Abstract
Arabidopsis thaliana, the first plant for which the entire genome sequence is available, was also among the first plant species from which Hsp100 proteins were characterized. The Athsp101 complementary DNA (cDNA) corresponds to the gene identification At1g74310 in the Arabidopsis genome sequence. Analysis of the genome revealed 7 additional proteins that are variably homologous with At1g74310 throughout the entire amino acid sequence and significant similarities or identities in the signature sequences conserved among Hsp100 proteins. Although AtHsp101 is cytoplasmic, 5 of the 7 related proteins have predicted plastidial localization signals. This complete description of the AtHsp100 family sets the stage for future research on expression and function.
THE Hsp100/Clp CHAPERONE FAMILY
The Hsp100 family of proteins has a wide distribution in both prokaryotes and eukaryotes (Singla et al 1998a; Katiyar-Agarwal et al 2001). Members of the Hsp100 family were described first as components of the 2-subunit bacterial Clp protease system (Gottesman et al 1990). The large-subunit ClpA represents an adenosine triphosphate (ATP)–dependent unfoldase, whereas the small-subunit ClpP is the protease. ClpA alone has no proteolytic activity, but it is able to prevent target proteins from aggregation. Interestingly, many ClpA-related proteins were characterized in bacteria and eukaryotes as stress-induced proteins, and hence, they are summarized as members of the Hsp100 family (Schirmer et al 1996; Gottesman et al 1997).
A proteolytic subunit (ClpP) is only found in bacteria associated mainly or exclusively with ClpA protein, whereas in eukaryotes, only the large subunit with chaperone function is observed. A peculiarity of the Hsp100 chaperones is their capability to promote dissociation of aggregated proteins in an ATP-dependent manner (Parsell et al 1994). Several researchers showed that Hsp104 protein is required for induced thermotolerance in yeast and that homologues of the yeast Hsp104 are induced during heat stress in Escherichia coli and HeLa cells (Sanchez and Lindquist 1990; Parsell et al 1991).
GENERAL STRUCTURE OF Hsp100/Clp PROTEINS
Based on the presence of 1 or 2 ATP-binding domains, Hsp100 proteins were divided into 2 major classes (Schirmer et al 1996): class I (Hsp100 types A–D) contains 2 ATP-binding domains and class II (Hsp100 types M, N, X, and Y) contains only 1 ATP-binding domain. The length of the polypeptides varies between 75 and 100 kDa because of the size of the nonconserved spacers between the 2 domains and additional sequences at the C- and N-terminus. Functionally active Hsp100 proteins form hexamers with 12 molecules of ATP bound to it.
Hsp100 proteins are composed of 5 specific domains: (1) amino (N)–-terminal domain, (2) nucleotide-binding domain (NBD) 1, (3) middle domain, (4) NBD2, and (5) carboxyl (C)–terminal domain (Fig 1). The conserved sequences within each domain (signature sequences) as defined by Schirmer et al (1996) are indicated in Figures 3 and 4 on top of the boxes with the corresponding sequences of the Arabidopsis members of the Hsp100 family. For explanations of the letter code, see the legend to Figure 3. The conserved sequences of the 5 domains are as follows:
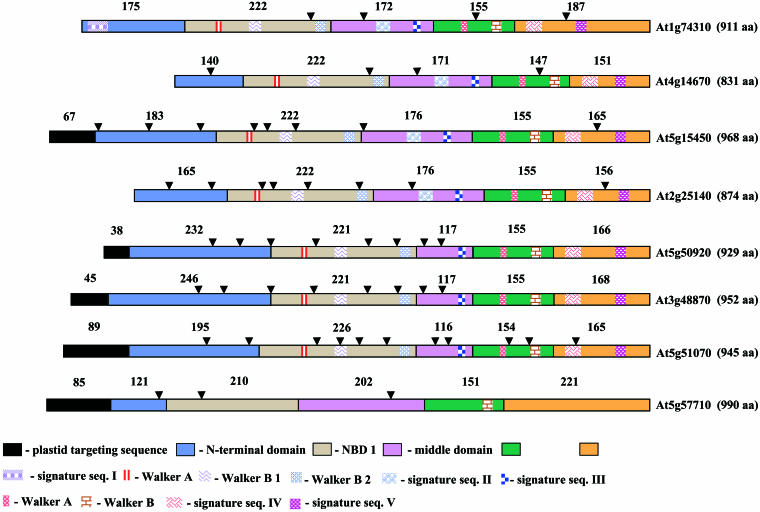
Fig 1.
Block diagrams of AtHsp100 and its homologues. Arrowheads show the positions of introns. Numbers given on top represent the length of a given domain (number of amino acid residues). The color code and pictographs are explained at the bottom (see text and Figs 3 and 4 for further details)
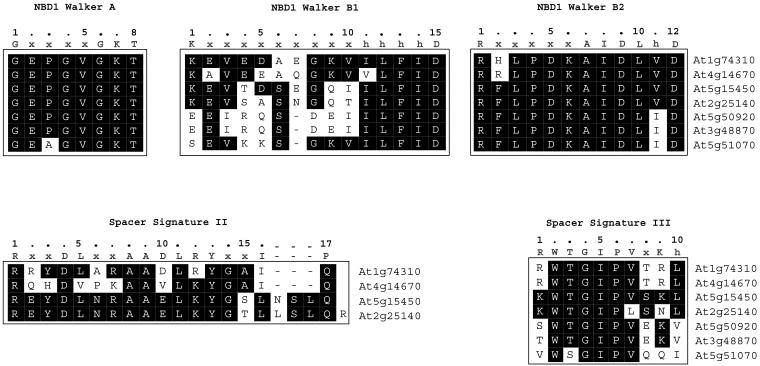
Fig 3.
Sequence alignments of the signature sequences present in NBD1 and spacer domain of Arabidopsis Hsp100 and related protein members. The consensus sequences described by Schirmer et al (1996) are indicated at the top of the boxes. Large case letters represent the corresponding amino acid residues in the 1-letter code; x is any amino acid residue, whereas h stands for any of the hydrophobic amino acid residues, ie, isoleucine, leucine, valine, methionine, phenylalanine, or tyrosine. Note that in some cases these consensus motifs do not reflect the reality of the Arabidopsis Hsp100 proteins (see examples of signature sequence II and Walker B motif in NBD2)
Signature sequence I in the N-terminal domain (consensus sequence xKFTx5ALAxAxxLAx4HxxhxPhHLAxALh), which is only found in Hsp101.
The Walker A, Walker B1, and Walker B2 sequences in the NBD1.
Signature sequence II and signature sequence III in the middle domain.
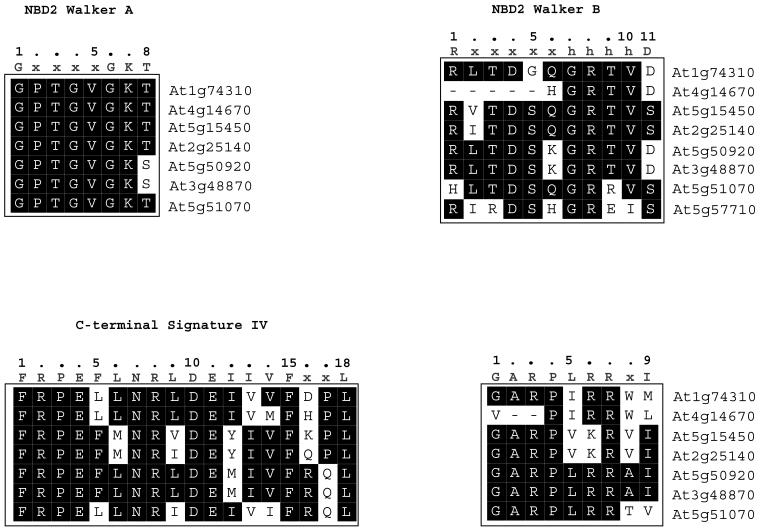
Walker A and Walker B sequences in NBD2.
Signature sequence IV and V in the C-terminal domain.
Hsp100 IN PLANTS
Singla and Grover (1993, 1994) noted that heat stress in rice cells caused accumulation of an approximately 100-kDa protein that cross-reacted with antibodies raised against yeast Hsp104. Subsequently, Hsp100 proteins have been detected and characterized from a range of plant species (Katiyar-Agarwal et al 2001). The results from Southern analyses suggest that the size of the plant hsp100 gene family varies, with a single copy detected in Arabidopsis (Schirmer et al 1994), rice (M. Agarwal et al, in preparation), and maize (Nieto-Sotelo et al 1999) and multiple copies observed in soybean (Lee at al 1994), tobacco, and wheat (D. R. Gallie, in preparation). To date, cDNAs encoding hsp100 have been isolated from Arabidopsis, soybean, tobacco, rice, maize, and wheat (Lee et al 1994; Schirmer et al 1994; Wells et al 1998; Nieto-Sotelo et al 1999; M. Agarwal et al, in preparation). Hsp100 expression is induced during heat stress of Arabidopsis (Schirmer et al 1994), soybean (Lee at al 1994), rice (Pareek et al 1995, M. Agarwal et al, in preparation), maize (Nieto-Sotelo et al 1999; T. E. Young et al, in preparation), tobacco, and wheat (D. R. Gallie, in preparation). The expression of Hsp100 is developmentally regulated in plants (Queitsch et al 2000). Singla et al (1998b) noted high levels of this protein in mature seeds of several plant species and localized it to the embryonal portion of the seeds in rice. In nonstressed maize, Hsp101 is expressed to a high level in the tassel at the premeiosis stage, the ear (including silks), and the developing endosperm and embryo (T. E. Young et al, in preparation).
THE ARABIDOPSIS Hsp100 FAMILY
The recent completion of the Arabidopsis genome sequence (The Arabidopsis Genome Initiative 2000) led us to search how many open reading frames (ORFs) are significantly homologous to the AtHsp101 (gene At1g74310) that was first reported by Schirmer et al (1994). Our analysis of the MIPS database (//mips.gsf.de/proj/thal/db/search_frame.html) shows that there are 7 other ORFs encoded by At4g14670, At5g15450, At2g25140, At5g50920, At3g48870, At5g51070, and At5g57710 variably homologous to At1g74310 over their entire length. The extent of identity of these members with respect to At1g74310 is as follows: 70% (At4g14670), 50% (At5g15450), 49% (At2g25140), 41% (At5g50920), 41% (At3g48870), 38% (At5g51070), and 21% (At5g57710). The closest homologue of At1g74310 protein in the present analysis turned out to be the At4g14670 protein. Hong and Vierling (2000) have reported earlier that a protein of 92.7 kDa (accession number D71409) is 74% identical to AtHsp101 (ie, At1g74310). Note that the ORF with identification At4g14670 is only 668 amino acids long, whereas the protein sequence deduced by Hong and Vierling (2000) has 831 amino acid residues. Both sequences are identical up to position 659; we assume that the truncated form in the MIPS database is the result of a sequencing error creating a frame shift, including a stop codon. Hence, we have included the ORF of D71409 under identification At4g14670 in our Figures 1 and 4 and Table 1.
Table 1.
Survey of members of the Arabidopsis Hsp100 family
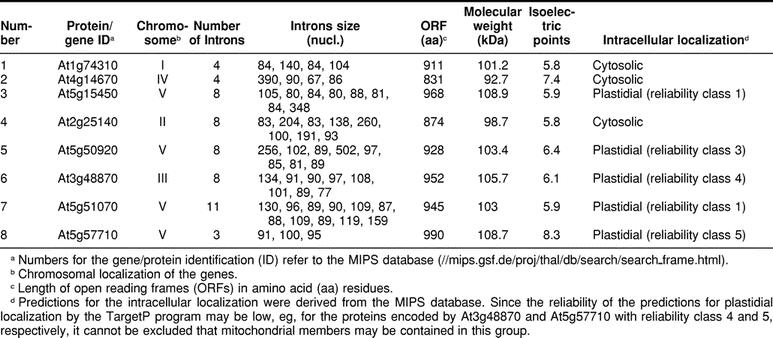
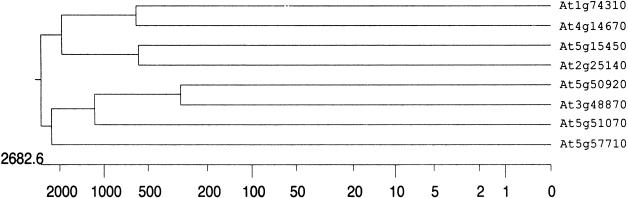
For the 8 proteins that are Hsp100 orthologs, the predicted features, such as molecular weight, isoelectric points, ORF length, subcellular localization of these proteins, and genomic organization, are compiled in Table 1. The comparative structural properties of these Hsp100 members, including the predicted positions of the introns, are presented in Figure 1. The relationship of these members in the form of an evolutionary tree is shown in Figure 2.
Fig 2.
Phylogenetic relationships among Arabidopsis Hsp100 and related proteins. The scale bar shows the correspondence between the unit length and the number of substitution events per position. The phylogenetic tree was constructed using the J. Hein method with residue weight set at PAM 250 in the Megalign program of DNaStar
The intracellular localization of the different AtHsp100 proteins was predicted using the details given in the MIPS database. The protein encoded by At5g15450 has a predicted plastidial targeting sequence of 67 amino acids. In contrast, At1g74310 appears to be cytosolic because it lacks any distinctive targeting sequence. Interestingly, another protein (encoded by At2g25140) has 82% identity to the Phaseolus lunatus plastidial Hsp100 (Keeler et al 2000) but lacked a predicted plastidial targeting sequence. The protein corresponding to At5g50920 has also a predicted plastidial targeting sequence of 35 amino acids. The protein encoded by At3g48870 was significantly homologous to ClpC proteins (Nakabayashi et al 1999), representing one type of the regulatory subunits of the Clp family, whereas the protein encoded by At5g51070 is closely related to Erd1, ie, to the group of ClpA/B subunits. Both proteins have predicted plastidial targeting sequences. Finally, the protein with the least similarity to Hsp101 is encoded by At5g57710 and has a predicted plastidial targeting sequence of 85 amino acids.
Critical for the assignment of an ORF to the Hsp100 family is the conservation of Hsp100 signature sequences, which have been defined by Schirmer et al (1996) on the basis of sequence comparison of members of the Hsp100 family from diverse organisms. Among these signature sequences are so-called Walker boxes, originally identified by Walker et al (1982) for ATP-binding proteins. We find that the NBD1 Walker A and Walker B2 sequences are highly conserved for all 8 members of the Arabidopsis Hsp100 family (Fig 3). The same is true for the NBD1 Walker B1 sequence with the exception of the first amino acid residue, which is a lysine in At1g74310, At4g14670, At5g15450, and At2g25140, glutamic acid in At5g50920 and At3g48870, and serine in At5g51070. Although At1g74310, At4g14670, At5g15450, and At2g25140 encoded proteins contain the middle domain signature II sequence (with some deviations from the consensus sequence), this signature sequence is lacking in the proteins encoded by At5g50920, At3g48870, and At5g51070. The middle domain signature III sequence is found in all members, albeit with some deviations from the consensus sequence (Fig 3). The Walker A and B sequences of the NBD2 domain are also well conserved (Fig 4), except for protein encoded by At4g14670 with a truncated NBD2 Walker B sequence. The C-terminal domain signature sequences IV and V are present in all proteins of the Arabidopsis Hsp100 family (Fig 4). Protein encoded by At5g57710 has only the NBD2 Walker B sequence.
Fig 4.
Sequence alignments of the signature sequences present in NBD2 and C-terminal domain of Arabidopsis Hsp100 and related protein members. For further explanations see legend to Fig 3
Database searches revealed the presence of expressed sequence tags (ESTs) for the different Athsp100 members (Table 2). Four ESTs were found for At1g74310, including 2 from developing seeds, 1 from the mixed library, and 1 from the untreated rosette tissue. The presence of ESTs from these tissues is expected, considering that AtHsp100 is known to be developmentally regulated with constitutive expression in seeds. No EST was found for At1g14670, which has been reported to encode a heat stress–inducible protein (Hong and Vierling 2000). The lack of EST is not surprising, since heat stress plants were not used for the construction of EST libraries available in the databases.
Table 2.
ESTs corresponding to different hsp100 homologs
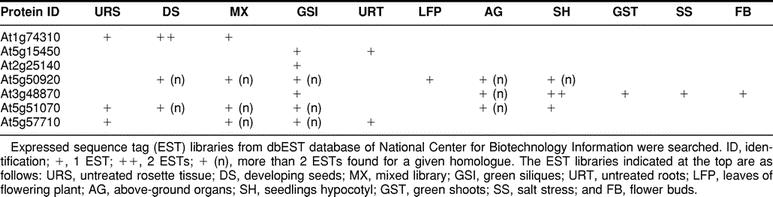
The only member of the Arabidopsis Hsp100 family, whose expression in response to heat stress is essential for thermotolerance, is the At1g74310 encoded Hsp101 (Hong and Vierling 2000, 2001; Queitsch et al 2000). In addition to this, there are 7 other proteins significantly homologous to the At1g74310 encoded protein. Because selected portions of the Arabidopsis genome represent duplication events (Vision et al 2000), it is possible that some of the genes revealed in this study might represent such duplication events that subsequently underwent divergence. It is important to establish how far the structurally divergent members are functionally similar and which genes are expressed under normal growth conditions and/or under heat stress conditions. Considerable insight into the range of the functional roles of Hsp100 and related proteins will be gained by mutational or reverse genetic approaches.
Finally, our BLAST searches revealed other ORFs that were somewhat homologous to the 8 members of the AtHsp100 family. They were not included into our considerations: (1) Five high-molecular-weight proteins encoded by At3g52490, At4g30350, At2g29970, At1g07200, and At2g40130 exhibit similarity to At1g74310 with respect to N-terminal plus NBD1 and NBD2 domains. (2) Six high-molecular-weight proteins encoded by At4g29920, At5g57130, At4g28000, At1g64110, At3g15120, and At5g47040 exhibit similarity to At1g74310 in limited portions of N-terminal, NBD1, or NBD2. (3) Six low-molecular-weight proteins encoded by At2g25030, At3g45450, At2g25040, At3g24530, At1g24290, and At1g50140 exhibit similarity to At1g74310 protein in small regions only. Some of these proteins have primitive signature sequences too. However, it is premature to define these proteins as members of the Hsp100 family, because their overall similarity in amino acid sequence is low and they lack many of the characteristic signature sequences.
Acknowledgments
A.G. is thankful to the Department of Biotechnology and National Agricultural Technology Project (NATP), Government of India, for financial support. M.A., S.K.A., and C.S. are thankful to the Council of Scientific and Industrial Research, Government of India, for fellowship awards. M.A. also thanks NATP for the award of Research Associateship award. Grants from the US Department of Agriculture (00–35301–9086) and National Science Foundation (MCB-9816657) to D.R.G. have supported work on Hsp101.
REFERENCES
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Squires C, Pichersky E, Carrington M, Hobbs M, Mattick JS. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci U S A. 1990;87:3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Wickner S, Maurizi MR. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci U S A. 2000;97:4392–4397. doi: 10.1073/pnas.97.8.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E 2001 Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J in press. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Gallie DR, Grover A. Search for the cellular functions of plant Hsp100/Clp family proteins. Crit Rev Plant Sci. 2001;20(3):277–295. [Google Scholar]
- Keeler SJ, Boettger CM, Haynes JG, Kuches KA, Johnson MM, Thureen DL, Keeler Jr CL, Kitto SL. Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol. 2000;123:1121–1132. doi: 10.1104/pp.123.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Nagao RT, Key JL. A soyabean 101-kD heat stress protein complements yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell. 1994;6:1889–1897. doi: 10.1105/tpc.6.12.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Ito M, Kiyosue T, Shinozaki K, Watanabe A. Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol. 1999;40:504–514. doi: 10.1093/oxfordjournals.pcp.a029571. [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Kannan KB, Segal MC. Characterization of a maize heat-shock protein 101 gene, HSP101, encoding a ClpB/Hsp100 protein homologue. Gene. 1999;230:187–195. doi: 10.1016/s0378-1119(99)00060-8. [DOI] [PubMed] [Google Scholar]
- Pareek A, Singla SL, Grover A. Immunological evidence for accumulation of two high-molecular-weight (104 and 90 kDa) HSPs in response to different stresses in rice and in response to high temperature stress in diverse plant genera. Plant Mol Biol. 1995;29:293–301. doi: 10.1007/BF00043653. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat stress protein 104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Sanchez Y, Stitzel JD, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong S-W, Vierling E, Lindquist S. Heat stress protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist S. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–295. [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S, Vierling E. An Arabidopsis heat stress protein complements a thermotolerance defect in yeast. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla SL, Grover A. Antibodies raised against yeast HSP104 cross-react with a heat- and abscisic acid-regulated polypeptide in rice. Plant Mol Biol. 1993;22:1177–1180. doi: 10.1007/BF00028989. [DOI] [PubMed] [Google Scholar]
- Singla SL, Grover A. Detection and quantification of a rapidly accumulating and predominant 104 kDa heat stress polypeptide in rice. Plant Sci. 1994;97:23–30. [Google Scholar]
- Singla SL, Pareek A, Grover A. Plant Hsp 100 family with special reference to rice. J Biosci. 1998a;23:337–345. [Google Scholar]
- Singla SL, Pareek A, Kush AK, Grover A. Distribution patterns of the 104 kDa stress-associated protein of rice reveal its constitutive accumulation in seeds and disappearance from the just-emerged seedlings. Plant Mol Biol. 1998b;37:911–919. doi: 10.1023/a:1006099715375. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in. Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DR, Tanguay RL, Le H, Gallie DR. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev. 1998;12:3236–3251. doi: 10.1101/gad.12.20.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]