Abstract
Sequencing of the Arabidopsis genome revealed a unique complexity of the plant heat stress transcription factor (Hsf) family. By structural characteristics and phylogenetic comparison, the 21 representatives are assigned to 3 classes and 14 groups. Particularly striking is the finding of a new class of Hsfs (AtHsfC1) closely related to Hsf1 from rice and to Hsfs identified from frequently found expressed sequence tags of tomato, potato, barley, and soybean. Evidently, this new type of Hsf is well expressed in different plant tissues. Besides the DNA binding and oligomerization domains (HR-A/B region), we identified other functional modules of Arabidopsis Hsfs by sequence comparison with the well-characterized tomato Hsfs. These are putative motifs for nuclear import and export and transcriptional activation (AHA motifs). There is intriguing flexibility of size and sequence in certain parts of the otherwise strongly conserved N-terminal half of these Hsfs. We have speculated about possible exon-intron borders in this region in the ancient precursor gene of plant Hsfs, similar to the exon-intron structure of the present mammalian Hsf-encoding genes.
INTRODUCTION
Heat stress transcription factors (Hsfs) are the terminal components of a signal transduction chain mediating the activation of genes responsive to both heat stress and a large number of chemical stressors (Wu 1995; Nover et al 1996; Morimoto 1998; Scharf et al 1998b; Schöffl et al 1998; Nakai 1999). They recognize palindromic binding motifs, so-called heat stress elements (HSEs; 5′-AGAAnnTTCT-3′) conserved in promoters of heat stress–inducible genes of all eukaryotes (Bienz and Pelham 1987; Nover 1987, 1991). The initial cloning and characterization of the yeast Hsf (Sorger and Pelham 1988; Wiederrecht et al 1988) were rapidly followed by cloning of the corresponding genes from Drosophila (Clos et al 1990), mammals (Rabindran et al 1991; Sarge et al 1991; Schuetz et al 1991), and tomato (Scharf et al 1990). The analysis of the tomato Hsf system exposed 2 interesting peculiarities. First, there are at least 4 different Hsfs (Scharf et al 1990, 1993; Treuter et al 1993; Bharti et al 2000) belonging to 2 classes, ie, class A with Hsfs A1, A2, and A3 and class B with HsfB1. Second, 2 of the 4 Hsfs (HsfA2 and B1) are heat stress–inducible proteins themselves. Although, with few exceptions, multiple Hsfs or Hsf-related proteins were subsequently found in other organisms as well (Nover et al 1996; Morimoto 1998; Nakai 1999), the peculiarities of an extended HR-A/B region in the class A Hsfs (Fig 1) and the heat stress–dependent expression are unique features of the plant Hsf system. The sequencing of the Arabidopsis thaliana genome now allows a more detailed discussion on the plant Hsf system based on the combination of sequence comparison and results from the fairly advanced functional analysis of tomato Hsfs.
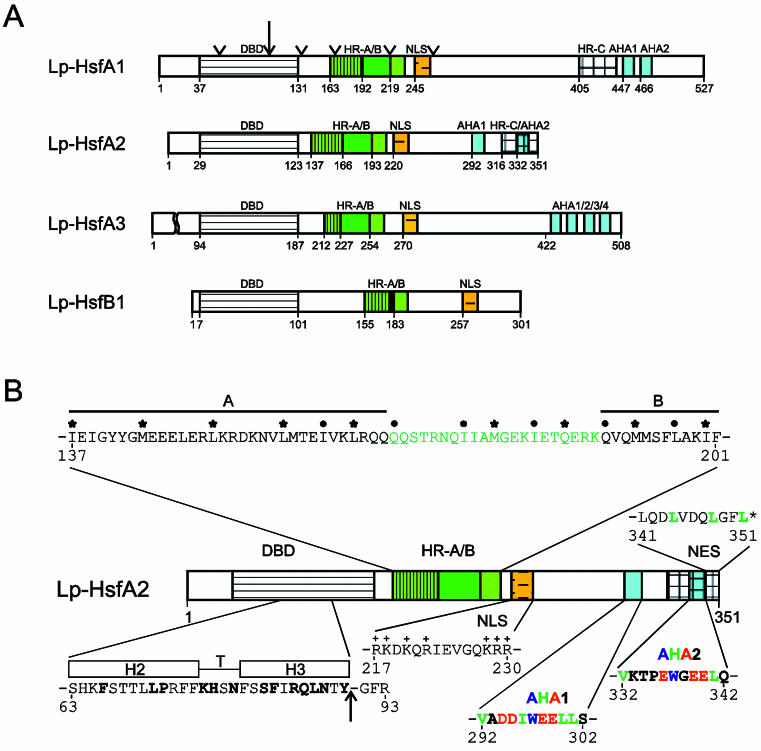
Fig 1.
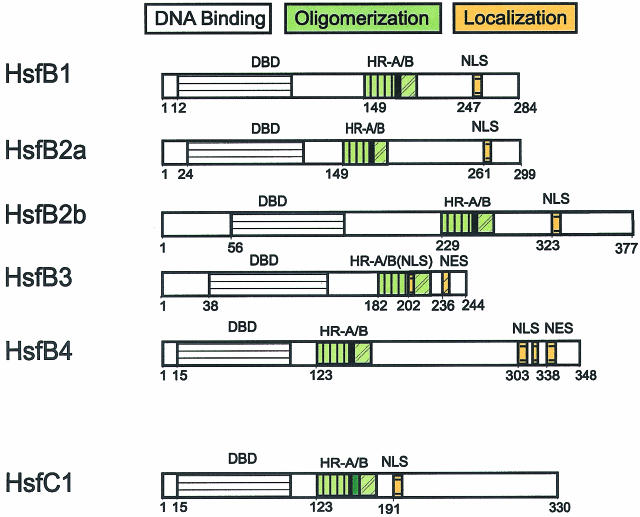
Basic structure of Hsfs. The block diagrams in part A represent tomato (Lycopersicon peruvianum [Lp]) Hsfs with their conserved functional domains. For abbreviations see part B. Arrowheads at the block diagram of Lp-HsfA1 indicate the positions of putative introns in the ancient Hsf gene. (B) Essential structural details are represented by the tomato HsfA2. (1) The central part of the DBD is the helix-turn-helix motif (H2-T-H3) with a considerable number of amino acid residues invariant among different organisms (boldfaced letters). The arrow indicates the position of the intron conserved in all plant Hsfs. (2) The oligomerization domain HR-A/B is characterized by the heptad pattern of hydrophobic residues (dots, asterisks). The insertion of additional 21 amino acid residues between parts A and B are marked in green. (3) The bipartite NLS represents a cluster of basic residues (K, R) recognized by the NLS receptor. (4) Central elements of the activator region are short motifs (AHA elements) rich in aromatic (W, Y, F), hydrophobic (L, I, V), and acidic amino acid residues (D, E). (5) A leucine-rich motif at the C-terminus functions as an NES
Basic structure and classification of 21 Arabidopsis Hsfs
Based on the presence of the conserved DNA-binding domain (DBD) plus the adjacent HR-A/B region (Fig 1), we identified 21 open reading frames in the Arabidopsis genome encoding putative Hsfs. Other sequences annotated as Hsf-like were not considered. Thus, the complexity of the plant Hsf system far exceeds that of any other organism whose genomic sequence is known. For comparison, there are a total of 4 Hsfs in vertebrates, only 1 Hsf in Drosophila and the nematode Caenorhabditis elegans, and 1 Hsf plus 3 Hsf-related proteins in yeast (Nover et al 1996; Nakai 1999).
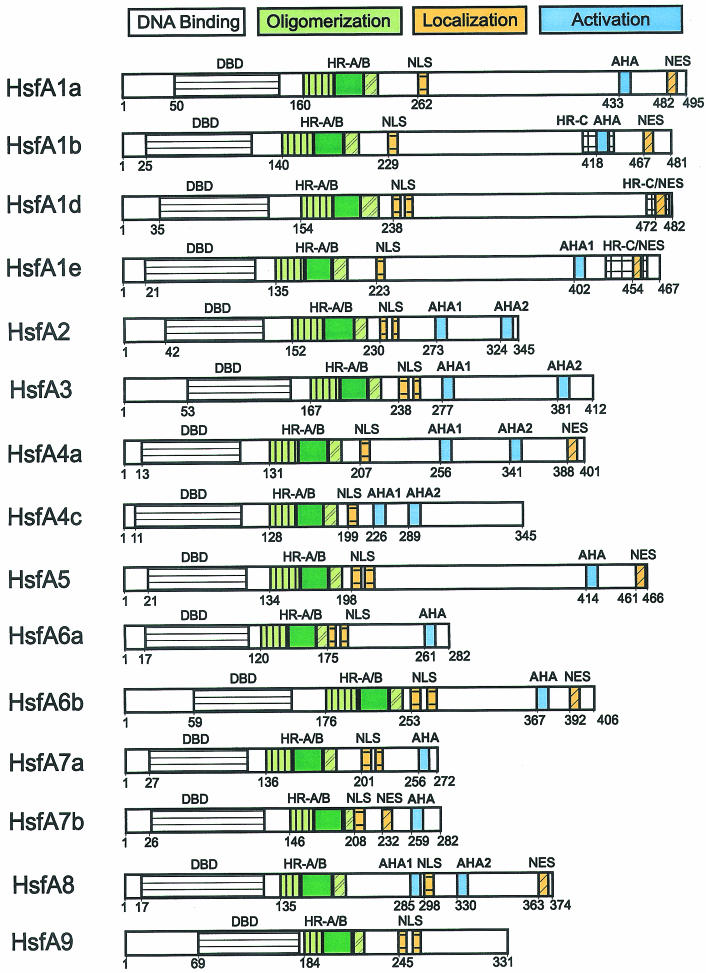
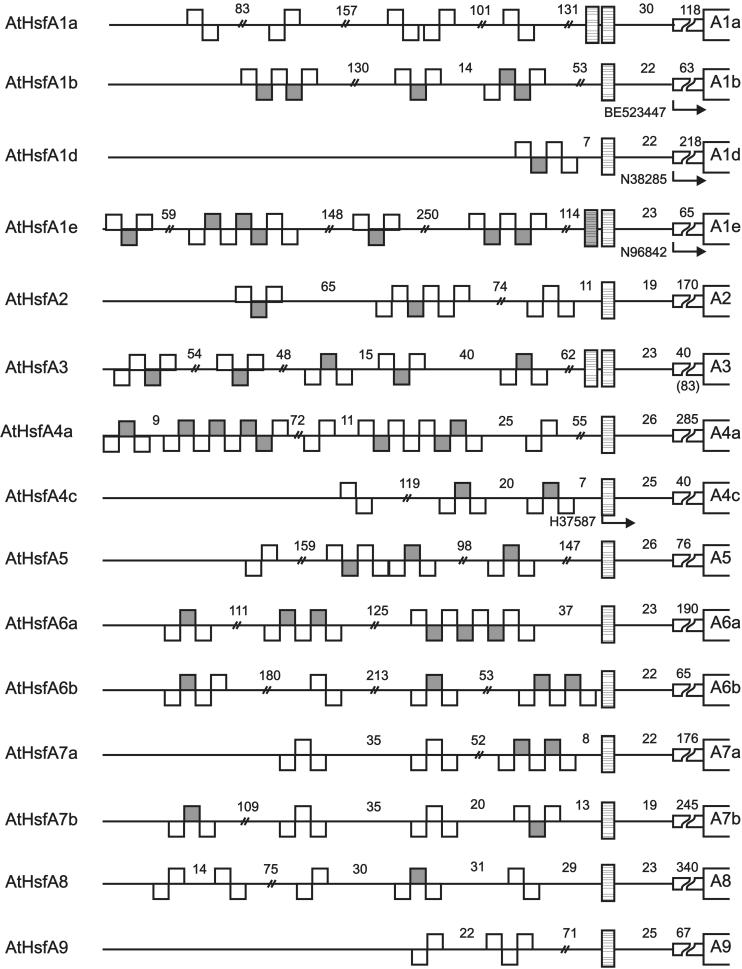
Similar to many other proteins regulating gene activity, Hsfs have a modular structure. Despite considerable variability in size and sequence, their basic structure is conserved among eukaryotes. This is exemplified by tomato HsfA2, with sequence details given for modules identified by mutation and functional analysis (Fig 1B). This knowledge helped to identify corresponding functional motifs for the Arabidopsis Hsfs compiled in Figure 2 and Table 1.
Fig 2.
Survey of Arabidopsis Hsfs. For explanations see legend to Figure 1
Table 1.
Survey of plant Hsfs
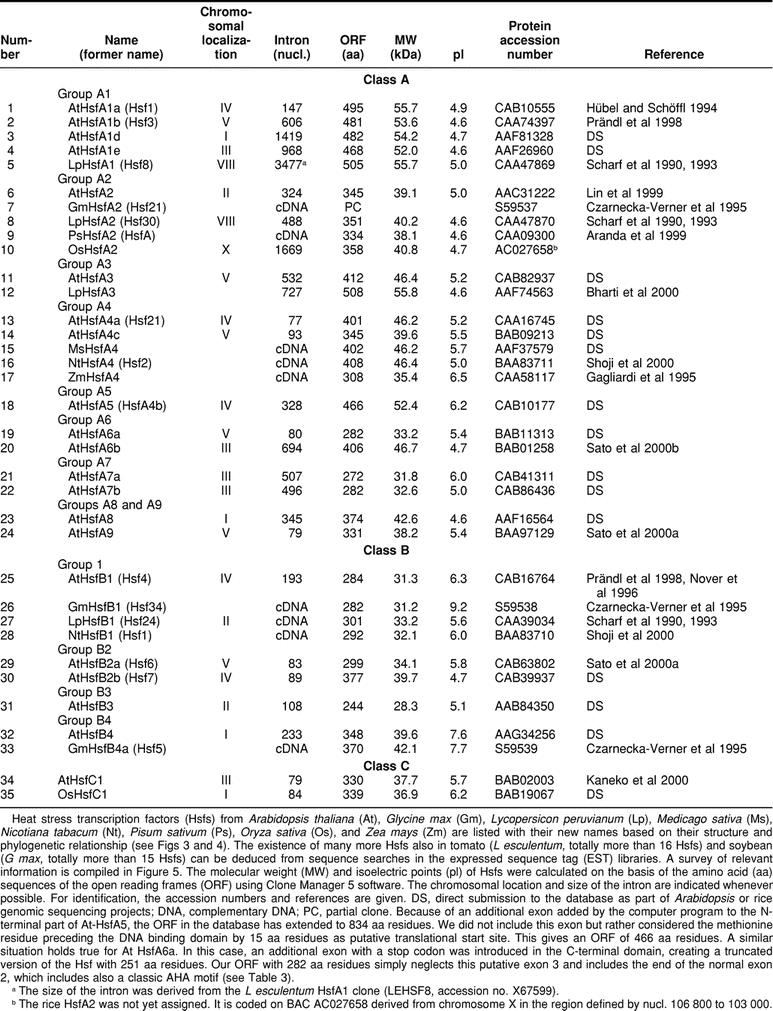
Close to the N-terminus, the highly structured DBD is the most conserved part of Hsfs. It consists of a 3-helical bundle (H1, H2, H3) and a 4-stranded antiparallel β-sheet (β1, β2, β3, β4). The hydrophobic core of this domain ensures the precise positioning of the central helix-turn-helix motif (H2-T-H3) required for specific recognition of the palindromic HSEs (Damberger et al 1994; Harrison et al 1994; Vuister et al 1994; Schultheiss et al 1996). The only crystal structure of a Hsf-DNA complex was reported for the DBD of the Kluyveromyces lactis Hsf (Littlefield and Nelson 1999). Interestingly, binding of 2 monomers of the DBD to the HSE motif involves protein-protein contacts mediated by the 10 amino acid residues of the loop (wing) between β3 and β4 strands, which is lacking in plant Hsfs. It will be interesting to elaborate the differences in the arrangement of DNA-bound Hsf subunits between plants and other organisms.
Similar to all known Hsf coding genes of other organisms, the DBD of plant Hsfs is encoded in 2 parts separated by the only intron, which is inserted immediately upstream of the coding part for the H2-T-H3 DNA binding motif (Fig 1). The position of the intron is identical in all cases, but the size is highly variable (Table 1). In the plant Hsf genes, the exon-intron borders are defined by the codons for the invariant Tyr residue (codons UAU or UAC) at the end of the HTH motif and the Gly residue (codons GGG, GGA, or GGT) in the following turn to β3 (intron sequence is indicated by the small case letters): 5′ TAT(C)-ag°°°°°°°°°°ag-GG(T, A, G). The arrow in Figure 1 indicates the position of the intron. Interestingly, the mammalian Hsf genes contain many additional introns (Zhang et al 1998; Manuel et al 1999). Based on structural similarities between mammalian and plant Hsfs, we marked the hypothetical positions of these introns by arrowheads at the block diagram of LpHsfA1 (Fig 1A). We will discuss arguments supporting a similar exon-intron organization in the ancient precursor gene encoding plant Hsfs.
The oligomerization domain (HR-A/B region) is connected to the characteristic cluster of basic amino acid residues at the C-terminus of the DBD by a flexible linker of 9 to 39 amino acid residues for class A, 50 to 78 amino acid residues for class B, and 14 to 49 amino acid residues for class C Hsfs (Fig 2, Table 2). From experiments with the yeast and mammalian Hsfs (Flick et al 1994; Liu and Thiele 1999), it is apparent that this linker or at least part of it is important for the oligomerization behavior. The heptad pattern of hydrophobic amino acid residues in the HR-A/B region (Fig 1B) suggest a coiled-coil structure similar to that reported also for leucine-zipper–type protein interaction domains (Peteranderl et al 1992, 1999).
Table 2.
Comparison of the linker/HR-A/B regions of Hsfs
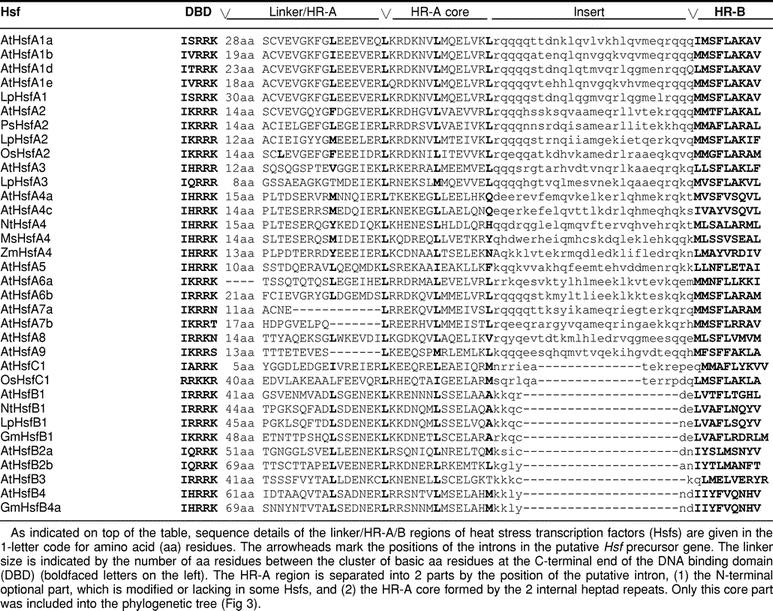
In plants, there are 3 classes in the Hsf protein family (classes A, B, and C), which are discriminated by peculiarities of their flexible linkers and HR-A/B regions (Figs 1 and 2 and Table 2). The HR-A/B region of class B Hsfs are similar to all nonplant Hsfs, whereas all class A and class C Hsfs have an extended HR-A/B region due to an insertion of 21 (class A) and 7 (class C) amino acid residues between the A and B parts (see sequence details in Table 2). It is remarkable that representatives of the third class (class C) were unnoticed so far. From analysis of expressed sequence tag (EST) libraries, they are evidently well expressed in tomato, soybean, potato, barley, and Arabidopsis, at least on the RNA level. This HsfC1 type is clearly separated from all others by sequence details of the DBDs and by the characteristics of the HR-A/B region. The significance of these extended oligomerization domains in class A and class C Hsfs for the coiled-coil structure and oligomerization behavior is not yet clear. However, there is evidence that the tomato HsfB1 exists as a dimer, whereas HsfA1 and HsfA2 are trimers.
The flexibility of size and sequence in this part of the Hsfs between the C-terminus of the DBD and the B part of the HR-A/B region is remarkable (Table 2). Because of the lack of sufficient experimental data, we can only speculate that the individual properties of Hsfs, ie, their oligomerization, their specific role and regulatory behavior in the Hsf network, and their interaction with other proteins, are dependent on sequence information in this region. In some cases the linker between DBD and the first repeat of the HR-A part is very short and/or the first repeat is lacking (see examples in Table 2, Lp-HsfA3, At-Hsfs A6a, A7a, A7b, and A9). In this context, the peculiarities of the HsfC1 in the linker/HR-A/B region are particularly striking.
In most cases, the nuclear localization signals (NLSs) of class A and class C Hsfs are monopartite or bipartite clusters of basic amino acid residues adjacent to the HR-A/B region. The corresponding motifs are marked with NLS in Figure 2 (for details of sequences and positions see Table 3). Because of the investigations on the nuclear import signals of tomato Hsfs A1 and A2 (Lyck et al 1997), it is clear that only these clusters, and not the clusters of conserved basic amino acid residues at the end of the DBD (Table 2), contribute to the nuclear import of Hsfs.
Table 3.
Functional motifs of plant Hsfs
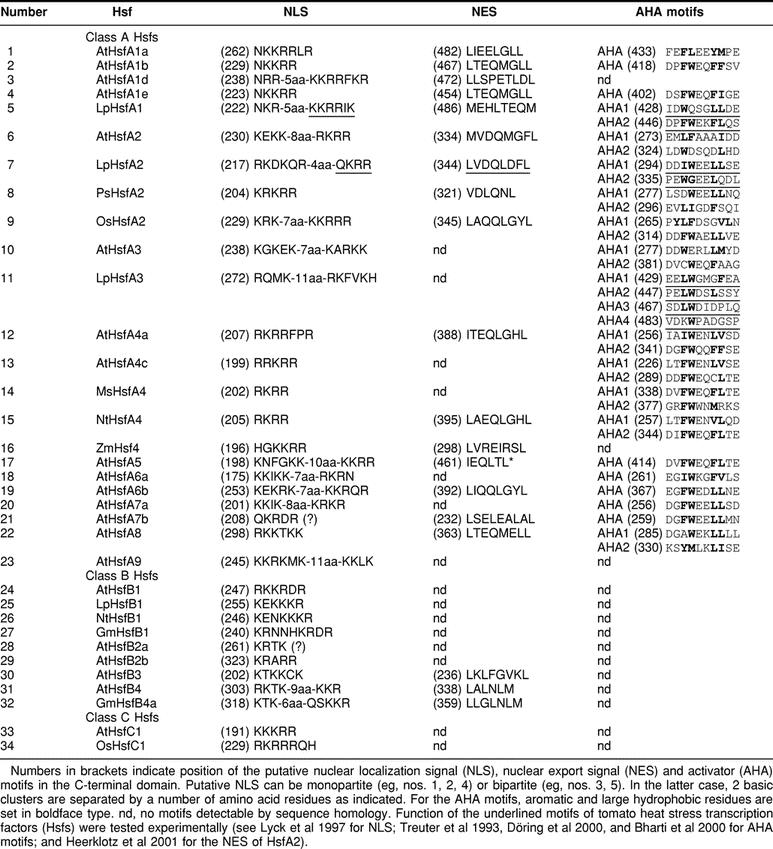
A cluster of arginine and lysine residues close to the C-terminus of tomato HsfB1 is responsible for its permanent nuclear localization (Scharf et al 1998a; Heerklotz et al 2001). Similar motifs are also found in other representatives of this group and in groups B2 and B4. An exception is At-HsfB3, which is the smallest of all Hsfs identified so far. In this case, the only cluster of basic amino acid residues, which might function as a NLS, is present in the linker region between HR-A and HR-B (Fig 2 and Table 3).
The nucleocytoplasmic distribution of proteins can be markedly influenced by nuclear export. Because of a leucine-rich export signal in the HR-C region (nuclear export signal [NES], see Fig 1 and Table 3 for sequence details), the tomato HsfA2 is mostly found in the cytoplasm unless complexed in hetero-oligomers with HsfA1 (Scharf et al 1998a; Heerklotz et al 2001). This phenomenon of changing nucleocytoplasmic distribution due to the balance of NLS and NES, which may be altered by protein modification or by interaction with other proteins, is crucial for many signaling pathways involving transcription factors (for references see Heerklotz et al 2001; Görlich and Kutay 1999). Inspection of the C-terminal parts of Arabidopsis Hsfs was shown in several cases to be similar to leucine-rich sequences, which might function as NESs (Fig 2 and Table 3).
The C-terminal activation domains (CTAD) of the Hsfs are the least conserved in sequence and size. For all class A Hsfs, the CTADs are acidic and enriched in proline, serine, threonine, glutamic acid, and aspartic acid residues. The function of Hsfs as transcription-activating proteins is evidently connected with short peptide motifs (AHA motifs, see Nover and Scharf 1997; Döring et al 2000), which are characterized by aromatic (W, F, Y), large hydrophobic (L, I, V), and acidic (E, D) amino acid residues (see examples given in Fig 1 and Table 3). With few exceptions, such AHA motifs are found in the center of the CTADs of most Arabidopsis class A Hsfs. Similar AHA motifs are also in the center of the activation domains of human, Drosophila, and yeast Hsfs and of many other transcription factors of mammals, eg, VP16, RelA, Sp1, Fos, Jun, steroid receptors, and the yeast, eg, Gal4 and Gcn4 (see references in Nover and Scharf 1997; Döring et al 2000). Most likely, they represent the essential sites of contact with subunits of the basal transcription complex as shown by pull-down experiments (Yuan and Gurley 2000; Döring, in preparation) .
It is remarkable that the C-terminal domains (CTDs) of class B Hsfs are completely different. For the group of the most conserved Hsfs in plants, ie, HsfB1, the CTD is positively charged, and AHA motifs are lacking. We have evidence that HsfB1 plays a special role in gene activation as a synergistic partner of HsfA1 (Bharti et al, unpublished data). The CTDs of Hsfs B2, B3, B4, and C1 are neutral, with clusters of basic amino acid residues interspersed. AHA motifs are not detectable. It will be interesting to test the function of these Hsfs in reporter assays alone and in combination with other Hsfs to find out which of them are synergistic coactivators (Bharti et al, in preparation) or repressors (Czarnecka-Verner et al 2000).
Four tomato Hsfs with their structural and functional identities
Although the picture is far from complete, the experimental data obtained with the tomato Hsfs indicate that the multiplicity may be connected with distinct functions in the Hsf network. The question of how many Hsfs we need cannot be answered at present. However, the multiplicity of regulatory effects in the Hsf system of tomato with only 4 Hsfs cloned and experimentally studied so far is surprising. It gives an idea of the real complexity of the Hsf network with 21 representatives in Arabidopsis. In fact, the overall complexity of Hsfs in tomato and other plants is comparable. Searching EST libraries, we found expression data for 15 new tomato Hsfs with representatives in practically all groups and classes defined for Arabidopsis (see additional information compiled in Fig 5 available in the online version only).
In the following, we will briefly summarize the relevant experimental data, indicating well-defined and nonredundant roles of the 4 tomato Hsfs in the Hsf network.
First, the constitutively expressed HsfA1 (527 aa residues) is the largest of the 4 tomato Hsfs studied so far. There is evidence that HsfA1 plays a central role for the heat stress response in general and for the expression and function of the other Hsfs. The CTAD harbors 2 AHA motifs (Table 3) that are essential for the activator function (Döring et al 2000).
Second, synthesis of HsfA2 (351 amino acid residues) is strictly heat stress dependent. It accumulates to fairly high levels in tomato cell cultures and different tomato tissues, especially in periods of repeated heat stress and recovery. Similar to HsfA1, HsfA2 is a strong transcription activator with 2 AHA motifs in its CTAD (Fig 1B, Table 3). In the course of a heat stress regimen, HsfA2 exists in 3 different forms characterized by their intracellular distribution and modes of protein interaction: (1) Nuclear form: Because of the strong C-terminal NES (see details in Fig 1B), significant nuclear retention of HsfA2 and activator function are found only in the presence of HsfA1, ie, in form of the HsfA1/A2 heterooligomer (Scharf et al 1998; Heerklotz et al 2001). (2) Cytoplasmic insoluble form: The ongoing accumulation of HsfA2 during long-term heat stress and its stability coincides with its interaction with Hsp17 class II (Scharf et al, unpublished data). During heat stress, both proteins are reversibly incorporated into the cytoplasmic chaperone complexes built of the heat stress granules (HSGs). (3) Cytoplasmic soluble form: After dissociation of the HSG complexes during the recovery, HsfA2 and small Hsps are found in soluble oligomers in the cytoplasm. Release of HsfA2 from the HSG complexes needs the activity of the Hsp90 chaperone machinery, ie, it is inhibited by geldanamycin (Scharf et al 1998a, unpublished results).
Third, HsfA3 (508 amino acid residues) is the least studied of the 4 tomato Hsfs. It is found constitutively expressed in cell cultures but is barely detectable in tomato leaves. It may represent a developmentally regulated Hsf with expression only in rapidly dividing cells. Four AHA motifs in the CTAD with a central Trp residue contribute to its activator function (Bharti et al 2000).
Fourth, HsfB1 (301 amino acid residues) is the only class B Hsf so far studied in tomato. Its very low level found in unstressed cell cultures or tissues is transiently increased several-fold after heat stress. HsfB1 is relatively short-lived and always found in the nucleus. In contrast to the class A Hsfs, HsfB1 probably has no activator function itself. This can be explained by the differences in the CTDs (see above). However, we have preliminary evidence that coexpression of low levels of HsfA1 with HsfB1 can result in strong synergistic effects in reporter gene activation (Bharti et al, unpublished data).
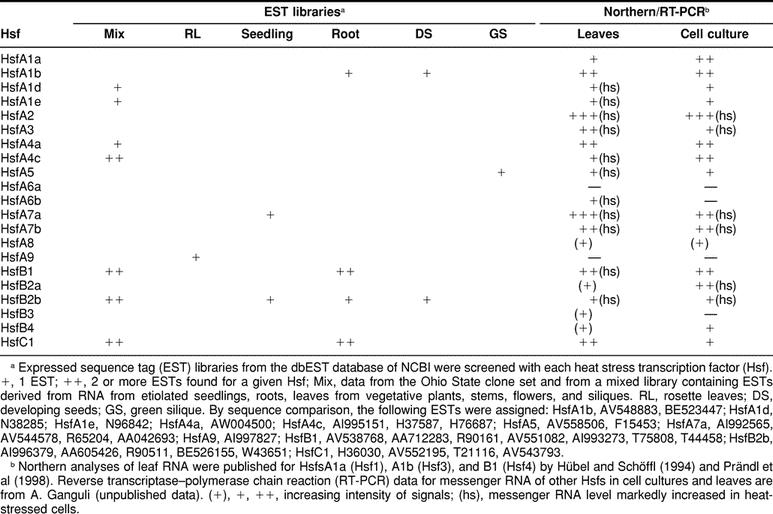
Expression of Arabidopsis Hsfs
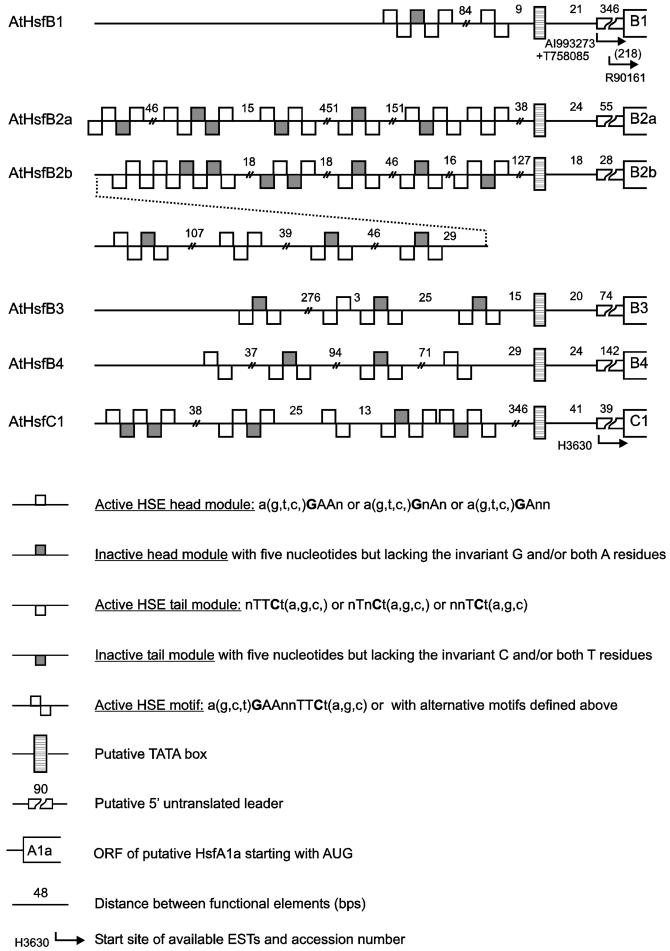
Expression data for Arabidopsis Hsfs are fragmentary and stem from different sources (see compilation in Table 4). First, a number of ESTs are found in the database. They were derived from RNAs isolated from different tissues of Arabidopsis (see details given in footnote b to Table 4). Second, Schöffl’s group published data from Northern analyses for Hsfs A1a, A1b, and B1 using control and heat stress leaves (Hübel and Schöffl 1994; Prändl et al 1998). Third, we did some preliminary studies using RNA from control and heat stress cell cultures and leaves (A. Ganguli, unpublished data). Among the 21 Hsfs, 15 are represented in Table 4, whereas Hsfs A3, A6a, A6b, A7b, B2a, and B3 could not be detected in any of the tissues or conditions analyzed so far. However, a severe drawback of the data from EST libraries is the lack of samples from heat stress tissues. This is best illustrated for HsfA2. Comparable to the situation with the tomato HsfA2, its messenger RNA was not detectable in control cells but was detected very strongly in heat stress cells. From this expression pattern, it is not surprising that no EST was found in the libraries created exclusively from RNA isolated from control tissues. Closer inspection of the putative promoter/5′-UTR regions indicate an intriguing pattern of HSE modules for all Hsf-encoding genes of Arabidopsis (see Fig 6 in the online version only). It was shown earlier (Treuter et al 1993; Czarnecka-Verner et al 2000) that, depending on the position of the HSE, upstream or downstream of the TATA box, Hsfs may activate or repress the transcription of the adjacent gene. It is tempting to speculate that the complex HSE patterns in the promoter/5′-UTR regions indicate a network of regulatory interactions between Hsfs and their encoding genes.
Table 4.
Expression of Arabidopsis Hsfs
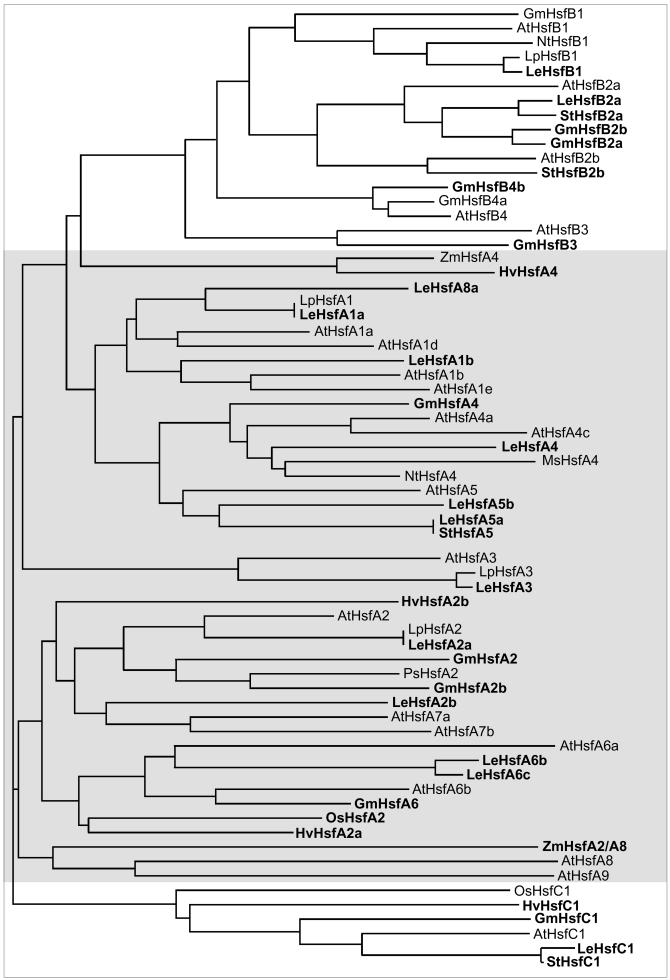
Phylogenetic analysis of plant Hsfs
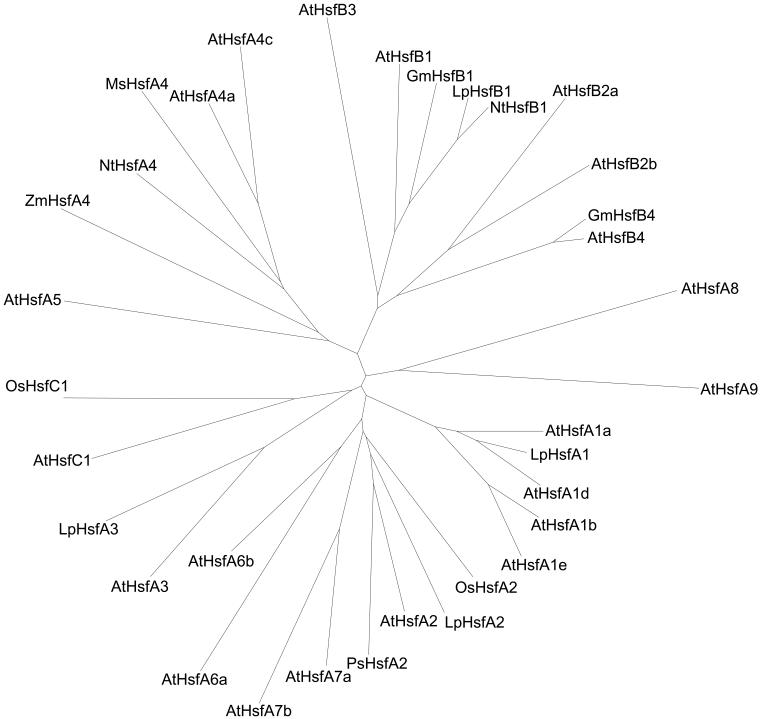
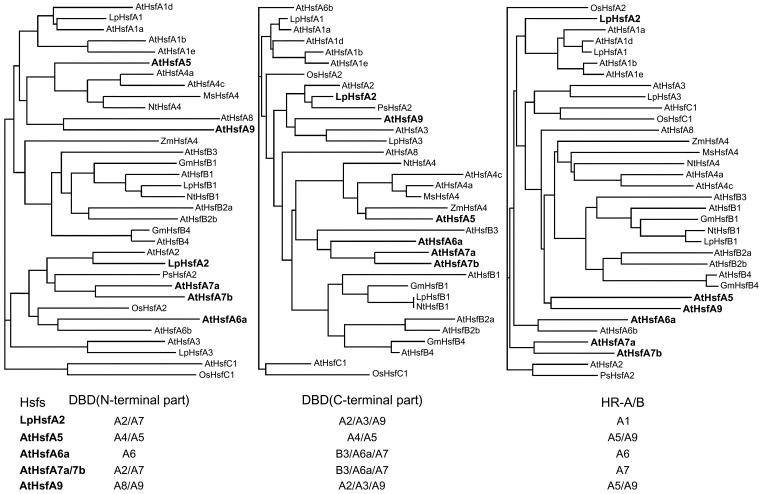
Analyses of the amino acid sequences in the conserved N-terminal half of the Hsfs revealed 2 remarkable features (Table 2): (1) Two highly variable parts, ie, the flexible linker between DBD and HR-A and the insert connecting the HR-A and HR-B parts, are interspersed between the very conserved DBD and the 2 parts of the oligomerization domain. (2) The positions of most of the Hsfs in the phylogenetic tree (Fig 3) are fixed irrespective of the sequence parts used for the generation, ie, the N-terminal or C-terminal parts of the DBD, the whole DBD, the HR-A/B region, or the DBD plus HR-A/B region as used for the generation of the tree presented in Figure 3. However, there are a few exceptions to this rule, ie, Hsfs changing their position because they are evidently mosaic proteins formed from different phylogenetic parts (see examples and explanations given in Fig 4).
Fig 3.
Phylogenetic relationship of Hsfs based on amino acid sequence comparison of the DBDs and HR-A/B regions (Clustal analysis). The consensus tree for all Hsfs was elaborated using the Clustalx 1–8_msw and Tree view software. For names, accession numbers, and identification of the Hsfs, see Table 1 and explanations given to this table. An extended version of this tree is available in the online version (Fig 5). It includes sequence information of many additional Hsfs from tomato, potato, barley, and soybean as derived from EST libraries
Fig 4.
Clustal analysis of the phylogenetic relationship based on the comparison of sequences of different parts of the N-terminal half of plant Hsfs. Three different conserved parts were used to create the phylogenetic trees: (1) the N-terminal part of the DBD until the position of the intron, (2) the C-terminal part of the DBD, and (3) the HR-A/B region, including 2 heptad repeats of HR-A, the insert, and HR-B (see borders defined in Table 2). Most Hsfs are found in identical groups irrespective of the sequence parts used for the analysis. However, a few Hsfs marked by boldfaced letters change their positions (see also the summary given at the bottom of the figure). We assume that, similar to the situation in the present mammalian Hsf genes, these 3 parts were separated by introns in the ancient plant Hsf precursor gene, ie, exon shuffling could have generated mosaic Hsfs. For AtHsfs A7a and A7b, the following situation is envisaged. The N-terminal part of the DBD and the HR-A/B region are derived from the putative A2/A7 precursor gene, whereas the C-terminal part of the DBD stems are from the B3/A7/A6a precursor gene
A possible explanation for this behavior can be found by comparing the exon-intron organization of mammalian and plant Hsf genes. As mentioned already, there is only one intron in the coding part of the DBD in all plant Hsfs. However, besides this conserved intron in the DBD, there are 11 additional introns in the mouse hsf1 and hsf2 genes separating structural and functional modules (Zhang et al 1998; Manuel et al 1999). This exon-intron organization is particularly striking for the region encoding the DBD/HR-A/B part of these Hsfs. It is tempting to speculate that the position of introns in an ancient precursor gene of plant Hsfs is similar to the present mammalian genes. At least insertion of these hypothetical borders in the block diagram for the Lp-HsfA1 (Fig 1A) indicates that separate exons may have generated the C-terminal part of the DBD, the flexible linker, the HR-A part plus insertion, and the HR-B part plus adjacent NLS region. This hypothetical exon-intron structure helps to understand the variability of sequence and length of the linker and of the insertion between HR-A and HR-B, making 6 amino acid residues for all class B Hsfs, 6 + 7 amino acid residues for HsfC1, and 6 + 21 amino acid residues for all class A Hsfs (Table 2). We further hypothesize that a type of exon shuffling and elimination of most of the introns in early times of the evolution of plant hsf genes contributed to the fixation of the present state and the generation of the mosaic type of Hsfs exemplified in Figure 4 for Lp-HsfA2 and At-Hsfs A5, A6a, A7a/A7b, and A9.
Fig 5.
Extended version of phylogenetic relationships of plant Hsfs. To emphasize the complexity of the plant Hsf family in general, an extended version of the phylogenetic tree was created by Clustal analysis based on the N-terminal parts of the DBD of those Hsfs contained in Fig 3 plus a considerable number of additional partial clones derived from the database, mostly from EST libraries of tomato (Lycopersicon esculentum), potato (Solanum tuberosum), soybean (Glycine max), and barley (Hordeum vulgare). For other abbreviations see Fig 3 and Table 1. New entries in the figure are marked by boldfaced letters. Information was derived from the following ESTs and accession numbers: L esculentum HsfA1a (AW933448, AW399336, AW223123), HsfA1b (BE354387), HsfA2a (AW034874), HsfA2b (AW930998), HsfA3 (BE433610, AI895834, AW034135, AW035854, AW035844, AW030642, AW033013; all ESTs represent incompletely spliced messenger RNAs), HsfA4 (AW038959, AW933529), HsfA5a (AW217982, AW041695, AW030725, BF096782), HsfA5b (AW034402), HsfA6b (AW036683, AW932142, AW222011, BE 434585, BE433803, AI895294, AI489721, BG132247), HsfA6c (BG351853), HsfA8a (AW738023, mosaic Hsf, whose annotation to the HsfA8 group depends on the C-terminal part of the DBD and the HR-A/B region), HsfA8b (AW931892, not included, because only C-terminal part with HR-A/B region available), HsfB1 (BF097217, BG134658, AI895934), HsfB2a (AW931781, AW220758, AW931176), HsfC1 (AW738534, AW979619, BE451302, AW649243); G max HsfA2a (Hsf21 fragment, Z46952), HsfA2b (AW164509), HsfA4 (BE611683, AW756148, BG405291, BE330669), HsfA6 (BG041837), HsfB2a (BE346810, BF067962), HsfB2b (AW703969), HsfB3 (BE019974), HsfB4b (BF597135), HsfC1 (BE347442, AW596493, BG352891); S tuberosum HsfA5 (BF459947), HsfB2a (BE473183), HsfB2b (BF052865), HsfC1 (BF459641); H vulgare HsfA2a (BE603513), HsfA4 (BE216310), HsfA2b (BF264338), HsfC1 (BF616419); Zea mays: ZmHsfA2/A8 (Hsfa fragment, S61458)
Fig 2.
Continued
Fig 6.
Promoter architecture of Arabidopsis Hsf genes with respect to the positioning of HSE clusters. For orientation and positioning of the putative TATA box, we used the well-defined start site of the open reading frames, data for genomic clones with experimentally defined transcription start sites, and the starting points of the indicated ESTs. Symbols are explained at the bottom of the figure. Numbers on the left refer to the corresponding numbers in Table 1
Fig 6.
Continued
Acknowledgments
Experimental work in our group was supported by grants from the Deutsche Forschungsgemeinschaft and by the Fonds der Chemischen Industrie.
REFERENCES
- Aranda MA, Escaler M, Thomas CL, Maule AJ. A heat stress transcription factor in pea is differentially controlled by heat and virus replication. Plant J. 1999;20:153–161. doi: 10.1046/j.1365-313x.1999.00586.x. [DOI] [PubMed] [Google Scholar]
- Bharti K, Schmidt E, Lyck R, Bublak D, Scharf KD. Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 2000;22:355–365. doi: 10.1046/j.1365-313x.2000.00746.x. [DOI] [PubMed] [Google Scholar]
- Bienz M, Pelham HRB. Mechanisms of heat-shock gene activation in higher eukaryotes. Adv Genet. 1987;24:31–72. doi: 10.1016/s0065-2660(08)60006-1. [DOI] [PubMed] [Google Scholar]
- Clos J, Westwood JT, Becker PB, Wilson S, Lambert U, Wu C. Molecular cloning and expression of a heaxameric Drosophila heat stress factor subject to negative regulation. Cell. 1990;63:1085–1097. doi: 10.1016/0092-8674(90)90511-c. [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Yuan CX, Fox PC, Gurley WB. Isolation and characterization of six heat stress transcription factor cDNA clones from soybean. Plant Mol Biol. 1995;29:37–51. doi: 10.1007/BF00019117. [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Yuan CX, Scharf KD, Englich G, Gurley WB. Plants contain a novel multi-member class of heat stress factors without transcriptional activator potential. Plant Mol Biol. 2000;43:459–471. doi: 10.1023/a:1006448607740. [DOI] [PubMed] [Google Scholar]
- Damberger FF, Pelton JG, Harrison CJ, Nelson HCM, Wemmer DE. Solution structure of the DNA-binding domain of the heat stress transcription factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Protein Sci. 1994;3:1806–1821. doi: 10.1002/pro.5560031020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring P, Treuter E, Kistner C, Lyck R, Chen A, Nover L. Role of AHA motifs for the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell. 2000;12:265–278. [PMC free article] [PubMed] [Google Scholar]
- Flick KE, Gonzalez L, Harrison CJ, Nelson HCM. Yeast heat stress transcription factor contains a flexible linker between the DNA-binding and trimerization domains—implications for DNA binding by trimeric proteins. J Biol Chem. 1994;269:12475–12481. [PubMed] [Google Scholar]
- Gagliardi D, Breton C, Chaboud A, Vergne P, Dumas C. Expression of heat stress factor and heat stress protein 70 genes during maize pollen development. Plant Mol. Biol. 1995;29:841–856. doi: 10.1007/BF00041173. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HCM. Crystal structure of the DNA binding domain of the heat stress transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. The balance of nuclear import and export determines the intracellular distribution of tomato heat stress transcription factor HsfA2. Mol Cell Biol. 2001;21:1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübel A, Schöffl F. Arabidopsis heat stress factor: isolation and characterization of the gene and the recombinant protein. Plant Mol Biol. 1994;26:353–362. doi: 10.1007/BF00039545. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Katoh T, Sato S, Nakamura A, Asamizu E, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 3, II: sequence features of the 4,251,695 bp regions covered by 90 P1, TAC and BAC clones. DNA Res. 2000;7:217–221. doi: 10.1093/dnares/7.3.217. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, and Rounsley SD. et al. . 1999 Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 402:761–768. [DOI] [PubMed] [Google Scholar]
- Littlefield O, Nelson HCM. A new use for the ‘wing’ of the “winged” helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- Liu PCC, Thiele DJ. Modulation of human heat stress factor trimerization by the linker domain. J Biol Chem. 1999;274:17219–17225. doi: 10.1074/jbc.274.24.17219. [DOI] [PubMed] [Google Scholar]
- Lyck R, Harmening U, Höhfeld I, Treuter E, Scharf KD, Nover L. Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta. 1997;202:117–125. doi: 10.1007/s004250050110. [DOI] [PubMed] [Google Scholar]
- Manuel M, Sage J, Mattei MG, Morange M, Mezger V. Genomic structure and chromosomal localization of the mouse Hsf2 gene and promoter sequences. Gene. 1999;232:115–124. doi: 10.1016/s0378-1119(99)00092-x. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat stress transcriptional response: cross talk between family of heat stress factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Nakai A. New aspects in the vertebrate heat stress factor system: HsfA3 and HsfA4. Cell Stress Chaperones. 1999;4:86–93. doi: 10.1379/1466-1268(1999)004<0086:naitvh>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L. Expression of heat stress genes in homologous and heterologous systems. Enzyme Microb Technol. 1987;9:130–144. [Google Scholar]
- Nover L 1991 Heat Shock Response. CRC Press, Boca Raton, FL. [Google Scholar]
- Nover L, Scharf KD. Heat stress proteins and transcription factors. Mol Cell Life Sci. 1997;53:80–103. doi: 10.1007/PL00000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteranderl R, Nelson HCM. Trimerization of the heat stress transcription fact factor by a triple-stranded alpha-helical coiled-coil. Biochemistry. 1992;31:12272–12276. doi: 10.1021/bi00163a042. [DOI] [PubMed] [Google Scholar]
- Peteranderl R, Rabenstein M, Shin Y, Liu CW, Wemmer DE, King DS, Nelson HCM. Biochemical and biophysical characterization of the trimerization domain from the heat stress transcription factor. Biochemistry. 1999;38:3559–3569. doi: 10.1021/bi981774j. [DOI] [PubMed] [Google Scholar]
- Prändl R, Hinderhofer K, Eggers-Schumacher G, Schöffl F. Hsf3, a new heat stress factor from Arabidopsis thaliana, derepresses the heat stress response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen Genet. 1998;258:269–278. doi: 10.1007/s004380050731. [DOI] [PubMed] [Google Scholar]
- Rabindran SK, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat stress factor. Proc Natl Acad Sci U S A. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat stress factors with distinct inducible and constitutive DNA binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Katoh T, Asamizu E, Kotani H, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5, X: sequence features of the regions of 3,076,755 bp covered by sixty P1 and TAC clones. DNA Res. 2000a;7:31–63. doi: 10.1093/dnares/7.1.31. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Katoh T, Asamizu E, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 3, I: sequence features of the regions of 4,504,864 bp covered by sixty P1 and TAC clones. DNA Res. 2000b;7:131–135. doi: 10.1093/dnares/7.2.131. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol. 1998a;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Höhfeld I, Nover L. Heat stress response and heat stress transcription factors. J Biosci. 1998b;23:313–329. [Google Scholar]
- Scharf KD, Rose S, Thierfelder J, Nover L. Two cDNAs for tomato heat stress transcription factors. Plant Physiol. 1993;102:1355–1356. doi: 10.1104/pp.102.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Rose S, Zott W, Schöffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F, Prändl R, Reindl A. Regulation of the heat stress response. Plant Physiol. 1998;117:1135–1141. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. Isolation of a cDNA for HSF2: evidence for two heat stress factor genes in humans. Proc Natl Acad Sci U S A. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss J, Kunert O, Gase U, Scharf KD, Nover L, Rüterjans H. Solution structure of the DNA-binding domain of the tomato heat stress transcription factor HSF24. Eur J Biochem. 1996;236:911–921. doi: 10.1111/j.1432-1033.1996.00911.x. [DOI] [PubMed] [Google Scholar]
- Shoji T, Kato K, Sekine M, Yoshida K, Shinmyo A. Two types of heat stress factors in cultured tobacco cells. Plant Cell Rep. 2000;19:414–420. doi: 10.1007/s002990050749. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HRB. Yeast heat stress factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Treuter E, Nover L, Ohme K, Scharf KD. Promoter specificity and deletion analysis of three heat stress transcription factors of tomato. Mol Gen Genet. 1993;240:113–125. doi: 10.1007/BF00276890. [DOI] [PubMed] [Google Scholar]
- Vuister GW, Kim SJ, Orosz A, Marquardt J, Wu C, Bax A. Solution structure of the DNA-binding domain of Drosophila heat stress transcription factor. Nat Struct Biol. 1994;1:605–614. [PubMed] [Google Scholar]
- Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat stress transcription factor. Cell. 1988;54:841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- Wu C. Heat stress transcription factors. Annu Rev Cell Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Yuan CX, Gurley WB. Potential targets for HSF1 within the preinitiation complex. Cell Stress Chaperones. 2000;5:229–242. doi: 10.1379/1466-1268(2000)005<0229:ptfhwt>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Koushik S, Dai RJ, Mivechi NF. Structural organization and promoter analysis of murine heat stress transcription factor-1 gene. J Biol Chem. 1998;273:32514–32521. doi: 10.1074/jbc.273.49.32514. [DOI] [PubMed] [Google Scholar]