Abstract
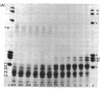
Exhaustive digestion of chromatin with trypsin leads to the cleavage of only 20-30 amino acids from each of histones III (f3), IV (f2a1), IIb2 (f2b), and IIb1 (f2a1), the remainder of these chains being resistant. This resistance is not altered by removing the histones from the DNA with 2 M NaCl, but is dramatically reduced in 6 M urea. Histones III, IV, IIb2, and possibly IIb1 are cleaved at their N-termini. Histones I and V and the nonhistone proteins are the first to be attacked by trypsin and have no detectable trypsin-resistant fragments. The arginine rich histones, III and IV, are then cleaved as a pair, followed by most of IIb2 and IIb1, also as a pair. This data is consistent with a model in which basic N-terminal “arms” extend from a trypsin-resistant histone complex. The structural arrangement of these arms relative to the trypsin-resistant histone complex may define the spatial coordinates of DNA binding sites and, consequently, the folding of the DNA fiber in the chromosome.
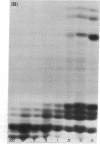
Accompanying the tryptic digestion of the N-terminals of histones III, IV, IIb2, and possibly IIb1, is an increased sensitivity of chromatin to staphylococcal nuclease. As analyzed by electrophoresis, untrypsinized chromatin is digested into eight discrete limit-digest fragments by nuclease. Trypsinization results in the nuclease digestion of some, but not all, of these DNA bands. Together with the information on the way trypsin cleaves histones in chromatin, the analysis of the resistant DNA suggests that histone N-terminals are associated with some DNA bands and histone C-terminals with other DNA bands. We propose that histones fold the chromosome by crosslinking the DNA corresponding to these bands.
Keywords: histones, fingerprint, cleavage-products, DNA electrophoresis, iodination
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartley J. A., Chalkley R. The binding of deoxyribonucleic acid and histone in native nucleohistone. J Biol Chem. 1972 Jun 10;247(11):3647–3655. [PubMed] [Google Scholar]
- Bray D., Brownlee S. M. Peptide mapping of proteins from acrylamide gels. Anal Biochem. 1973 Sep;55(1):213–221. doi: 10.1016/0003-2697(73)90306-0. [DOI] [PubMed] [Google Scholar]
- Clark R. J., Felsenfeld G. Structure of chromatin. Nat New Biol. 1971 Jan 27;229(4):101–106. doi: 10.1038/newbio229101a0. [DOI] [PubMed] [Google Scholar]
- Danna K., Nathans D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2913–2917. doi: 10.1073/pnas.68.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A. J., Candido E. P., Dixon G. H. Enzymatic modifications and their possible roles in regulating the binding of basic proteins to DNA and in controlling chromosomal structure. Cold Spring Harb Symp Quant Biol. 1974;38:803–819. doi: 10.1101/sqb.1974.038.01.084. [DOI] [PubMed] [Google Scholar]
- Platt T., Files J. G., Weber K. Lac repressor. Specific proteolytic destruction of the NH 2 -terminal region and loss of the deoxyribonucleic acid-binding activity. J Biol Chem. 1973 Jan 10;248(1):110–121. [PubMed] [Google Scholar]