozyme activities. J Mol Biol. 1970 Dec 14;54(2):219–246. doi: 10.1016/0022-2836(70)90428-6. [DOI] [PubMed] [Google Scholar]
- Richards F. M. The matching of physical models to three-dimensional electron-density maps: a simple optical device. J Mol Biol. 1968 Oct 14;37(1):225–230. doi: 10.1016/0022-2836(68)90085-5. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Alden R. A., Birktoft J. J., Kraut J., Powers J. C., Wilcox P. E. An x-ray crystallographic study of the binding of peptide chloromethyl ketone inhibitors to subtilisin BPN'. Biochemistry. 1972 Jun 20;11(13):2439–2449. doi: 10.1021/bi00763a009. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Tsugita A., Inouye M. Complete primary structure of phage lysozyme from Escherichia coli T4. J Mol Biol. 1968 Oct 14;37(1):201–212. doi: 10.1016/0022-2836(68)90083-1. [DOI] [PubMed] [Google Scholar]
- WEIDEL W., PELZER H. BAGSHAPED MACROMOLECULES--A NEW OUTLOOK ON BACTERIAL CELL WALLS. Adv Enzymol Relat Areas Mol Biol. 1964;26:193–232. doi: 10.1002/9780470122716.ch5. [DOI] [PubMed] [Google Scholar]
- Wright C. S., Alden R. A., Kraut J. Structure of subtilisin BPN' at 2.5 angström resolution. Nature. 1969 Jan 18;221(5177):235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]



