Abstract
A total of 89 J-domain proteins were identified in the genome of the model flowering plant Arabidopsis thaliana. The deduced amino acid sequences of the J-domain proteins were analyzed for an assortment of structural features and motifs. Based on the results of sequence comparisons and structure and function predictions, 51 distinct families were identified. The families ranged in size from 1 to 6 members. Subcellular localizations of the A thaliana J-domain proteins were predicted; species were found in both the soluble and membrane compartments of all cellular organelles. Based on digital Northern analysis, the J-domain proteins could be separated into groups of low, medium, and moderate expression levels. This genomics-based analysis of the A thaliana J-domain proteins establishes a framework for detailed studies of biological function and specificity. It additionally provides a comprehensive basis for evolutionary comparisons.
INTRODUCTION
DnaJ was originally characterized from Escherichia coli as a 41-kDa heat shock protein (Georgopoulos et al 1980). It was subsequently demonstrated in both biochemical and genetic experiments that DnaJ interacts directly with DnaK and GrpE (Liberek et al 1991; Scidmore et al 1993; Goffin and Georgopoulos 1998), constituting a molecular chaperone machine (Bukau and Horwich 1998; Miernyk 1997, 1999). Additionally, there is evidence that DnaJ can act independently as a chaperone (Laufen et al 1999). DnaJ binds to the adenosine triphosphate (ATP)–ligated form of DnaK and stimulates hydrolysis to adenosine 5′-diphosphate plus inorganic phosphate (Pi) (Liberek et al 1991).
DnaJ can be envisioned as having a linear, modular sequence consisting of the J-domain, a proximal G/F-domain, and a distal zinc finger (CxxCxGxG)4 domain, followed by less conserved C-terminal sequences (Caplan et al 1993; Silver and Way 1993). The J-domain consists of approximately 75 conserved amino acid residues that comprise 4 α-helices. The invariant tripeptide, HPD, which is both characteristic of and absolutely essential for the biological function of J-domains, is located between helices II and III. The G/F-domain, a sequence rich in Gly and Phe residues, comprises a flexible linker region that helps to convey specificity of interactions among DnaK, DnaJ, and target polypeptides (Wall et al 1995; Yan and Craig 1999). The zinc finger domain is believed to mediate protein-protein interactions among DnaK, DnaJ, and target polypeptides (Banecki et al 1996; Szabo et al 1996).
In recent years, a large number of DnaJ-related proteins have been nonsystematically characterized from a variety of different organisms. The relatively small genome size (The Arabidopsis Genome Initiative 2000), coupled with an abundance of well-defined mutants (Koncz et al 1992) and the ease of genetic manipulation (Redei 1975), has resulted in extensive study of Arabidopsis thaliana as a model flowering plant (Meinke et al 1998). Herein, a genomics approach to analysis of all the J-domain proteins from the simple flowering plant A thaliana is presented.
MATERIALS AND METHODS
Database analysis
The publication version of the A thaliana genome sequence was accessed via the San Diego Supercomputer Center (http://www.sdsc.edu/index.html). Using the National Center for Biotechnology Information (NCBI) Web site (http://www.ncbi.nlm.nih.gov/BLAST/), an iterative BLAST search was conducted using the previously cloned J-domain sequences (Zhou et al 1995; Kroczyńska et al 1996, 2000; Lin and Lin 1997; Zhou and Miernyk 1999; Miernyk and Coop 2000) as the search terms.
Nomenclature
Eukaryotic proteins related to DnaJ were initially referred to as DnaJ homologues. As the sequences of an increasing number of divergent proteins accumulated, a more systematic nomenclature became necessary. The initial attempt separated protein sequences into groups I, II, and III (Cheetham and Caplan 1998). Type I sequences would contain the J-, G/F-, and zinc finger domains, type II sequences would have the J- plus either a G/F- or zinc finger domain, and type III sequences have only the J-domain. It was subsequently noted that Roman numerals are not allowed in gene names, and it was proposed that A, B, and C be substituted for I, II, and III (Ohtsuka and Hata 2000). Furthermore, a complete nomenclature was proposed: 2 lowercase letters for the genus and species, Dj, A, B, or C, plus an Arabic numeral, indicating chronology (Ohtsuka and Hata 2000). In this system the first of the A thaliana J-domain proteins is designated atDjB1. This nomenclature has been used with one minor modification. The original proposals required the (CxxCxGxG)4 motif, which mediates protein-protein interactions, for type III/C sequences. Herein, any of the well-defined sequence motifs that mediate protein-protein interactions can be substituted for the DnaJ zinc-binding domain (other types of zinc finger sequences, coiled-coil motifs, tetratricopeptide repeat sequences).
J-domain sequence comparisons
Because the primary sequence lengths of the A thaliana J-domain proteins varied from as small as 112 to as large as 2535 amino acids in length, it seemed unlikely that overall group comparisons would yield meaningful information. Thus, only the J-domains were compared. The sequences were manually aligned using the absolutely conserved tripeptide HPD as the center point. The alignments were analyzed using SEQBOOT, and a tree constructed using PHYLIP (Felsenstein 1989).
In silico protein localization
Predictions of subcellular location of the A thaliana J-domain proteins used the following algorithms to search the J-domain protein amino acid sequences: PSORT (http://psort.nibb.ac.jp/form.html) (Nakai and Horton 1999); TargetP (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al 2000); Predotar (http://www.inra.fr/Internet/Produits/Predotar/index.html); and MITOPROT (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter) (Claros and Vincens 1996) plus direct visual examination.
Digital analysis of gene expression
The advanced BLAST algorithm was used to search the NCBI dbEST for A thaliana J-domain protein sequences (31 014 entries). The number of expressed sequence tag (EST) clones for each of the J-domain proteins was used for digital Northern analysis (Audic and Claverie 1997; Ewing et al 1999). Only EST clones isolated from nonnormalized libraries and sequenced from the 5′-end were scored (Audic and Claverie 1997; Mekhedov et al 2000; Ohlrogge and Benning 2000).
RESULTS AND DISCUSSION
The A thaliana J-domain proteins
An iterative BLAST search of the publication version of the A thaliana genome sequence (The Arabidopsis Genome Initiative 2000), using the deduced amino acid sequences of the previously analyzed J-domains as the search terms, unveiled a total of 89 proteins. Analyses aimed at reducing this final number, by exposing potential duplicity arising from errors in sequencing and gene identification, were not productive. The complete annotation of all 89 deduced amino acid sequences can be downloaded from http://www.agron.missouri.edu/ars_columbia/miernyk.html.
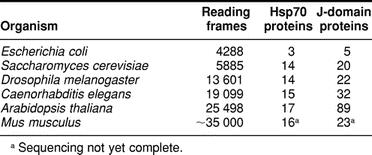
The relatively large number of A thaliana J-domain proteins was unexpected. Results from analyses of the yeast (Goffeau et al 1996), worm (The C. elegans Sequencing Consortium 1998), and fly (Adams et al 2000) genomes suggested a relatively constant relationship between the total number of 70-kDa stress proteins and J-domain proteins (Table 1). Taking into account the presence of the plastid class of organelles in plant cells, the number of 70-kDa stress proteins is similar to those of the other organisms (Lin et al 2001). Even considering what might be anticipated as additional plastidial J-domain proteins, the total number is unexpectedly large.
Table 1.
Relationships between genome size and the compliment of Hsp70 and J-domain chaperone proteins
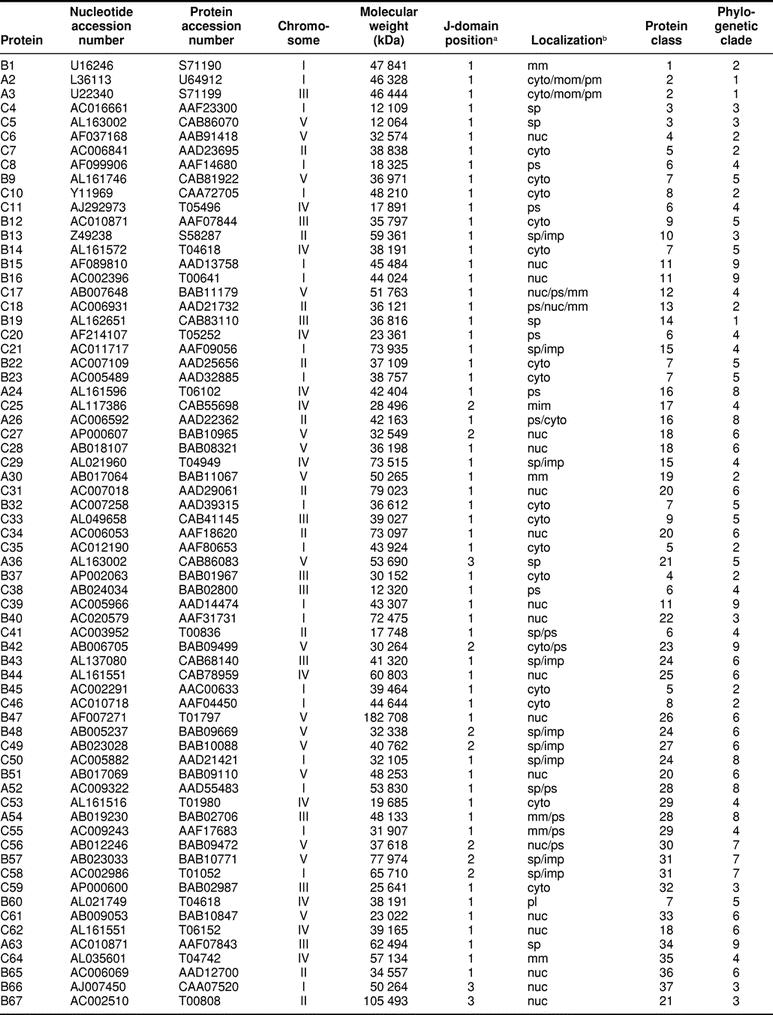
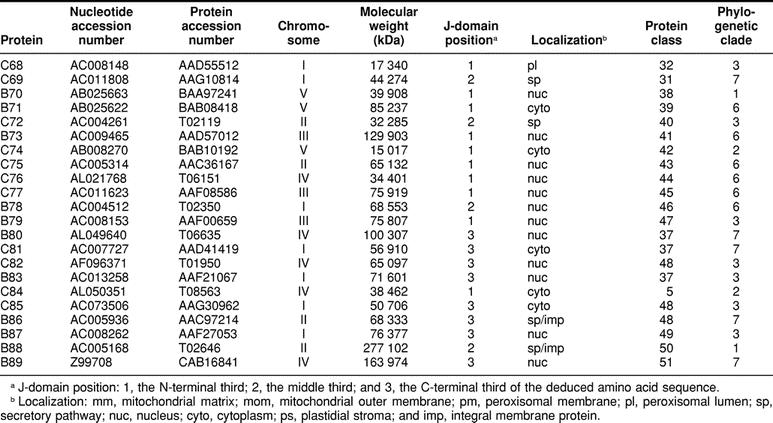
The 89 genes were distributed among the A thaliana chromosomes; I, 27; II, 14; III, 13; IV, 16; and V, 19 (Table 2). Of the 89 reading frames, 9 encode type A J-domain proteins, 35 type B, and 45 type C. There is no obvious position or location relationship among the genes, nor are there any structural or functional grouping of the encoded proteins.
Table 2.
The Arabidopsis thaliana J-domain proteins
Protein families
The deduced amino acid sequences of the A thaliana J-domain proteins were subjected to an extensive set of analyses to reveal the occurrence of structural and functional domains and motifs. The results from these analyses were a major consideration for division of the total into a series of families. The results from BLAST searches of the genomes of other organisms also played a role in defining the protein families. In total, 51 families were distinguished (http://www.agron.missouri.edu/ars_columbia/miernyk.html) (Table 2). The families vary in size from 1 to 6 members. Both median and mode values for the population are 1.
Several of the A thaliana J-domain protein families are distinguished by the relationship of the members with proteins previously characterized from other organisms. Family 2 (proteins A2 and A3), family 5 (C7, C35, B45, and C84), and family 15 (C21 and C29) are orthologous with the Saccharomyces cerevisiae YDJ1, DJP1/CAJ1, and SEC63 families, respectively. Families 7 (B9, B14, B22, B23, B32, and B60) and 9 (C12 and C33) contain paralogous members of the Hsp40 superfamily. Family 14 includes a single protein, atDjB19, related to members of the Homo sapiens hDj9/ERj3/HEDJ family. The family 21 (A36 and B37), 24 (B43, B48, and C50), and 32 (C59 and C68) proteins are orthologous with members of the mouse mmDj11, mmDj10, and mmDj6 families, respectively (Ohtsuka and Hata 2000). Families 37 (B66, C80, C81, C83) and 48 (C82, C85, C86) comprise 2 variants of the auxilin family of J-domain proteins (Quesada et al 1997; Umeda et al 2000). The atDjB88 protein, the only member of family 50, is very large, at 2535 amino acid residues. Initially, it seemed likely that a stop codon had been missed during gene annotation. Recently, however, an almost equally large homolog, RME-8, was identified from Caenorhabditis elegans (Zhang et al 2000).
A few of the J-domain protein families have been defined during genetic studies of A thaliana or other plants. The atDjB13 protein, the sole member of family 10, was identified in a study of plant genes that allowed yeast to survive exposure to the thiol-oxidizing agent diamide (Kushnir et al 1995). The family 11 proteins (B15, B16, and C39) were initially characterized during studies of the altered response to gravity A thaliana mutants (Sedbrook et al 1999). The members of family 31 (B57, C58, C69) are very closely related to the Brassica napus S-locus protein 5 family. The atDjB65 protein, family 36 is orthologous with the AHM1 protein from wheat. The wheat protein is a nuclear matrix–localized, MAR-binding protein that, in addition to the J-domain, contains an AT-hook motif (Morisawa et al 2000).
Additionally, it can be assumed that the type A J-domain proteins that are localized within mitochondria or plastids (families 19 and 28) act as general chaperones in these organelles. This leaves 31 families (44 proteins) that cannot be assigned a function based on analogy. The only known role for J-domain proteins is in association with the 70-kDa stress proteins as molecular chaperones (Greene et al 1998). The very large number of A thaliana J-domain proteins with unassigned functions suggests possible roles in plant-specific cellular processes and signal transduction pathways, possible association with partners other than the AtHsp70 proteins, or both.
The abundance of very short J-domain proteins from A thaliana is particularly noteworthy. In many cases, these proteins are little more than a J-domain. For example, the atDjC4/5 proteins are only 112 amino acids long, including an presumptive N-terminal targeting peptide of approximately 20 amino acids. This type of J-domain protein has not previously been reported in other systems. Functional analysis of one of these proteins, atDjC8, is currently in progress. Although the recombinant protein stimulates ATPase activity of the cognate 70-kDa stress protein, it does not stimulate chaperone activity (JA Miernyk, in preparation).
J-domain sequence comparisons
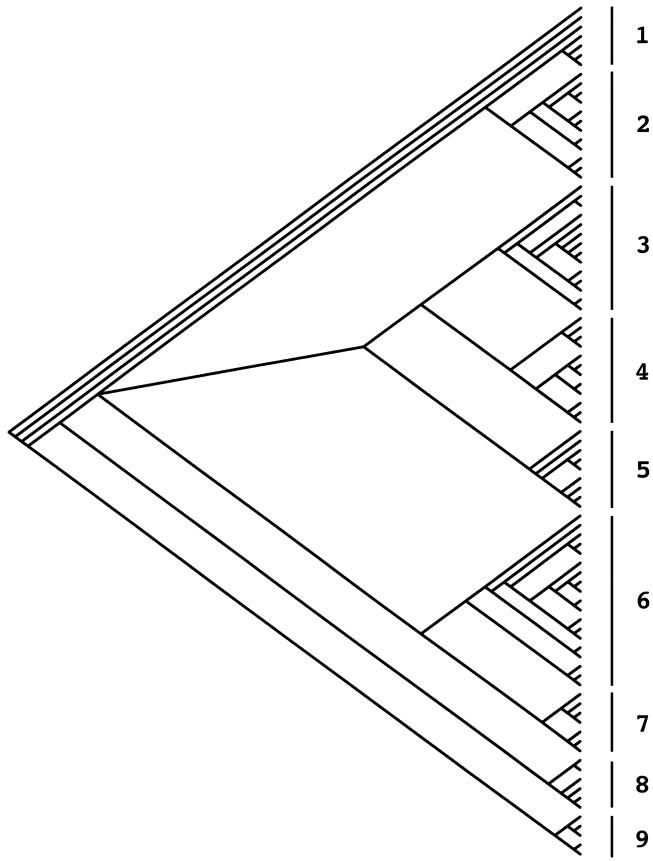
The relationships among the A thaliana J-domain sequences that were apparent after 500 rounds of bootstrapping are presented as a cladogram (Fig 1). The sequences have been resolved into 9 clades containing 5 to 18 members. There is excellent agreement between the inclusion of protein species into a family defined by the deduced amino sequences exclusive of the J-domain and the within-clade grouping. Nearly all members of any given family are located within the same clade. The most notable exception is with members of the 2 classes of auxilin-related sequences, which are more widely dispersed throughout the cladogram.
Fig 1.
Phylogenetic relationships among the J-domain sequences of the A thaliana J-domain proteins. The sequences were manually aligned, then analyzed using SEQBOOT. The resulting cladogram was constructed using the PHYLIP program. Members of the clades, from top to bottom, are as follows: 1, A3, A2, B19, C29-2, B70, C21-2, B88; 2, C18, C35, C10, C46, B45, C7, C84, A30, B1, C74, B37, C6; 3, B13, B40, C72, B79, B66, C85, B87, B83, C82, B67, C4, C5, C59, C68; 4, C25, C20, C11, C41, C38, C8, C17, C21-1, C29-1, C64, C53, C55; 5, A36, B9, B60, B14, B22, C12, B23, B32, C33; 6, C61, C75, C31, B51, C34, B71, C77, B47, B73, B44, C76, C62, C27, C28, B78, B65, C49, B43, B48; 7, B86, B89, B80, C81, B57, C69, C58; 8, C50, C56, A52, A54, A26, A24; 9, B15, B16, C39, A63, B42
The bases of the interrelationships among the clades are less apparent. In some instances, adjacent clades contain species predicted to have the same subcellular localization (Fig 1, clades 1 and 2). There seem, however, to be as many instances where this is not the case. It is likely that including still more distinguishing characteristics in the comparisons will help clarify the phylogenetic organization of this protein family.
Hennessy et al (2000) have presented an extensive analysis of the relationship between J-domain sequence and the class of the J-domain protein (I/A, II/B, III/C). They observed that the J-domain sequence of a type A protein is more likely to be closely related to other type A proteins, even from different species or kingdoms, than to type B or C sequences. Herein, the results of analysis of all of the members of a single complex organism are presented for the first time. In this study, no clear grouping of type A or B sequences is apparent (Fig 1). It should be noted, however, that this analysis includes a relatively small number of type A sequences.
Protein localization
Each of the 89 A thaliana J-domain protein sequences was subjected to analysis using multiple localization prediction algorithms. Examples of both soluble and integral membrane species were identified for virtually every subcellular compartment (Table 2). For the analysis presented herein, all proteins with a predicted localization in the secretory pathway downstream of the endoplasmic reticulum (Golgi apparatus, vacuole/lysosome, plasma membrane, extracellular) were combined under the heading “secretory pathway.” After some refinement through direct examination, a consensus for protein localization was reached for 80 of the 89. For the remaining 9 proteins, there are 2 to 3 contradictory predictions.
By far the most abundant predicted sites of J-domain protein localization are the cytoplasm and the nucleus (Table 2). Such a large proportion (27/89) of nuclear-localized chaperones is unprecedented. The most immediate question arising from the nuclear localization predictions is the identity of potential interacting proteins. Hsp70 is the only know partner for J-domain proteins (Greene et al 1998). Of the 17 AtHsp70 proteins (Lin et al 2001), 7 contain potential nuclear localization signal (NLS) sequences (Athsp70-1, -2, -3, -4, -5, -14, and -15) and are likely in vivo partners.
Six of the 9 instances of multiple location predictions involved potential N-terminal organelle-targeting peptides. The algorithms are apparently not yet sufficiently robust that it is always possible to make a high-probability distinction among a signal sequence, a transit peptide, and a mitochondrial-targeting peptide. The smallest group of predicted localization sites are those for peroxisomal proteins (Table 2), and the instances of peroxisome-cytoplasm predictions are suspect. These predictions are from PSORT, and this algorithm considers internal PTS1 sequences that are likely not functional in vivo (Subramani 1998). The only other instance of a predicted peroxisomal localization (atDjB66) is from visual examination. The N-terminus of this sequence contains a PTS2 motif (RLX5HL): A11ERLLGIAEKLL22. In vivo, however, there are good data supporting a transient association of members of the YDJ1-type (class 2) J-domain proteins with the outer surface of the peroxisome boundary membrane, mediated by protein prenylation (Preisig-Muller et al 1994; Diefenbach and Kindl 2000).
In the few instances to date where localization of A thaliana J-domain proteins has been determined, there is agreement between the computer predictions and experimental data (cf atDjB1, Kroczyńska et al 1996). However, in no case is there a published study of a J-domain protein with a controversial location. The subcellular location of atDjC8, variously predicted as plastidial, as mitochondrial, or in the secretory pathway (Table 2), has now been experimentally determined to be plastidial (JA Miernyk, in preparation). Thorough analyses of the subcellular localization of other A thaliana J-domain proteins are in progress, using green fluorescent protein (GFP) chimera. An unequivocal knowledge of cellular localization will substantially enhance subsequent studies of protein function.
Digital Northern analysis
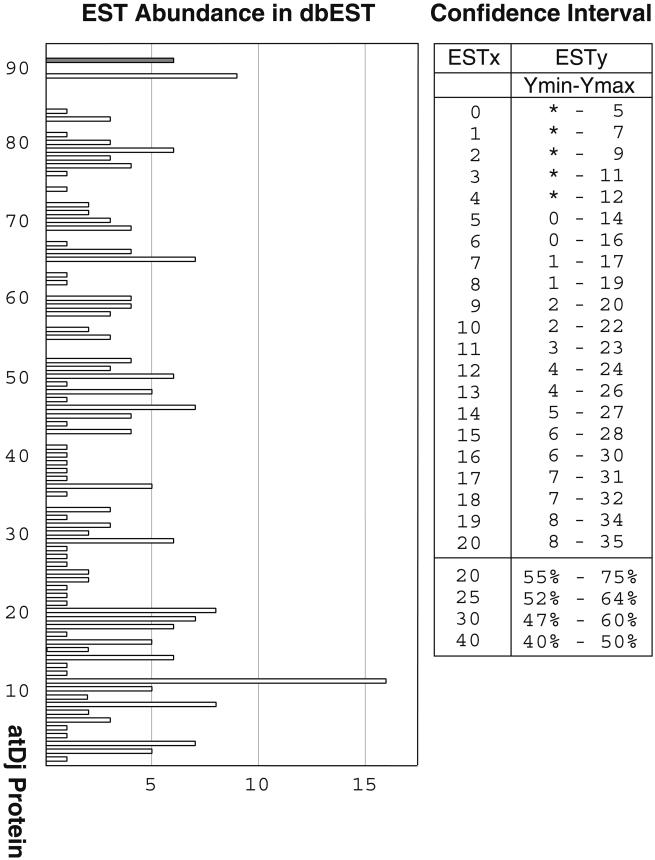
The concept of digital Northern analysis is based on the assumption that the number of EST clones will be a direct reflection of the abundance of messenger RNA (mRNA) in the population used to prepare the library (Audic and Claverie 1997; Ewing et al 1999; Mekhedov et al 2000). Just as with analog Northern data, it is equally accepted that there will be variance between “signal strength” and actual protein levels due to posttranslational regulation, mRNA, and protein turnover. The results from digital Northern analysis do, however, provide a first approximation of protein abundance.
Quantitation of the A thaliana J-domain protein ESTs in dbEST is presented in Figure 2. The EST clones can be separated into 3 classes: (1) those not significantly different (at the 95% confidence interval) from 0; (2) those that fall between 5 and 7; and (3) those with an abundance greater than 7. Seventy of the 89 proteins fall into class 1, 15 into class 2, and only 4 into class 3. It can be concluded that 19 of the A thaliana J-domain proteins are expressed at a relatively high level. Within this group, the number of ESTs for atDjC11 (16) is significantly larger than that for any other protein, suggesting a very high level of expression (Fig 2). The number of ESTs for A thaliana plastidial dihydrolipoyl acetyltransferase, a component of the pyruvate dehydrogenase complex that participates in fatty acid biosynthesis (Mooney et al 1999), is included for the sake of comparison (White et al 2000).
Fig 2.
Digital Northern analysis of the A thaliana J-domain proteins. The EST abundance in dbEST, as of January 2001, is presented. The histograms are, from bottom to top, atDjB1 to atDjC89. The shaded histogram, included for the sake of comparison, is for the plastidial enzyme dihydrolipoyl acetyltransferase. The differences between numbers, at the 95% confidence interval, are also presented. For a given EST number, indicated in the first column, the number of ESTs immediately beyond the confidence interval is indicated in the second column (Audic and Claverie 1997; Mekhedov et al 2000)
There is no obvious pattern of grouping or association (localization, type, size) among the J-domain proteins relative to gene expression. It will be necessary to validate the digital Northern results with those from analog Northern results. To date, the analog data are available for only a few proteins (Kroczyńska et al 1996, 2000; Zhou and Miernyk 1999). The analog Northern data from atDj1, atDj3, and atDj6 are all consistent with constitutive expression. This is in marked contrast to E coli DnaJ, which is a classic heat shock protein (Georgopoulos et al 1980). In addition to making comparisons among the 89 species of A thaliana J-domain proteins, it will be both interesting and informative to determine which is up- or down-regulated in response to changes in environmental conditions. For example, experimental results suggest that the mRNA levels for atDj8, a small plastidial protein, are responsive to light (JA Miernyk, in preparation), whereas those of atDj11, another small plastidial protein, are not (W. Orme and J. C. Gray, personal communication). Expression of atDjC8 is also circadian and diurnally regulated (Schaffer et al 2001). Furthermore, it has been recently reported that expression of atDjB19 was increased nearly 3-fold by a transient water stress, but was largely unaffected by cold temperature treatment (Seki et al 2001).
The J-domain proteins from the model flowering plant A thaliana comprise a family of chaperones unprecedented in both size and complexity. It is likely that this has resulted from the necessity for plants to continuously respond to a myriad of environmental insults (Nover and Miernyk 2001). The J-domain protein catalog provided herein should provide a useful framework for future studies that will append “functional” to the extant genomics.
Table 2.
Continued
Acknowledgments
F. Marsilio conducted the phylogenetic analyses presented in Figure 1, and B. P. Mooney assisted with preparation of Figure 2. Professor W. H. Miernyk provided a lucid explanation of the Poisson distribution and Bayesian statistics necessary for analysis of the digital Northern data. The efforts and patience of K. L. O'Donnell in explaining the bases of phylogenetics are greatly appreciated.
REFERENCES
- Adams MD, Celniker SE, and Holt RA. et al. 2000 The genome sequence of Drosophila melanogaster. Science. 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Banecki B, Liberek K, Wall D, Wawrzynow A, Georgopoulos C, Bertoli E, Tanfani F, Zylicz M. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J Biol Chem. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- The C. elegans Genome Initiative. Sequence and analysis of the genome of C. elegans. Science. 2000;282:2012–2018. [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homologues of Escherichia coli DnaJ: a diverse protein family that functions with hsp70 stress proteins. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Diefenbach J, Kindl H. The membrane-bound DnaJ protein located at the cytosolic site of glyoxysomes specifically binds the cytosolic isoform 1 of Hsp70 but not other Hsp70 species. Eur J Biochem. 2000;267:746–754. doi: 10.1046/j.1432-1327.2000.01053.x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Kahla AB, Poirot O, Lopez F, Audic S, Claverie JM. Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res. 1999;9:950–959. doi: 10.1101/gr.9.10.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP-Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Georgopoulos CP, Lundquist-Heil A, Yochem J, Feiss M. Identification of the E. coli dnaJ gene product. Mol Gen Genet. 1980;178:583–588. doi: 10.1007/BF00337864. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Burrell BG, and Bussey H. et al. 1996 Life with 6000 genes. Science. 274:563–567. [DOI] [PubMed] [Google Scholar]
- Goffin L, Georgopoulos C. Genetic and biochemical characterization of mutations affecting the carboxy-terminal domain of the Escherichia coli molecular chaperone DnaJ. Mol Microbiol. 1998;30:329–340. doi: 10.1046/j.1365-2958.1998.01067.x. [DOI] [PubMed] [Google Scholar]
- Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci U S A. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Cheetham MSDE, Dirr HW, Black GL. Analysis of the levels of conservation of the J domain among various types of DnaJ-like proteins. Cell Stress Chaperones. 2000;5:347–358. doi: 10.1379/1466-1268(2000)005<0347:aotloc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Nemeth K, Redei GP, Schell J. T-DNA insertional mutagenesis in Arabidopsis. Plant Mol Biol. 1992;20:963–976. doi: 10.1007/BF00027166. [DOI] [PubMed] [Google Scholar]
- Kroczyńska B, Coop NE, Miernyk JA. AtJ6, a unique J-domain protein from Arabidopsis thaliana. Plant Sci. 2000;151:19–27. [Google Scholar]
- Kroczyńska B, Zhou R, Wood C, Miernyk JA. AtJ1, a mitochondrial homologue of the Escherichia coli DnaJ protein. Plant Mol Biol. 1996;31:619–629. doi: 10.1007/BF00042234. [DOI] [PubMed] [Google Scholar]
- Kushnir S, Babiychuk E, Kampfenkel K, Belles-Boix E, Van Montagu M, Inze D. Characterization of Arabidopsis thaliana cDNAs that render yeasts tolerant toward the thiol-oxidizing drug diamide. Proc Natl Acad Sci U S A. 1995;92:10580–10584. doi: 10.1073/pnas.92.23.10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Moor A, Reinstein J, Bakau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Żylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Lin B-L. AtJ10, an Arabidopsis dnaJ homologue resembling calmodulin-binding CAJ1 in yeast. Plant Physiol. 1997;115:863. [Google Scholar]
- Lin B-L, Wang J-S, Liu H-C, Chen R-W, Meyer Y, Barakat A, and Delseny M 2001 Genomic analysis of the HSP70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282:679–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- Mekhedov S, de Ilárduya OM, Ohlrogge J. Towards a functional catalog of the plant genome: a survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–401. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA. The 70 kDa stress-related proteins as molecular chaperones. Trends Plant Sci. 1997;2:180–187. [Google Scholar]
- Miernyk JA. Protein folding in the plant cell. Plant Physiol. 1999;121:695–703. doi: 10.1104/pp.121.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA, Coop NE. AtJ20 (accession No. AF214107), a plastid-localized type III J-domain protein from Arabidopsis thaliana (PGR00–027) Plant Physiol. 2000;122:619. [Google Scholar]
- Miller SM, Kirk DL. glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development. 1999;126:649–658. doi: 10.1242/dev.126.4.649. [DOI] [PubMed] [Google Scholar]
- Mooney B, Miernyk JA, Randall DD. Cloning and characterization of the dihydrolipoamide S-acetyltransferase (E2) subunit of the plastid pyruvate dehydrogenase complex from Arabidopsis thaliana. Plant Physiol. 1999;120:443–452. doi: 10.1104/pp.120.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisawa G, Han-yama A, Moda I, Tamai A, Iwabuchi M, Meshi T. AHM1, a novel type of nuclear matrix-localized, MAR binding protein with a single AT hook and a J domain-homologous region. Plant Cell. 2000;12:1903–1916. doi: 10.1105/tpc.12.10.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nover L, Miernyk JA. A genomics approach to the chaperone network of Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:175–176. doi: 10.1379/1466-1268(2001)006<0175:agattc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Benning C. Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol. 2000;3:224–228. [PubMed] [Google Scholar]
- Ohtsuka K, Hata M. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–109. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig-Muller R, Muster G, Kindl H. Heat shock enhances the amount of prenylated Dnaj protein at membranes of glyoxysomes. Eur J Biochem. 1994;219:57–63. doi: 10.1111/j.1432-1033.1994.tb19914.x. [DOI] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL. OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett. 1999;461:101–106. doi: 10.1016/s0014-5793(99)01426-x. [DOI] [PubMed] [Google Scholar]
- Redei GP. Arabidopsis as a genetic tool. Annu Rev Genet. 1975;9:111–127. doi: 10.1146/annurev.ge.09.120175.000551. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore MA, Okamura HH, Rose MD. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JA, Chen R, Masson PH. ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc Natl Acad Sci U S A. 1999;96:1140–1145. doi: 10.1073/pnas.96.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PA, Way JC. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- Umeda A, Meyerholz A, Ungewickell E. Identification of the universal cofactor (auxilin 2) in clathrin coat dissociation. Eur J Cell Biol. 2000;79:336–342. doi: 10.1078/S0171-9335(04)70037-0. [DOI] [PubMed] [Google Scholar]
- Wall D, Zylicz M, Georgopoulos C. The conserved G/F motif of the DnaJ chaperone is necessary for the activation of the substrate binding properties of the DnaK chaperone. J Biol Chem. 1995;270:2139–2144. doi: 10.1074/jbc.270.5.2139. [DOI] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, et al. A new set of Arabidopsis ESTs from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Grant B, and Hirsh D 2000 RME-8, a conserved DnaJ domain containing protein, functions in the late endosome in C. elegans. Mol Biol Cell Suppl P-139. [Google Scholar]
- Zhou R, Kroczyńska B, Hayman GT, Miernyk JA. AtJ2, an Arabidopsis homologue of Escherichia coli dnaJ. Plant Physiol. 1995;108:821–822. doi: 10.1104/pp.108.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Miernyk JA. Cloning and analysis of AtJ3 gene in Arabidopsis thaliana. Acta Bot Sinica. 1999;41:597–602. [Google Scholar]