Abstract
Vertebrate cells contain at least 12 different genes for Hsp70 proteins, 3 of which are encoded in the major histocompatibility complex (MHC) class III region. In the human MHC, these are named Hsp70-1, -2, and -Hom. To characterize these proteins, we have determined their substrate binding specificity, their cellular and tissue distribution, and the regulation of their expression. We show for the first time (1) peptide binding specificity of Hsp70-Hom; (2) endogenous expression of Hsp70-Hom in human cell lines; (3) cytoplasmic location of Hsp70-Hom protein under basal conditions and concentration in the nucleus after heat shock; (4) unique RNA expression profiles in human tissues for each of the MHC-encoded Hsp70s, significantly different from that for the constitutive Hsc70; (5) a relative increase in levels of Hsp70-Hom protein, compared with other Hsp70s, in response to interferon gamma; and (6) a specific increase on lipopolysaccharide (LPS) treatment of in vivo messenger RNA levels for the MHC-encoded Hsp70s and the DnaJ homologue, hdj2, relative to other chaperones. The unique tissue distributions and specific up-regulation by LPS of the MHC-encoded Hsp70s suggest some specialization of functions for these members of the Hsp70 family, possibly in the inflammatory response.
INTRODUCTION
Hsp70 molecular chaperones comprise a highly conserved family of both constitutive and stress-induced proteins. They perform diverse cellular roles, such as binding and stabilization of nascent protein chains, maintenance of translocation-competent conformations for protein import into subcellular compartments, assembly and disassembly of protein complexes, and targeting of proteins for lysosomal degradation under certain conditions (Gething and Sambrook 1992; McKay 1993; Becker and Craig 1994; Hartl 1996). All these functions require recognition and binding of the Hsp70 proteins to exposed peptide regions in their target proteins. Although Hsp70 molecular chaperones are evolutionarily conserved, individual members display significant functional diversity that may be related to their exclusive substrate binding specificities (Fourie et al 1994; Gragerov and Gottesman 1994) and/or interactions with specific DnaJ homologues (Cyr and Douglas 1994).
In Escherichia coli, only 3 members of the Hsp70 family have been found (Bardwell and Craig 1984; Lelivelt and Kawula 1995; Itoh et al 1999), whereas in yeast and mammalian cells at least 12 different genes (Craig et al 1995; Tavaria et al 1996) coding for proteins from this family have been found. This divergence in eukaryotic cells suggests additional functional evolutionary pressure besides the need for a member in each subcellular compartment. Three of the genes for human Hsp70s are found in the major histocompatibility complex (MHC) class III region. These are Hsp70-1 and -2, which code for identical, heat-inducible proteins, and Hsp70-Hom, which exhibits low but constitutive RNA expression unaffected by heat shock (Milner and Campbell 1990). Hsp70-1/2 transcription and translation are known to be induced by heat shock and other stresses (Wu et al 1985), and increased levels of this protein have been shown to confer thermotolerance to cells (Li et al 1991; Kampinga et al 1997). However, little is known about the substrate specificity of Hsp70-1/2. Hsp70-Hom, on the other hand, has not been characterized at all at the protein level and nothing is known about its function or substrate specificity.
To characterize the substrate specificities and expression of Hsp70-Hom and Hsp70-1/2, in comparison to other Hsp70 chaperones, we raised antibodies specific for these MHC-encoded Hsp70 proteins and established stable cell lines expressing epitope-tagged human Hsp70-1/2 and Hsp70-Hom. Our studies show unique tissue distributions for the MHC-encoded Hsp70s and specific up-regulation of their messenger RNA (mRNA) levels by lipopolysaccharide (LPS) treatment, suggesting some specialization of function, possibly in the inflammatory response.
MATERIALS AND METHODS
Antibodies, proteins, and peptides
The mouse monoclonals αHsc70 (MA3-014) and M2α-Flag were obtained from Affinity Bioreagents and Sigma, respectively. Polyclonal antisera, αHsp70-C, and αHsp70-Hom-C were generated by immunizing rabbits with bovine serum albumin–conjugated peptides corresponding to sequences near the C-termini of Hsp70-1/2 and Hsp70-Hom. The peptides used were CGPGPGGFGAQGPKGGS, corresponding to amino acids 553–567 from Hsp70-1/2, and CSVVSDEGLKGKISES, corresponding to amino acids 553–567 from Hsp70-Hom. The resulting antisera were subjected to affinity purification using the same peptides conjugated to AffiGel. Polyclonal anti-C3, anti-LMP7, and anti–MHC class I have been described previously (Frueh et al 1992). Anti-Gp96 was obtained from Stressgen. The sources of purified bovine Hsc70 and DnaK were Stressgen and Epicentre Technologies, respectively. Peptides P17G (19 mer), T6L (8 mer), S2V10 (10 mer), S16D (18 mer), and P45 (20 mer) correspond to sequences from p53 (Fourie et al 1997) and were previously shown to bind to the Hsp70 proteins, Hsc70, DnaK, and immunoglobulin binding protein (BiP) (Fourie et al 1994, 1997). The synthesis, N-terminal biotinylation and sources of the peptides were as previously described (Blond-Elguindi et al 1993b; Fourie et al 1997).
Cloning of complementary DNAs coding for MHC-encoded Hsp70 proteins
The coding regions for Hsp70-1 and Hsp70-Hom were amplified by polymerase chain reaction (PCR) from lymphoblastoid cell and testis complementary DNA (cDNA) libraries, respectively, using oligonucleotides corresponding to the 5′ and 3′ ends of the published sequences (Milner and Campbell 1990). The PCR fragments were cloned, together with a cassette encoding an N-terminal Flag epitope, into the pUHD10-1 expression vector. The resulting constructs were fully sequenced and shown to correspond to the published sequences (Milner and Campbell 1990), apart from 2 differences in the Hsp70-Hom sequence. Sequencing of 3 independent clones amplified from testis cDNA revealed that nucleotide position 2182 was C instead of T as published, resulting in the codon for alanine as opposed to valine in amino acid position 408. Similarly, nucleotide position 2229 was A (not C as published), resulting in the codon for threonine as opposed to proline in amino acid position 424. Similar results were obtained for 2 independent clones amplified from spleen cDNA. Since alanine and threonine at these 2 positions, respectively, are conserved in 3 other human cytoplasmic Hsp70 proteins (Milner and Campbell 1990), it seems likely that the designation of valine and proline at these positions in Hsp70-Hom may have resulted from sequencing errors. Previous haplotype analysis (Milner and Campbell 1992) revealed that amino acid position 493 in Hsp70-Hom is polymorphic, with 9 of 62 cell lines containing the codon for threonine at that position, whereas the remainder contained the codon for methionine. All clones of Hsp70-Hom sequenced contained T at position 2437, resulting in the more common haplotype of methionine at position 493.
Overexpression and purification of Flag-tagged MHC-encoded Hsp70-Hom and Hsp70-1/2 proteins in HeLa cells
HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM), which contained 10% heat-inactivated fetal bovine serum, 100 U/mL of penicillin, 100 mg/mL of streptomycin, and 2 mM glutamine. The cDNA constructs described herein were used to transfect the HeLa cells using calcium phosphate precipitation as described previously (Yang et al 1992). Stable clones were established by treatment with 1 μM ouabain for which a resistance plasmid had been cotransfected (Yang et al 1995) and limiting dilution to isolate uniform clones. The clones expressing Hsp70-Hom and Hsp70-1/2 were identified by immunoblotting analysis.
HeLa cells (approximately 109) stably transfected with Flag-tagged Hsp70-Hom and Hsp70-1/2 were freeze-thawed and homogenized in hypotonic buffer (20 mM N-2-hydroxyethylpiperazine-N′2-ethane-sulfonic acid [HEPES], pH 7.2, 10 mM potassium chloride [KCl], 5 mM magnesium chloride (MgCl2)). Cell debris were pelleted and the supernatant subjected to ultracentrifugation at 100 000 × g. The S100 supernatant was adjusted to 150 mM sodium chloride (NaCl), and 1–2 mL of prewashed and equilibrated M2α-Flag agarose was added, followed by incubation for 3 hours at 4°C. The agarose was washed extensively in NET (150 mM NaCl, 5 mM ethylenediamine-tetraacetic acid, 50 mM Tris hydrochloride, pH 7.5), bound protein was eluted with 0.1 M glycine (pH 3.5), and each fraction was immediately neutralized. Fractions were concentrated using Centricon 30 concentrators and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting for quantity and purity of protein. The purity of recombinant Flag-tagged Hsp70s isolated as described from stably transfected HeLa cell lines was typically 90%.
Assays of peptide binding to Hsp70 proteins
Direct binding of N-terminally biotinylated peptides or competition of biotinylated peptide binding by excess unmodified peptides was measured as follows. Purified Hsp70 proteins (0.5 μg/assay) were incubated at 37°C for 15 minutes or 2 hours (as indicated in the figure legends) with biotinylated peptides in a final volume of 10 μL containing 40 mM HEPES (pH 7.0), 75 mM KCl, and 4.5 mM magnesium acetate (Mg(OAc)2). Binding assays were performed either in the absence (direct binding) or presence (competition) of a 20- to 200-fold excess of unmodified peptides. The Hsp70-peptide complexes were then separated from free peptides by nondenaturing PAGE (6% acrylamide) and transferred to nitrocellulose. Complexes of Hsp70 proteins and biotinylated peptides were visualized by incubation with peroxidase-conjugated streptavidin and the ECL detection system. The amount of each biotinylated peptide bound was quantitated by densitometric scanning of the bands corresponding to Hsp70 proteins.
Immunofluorescence analysis
HeLa cells expressing Flag-tagged Hsp70-1/2 or Hsp70-Hom were grown on coverslips at 37°C (control) or shifted to 43°C for 1.5 hours (heat shock) before formaldehyde fixation and permeabilization with 0.1% Triton X-100. Immunostaining was performed with rabbit polyclonal anti–αHsp70-C or mouse monoclonal M2α-Flag antibody, followed by fluorescein isothiocyanate–labeled goat–anti-rabbit or goat–anti-mouse immunoglobulin G, respectively.
Two-dimensional PAGE and immunoblotting
For analysis of endogenous Hsp70 expression, lysates were prepared from control and heat-shocked HeLa cells and from a B-cell line, JY, by lysis in 1% NP40 in phosphate-buffered saline (PBS) containing Complete protease inhibitors. The lysates were subjected to 2-dimensional electrophoresis isoelectric focussing (IEF) and SDS-PAGE; Yang et al 1992) and transferred to nitrocellulose, followed by immunoblotting with the polyclonal antibodies, αHsp70-C and αHsp70–Hom-C, against the MHC-encoded Hsp70s or a mouse monoclonal αHsc70. Hsp70 proteins were visualized by incubation with peroxidase-conjugated anti-mouse or anti-rabbit antibodies and the ECL detection system. The expression of the Hsp70 proteins, as well as C3 and LMP7 proteasomal subunits, MHC class I and Gp96, at different times during treatment of HeLa cells by interferon gamma (1000 U/mL), was determined by 1-dimensional SDS-PAGE, followed by immunoblotting, as described herein.
Northern blotting
Northern blots of RNA from 16 different human tissues (Clontech) were hybridized, according to the manufacturer's protocol, with phosphorus 32 (32P)–labeled oligonucleotides corresponding to 3′-untranslated regions of Hsc70, Hsp70-1, Hsp70-2, and Hsp70-Hom. As a control, hybridization to a 32P-labeled probe for actin mRNA was performed.
DNA microarray analysis of Hsp70 mRNA expression in vivo during LPS treatment
An in vivo study to analyze gene expression after systemic administration of LPS was conducted at Pharma Bio-Research International (The Netherlands) with 3 human subjects, who were healthy adult volunteers. Blood samples were taken at time zero, after which each individual received LPS intravenously by injection. Blood samples were collected at 2, 3, 6, 10, and 24 hours after injection and processed for white blood cell isolation and total RNA extraction. The RNA samples were amplified (Poirier et al 1997), and cDNAs labeled with cy5-deoxycytidine triphosphate (dCTP) were synthesized. The fluorescent cDNAs were then hybridized (Chambers et al 1999) to a DNA microarray chip containing partial sequences from approximately 3000 human genes, including molecular chaperones. The amount of fluorescent cDNA hybridized to each gene sequence was quantified by scanning with a Molecular Dynamics fluorescence scanner, after which the data were normalized and the mean of data from at least 3 individual DNA chips was calculated.
RESULTS
Binding of biotinylated peptides to purified Hsp70 proteins
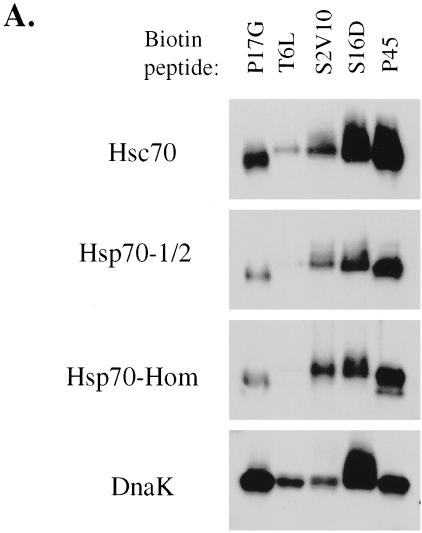
The Hsp70 proteins DnaK, Hsc70, and BiP are known to bind peptides, and previous studies have compared their peptide binding specificities (Fourie et al 1994; Gragerov and Gottesman 1994). However, little is known about the substrate specificity of the MHC-encoded Hsp70-1/2, and in the case of Hsp70-Hom, peptide or substrate binding has not yet been demonstrated. Therefore, direct binding assays of biotinylated peptides to purified Hsp70 proteins were performed to investigate these questions. Five biotinylated peptides (P17G, T6L, S2V10, S16D, and P45), ranging in length from 8 to 20 amino acids, with sequences corresponding to various segments of wild-type and mutant p53 (Fourie et al 1997), were tested for their ability to bind specifically to purified Hsc70, Hsp70-1/2, Hsp70-Hom, and DnaK. The biotinylated peptide binding assay has previously been characterized as representing specific peptide binding to the Hsp70 peptide binding domain (Fourie et al 1997). To compare the substrate specificities of the different Hsp70 proteins, the normalized ratios of binding by the different peptides to each protein were considered as an internally controlled “fingerprint” of substrate specificity.
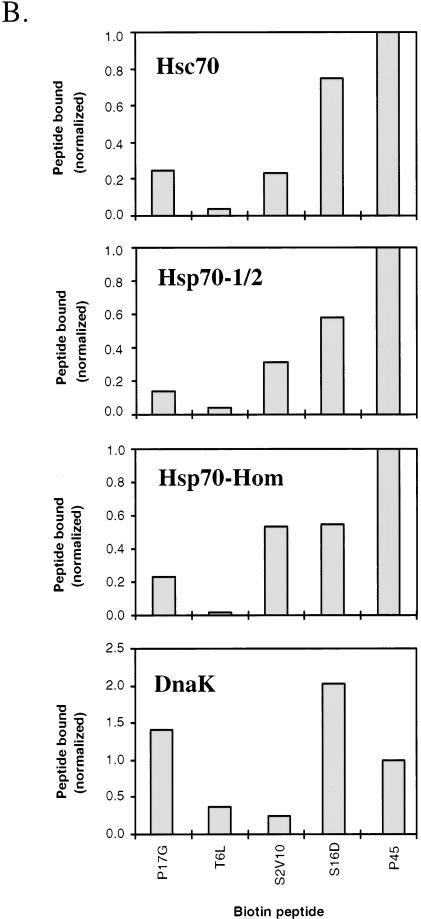
Results of a typical experiment are shown in Figure 1A and mean values of quantitation of results from 2 experiments in Figure 1B. As can be seen from the last lane in each panel of Figures 1 A,B, all 4 Hsp70 proteins bound the 19-mer peptide, P45, with relatively high affinity (normalized to a value of 1 in Fig 1B). Similarly to P45, peptide S16D (fourth lane in each case) bound with high affinity to all the Hsp70 proteins. Higher binding of S16D relative to P45 was observed for DnaK and similar binding for the 2 peptides to Hsc70, whereas for the other 2 Hsp70 proteins, S16D binding was significantly lower than for P45. Peptide S2V10 showed variable affinity for the different Hsp70 proteins. Compared with P45 binding (third lane vs last lane in each case), DnaK and Hsc70 had the lowest affinity of the Hsp70s for S2V10, whereas Hsp70-Hom bound with the highest apparent affinity to S2V10. Nonbiotinylated S2V10 was also more effective at competing with biotinylated P45 binding to Hsp70-Hom than to the other 3 proteins (results not shown). Peptide T6L (second lane) bound to DnaK with moderate affinity, similar to that for S2V10, and to Hsc70 with relatively low affinity. Binding of T6L to Hsp70-1/2 and Hsp70-Hom was almost undetectable relative to the other peptides, but could clearly be seen on longer exposures. Peptide P17G (first lane) bound with highest relative affinity to DnaK and with lower but significant affinity to Hsc70, Hsp70-1/2, and Hsp70-Hom.
Fig 1.
Direct binding of biotinylated peptides to Hsp70 proteins. Hsc70, DnaK, Hsp70-1/2, and Hsp70-Hom (0.5 μg per assay) were incubated for 15 minutes at 37°C with 50 μM biotinylated peptides P17G, T6L, S2V10, S16D, and P45. Free biotinylated peptides were then separated from Hsp70-peptide complexes by native PAGE, followed by transfer to nitrocellulose and detection with streptavidin-peroxidase and ECL. Panel A contains the results of 1 of 2 experiments that gave similar results, whereas panel B shows the mean values of densitometric quantitations of 2 such experiments. The quantitative binding data in B were normalized in each case relative to binding of peptide P45
Overall, the peptide substrate specificities of MHC-encoded Hsp70-Hom and Hsp70-1/2 were closer to that of Hsc70, another mammalian cytoplasmic Hsp70, than to that of the bacterial DnaK. However, some quantitative differences in specificity between the mammalian Hsp70s were observed, such as the higher binding of peptide S2V10 to Hsp70-Hom than to the other Hsp70s.
Sequence requirements for peptide binding by MHC-encoded Hsp70 proteins
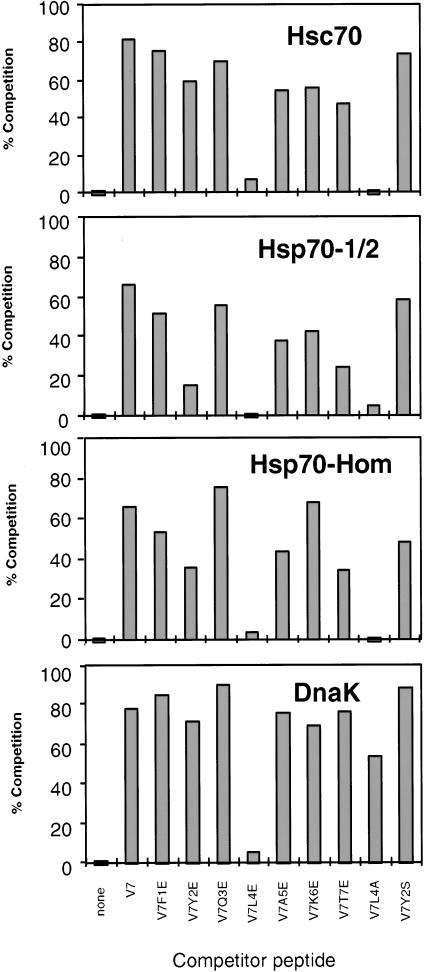
To enable a detailed analysis of the sequence requirements for peptide binding by Hsp70-Hom and Hsp70-1/2 relative to other Hsp70 proteins, the relative binding affinities of a series of heptapeptides (the minimum peptide length for optimum binding, results not shown) were investigated. The peptides used corresponded to peptide V7 (FYQLAKT) and amino acid–substituted derivatives thereof. Peptide V7 was previously shown to bind with moderate affinity to Hsc70 and with high affinity to DnaK and BiP (Fourie et al 1994). Previous studies (Flynn et al 1991; Blond-Elguindi et al 1993a; Fourie et al 1994; Gragerov et al 1994) have shown that the presence of acidic residues in a peptide was unfavorable for binding to Hsp70 proteins. Therefore, a glutamic acid scan of peptide V7 was performed to determine the positions where the negatively charged residue had the most detrimental effect, thus indicating the significant contribution of a particular residue to the binding affinity of the peptide for the Hsp70. Additional substitutions of serine in position 2 and alanine in position 4 were also tested. Binding affinities of the various heptapeptides for the Hsp70 proteins were estimated from their competition for binding of biotinylated P45 (2.5 μM for Hsc70 and DnaK and 25 μM for Hsp70-Hom and Hsp70-1/2). Higher concentrations of P45 were required to reach similar levels of binding to Hsp70-1/2 and Hsp70-Hom because of their lower affinity for this peptide relative to Hsc70 and DnaK (not shown).
The results in Figure 2 show the percentage of competition by each heptapeptide, at 0.5 mM concentration, measured relative to the amount of biotinylated peptide P45 bound in the absence of competitor (none). Excess V7 effectively competed approximately 80% of P45 binding to Hsc70 (top panel, second lane) and DnaK (bottom panel), consistent with previous findings using other assays (Fourie et al 1994). Similarly, V7 competed the binding of P45 to Hsp70-1/2 (second panel) and Hsp70-Hom (third panel), albeit to a somewhat lesser extent (approximately 65%).
Fig 2.
Competition of biotinylated P45 binding to Hsp70s by the peptide V7 and its substituted derivatives. Hsc70, DnaK, Hsp70-1/2, and Hsp70-Hom were incubated for 2 hours at 37°C with biotinylated P45 (2.5 μM for Hsc70 and DnaK and 25 μM for Hsp70-Hom and Hsp70-1/2) in the absence (none) or presence of 0.5 mM competing peptides (V7 and amino acid–substituted derivatives). The samples were then analyzed as described in the legend to Figure 1, and the results quantitated by densitometry. The data represent the means of 2 similar experiments for Hsc70 and DnaK or of 3 experiments for Hsp70-1/2 and Hsp70-Hom
Substitution of glutamic acid in positions 1 (V7F1E) and 3 (V7Q3E) had little or no effect on the affinity of the peptide for any of the Hsp70 proteins (compare third and fifth lanes to second). Differential effects were observed for substitution of glutamic acid at positions 2 (V7Y2E) and 7 (V7T7E). These substitutions had little effect on binding to DnaK or Hsc70, but caused significant decreases in the binding to Hsp70-1/2 and Hsp70-Hom. Substitution of serine in position 2 (V7Y2S) had a less detrimental effect than glutamic acid for binding to all the proteins (last lane in each panel). Replacement of the alanine in position 5 with glutamic acid (V7A5E) had no significant effect on binding to DnaK, but reduced the extent of competition for the other Hsp70 proteins by 30% to 40% relative to peptide V7. The charge-reversal substitution of glutamic acid in place of lysine in position 6 (V7K6E) had negligible effects on binding to DnaK and Hsp70-Hom, but decreased the competition of P45 binding to Hsc70 and Hsp70-1/2 by about 30%.
The most dramatic effects on the affinity of the heptamer peptide were observed for replacement of the central leucine in position 4 by glutamic acid. This substitution (V7L4E) reduced the competition of P45 binding from 70% to 80% to less than 10% in all cases (compare the sixth lane to the second lane in each panel). Furthermore, replacement of the leucine by the small hydrophobic residue, alanine (V7L4A), was equally detrimental for binding to Hsc70, Hsp70-1/2, and Hsp70-Hom, suggesting that a large hydrophobic residue is required to occupy the central pocket in the binding site (see 10th lane in each panel, Fig 8). However, in the case of DnaK, the effect of alanine was less dramatic than glutamic acid, although there was still a 30% reduction in the extent of competition under the conditions of the assay.
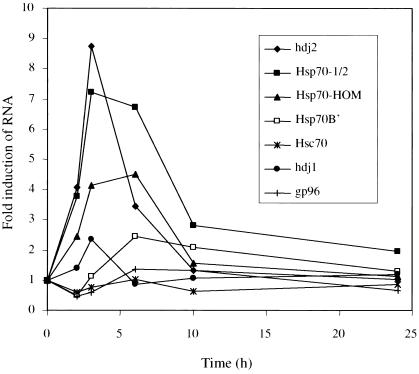
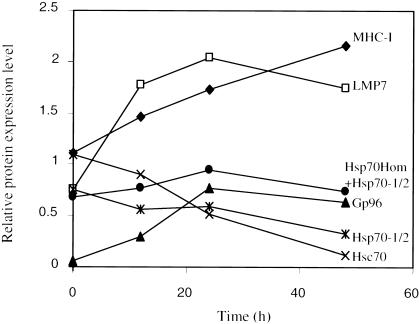
Fig 8.
Inducibility of MHC-encoded Hsp70s in vivo by LPS treatment. Blood samples were taken from 3 human subjects at time zero, after which each individual received LPS intravenously by injection. Blood samples were collected 2, 3, 6, 10, and 24 hours after injection and processed for white blood cell isolation and total RNA extraction. The RNA samples were amplified and cy5-dCTP–labeled cDNAs were synthesized. The fluorescent cDNAs were hybridized to a DNA microarray corresponding to human gene sequences, including hdj2, Hsp70-1/2, Hsp70-Hom, Hsp70-B, Hsc70, hdj1, and gp96, as indicated. The data points represent the average of results for 3 individual subjects, each analyzed in triplicate
Although numerous quantitative differences between the different Hsp70s were observed for the effects of substitutions at various positions, the overall pattern of specificity for the MHC-encoded Hsp70-Hom and Hsp70-1/2 was more similar to mammalian Hsc70 than to bacterial DnaK, in agreement with the results in Figure 1. Clearly, the most dramatic changes in affinity of the heptamer for all the Hsp70 proteins were obtained by the replacement of the central leucine, indicating that this is the most important residue contributing to the binding affinity.
Specificity of antipeptide antibodies raised against MHC-encoded Hsp70s
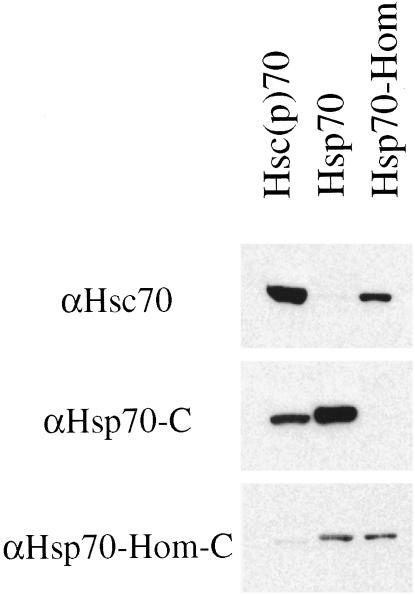
Polyclonal antibodies specific for the MHC-encoded Hsp70s were produced to distinguish their endogenous protein expression from other Hsp70 proteins. Rabbit antibodies, αHsp70-C, and αHsp70–Hom-C were raised against peptides corresponding to sequences near the C-termini of human Hsp70-1/2 and Hsp70-Hom, respectively. The specificity of these antibodies was determined by using equal quantities (0.5 μg) of purified bovine Hsc70 or Flag-tagged human Hsp70-1/2 and Hsp70-Hom (purified as described in Materials and Methods). The proteins were subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted with either a mouse monoclonal αHsc70 or one of the polyclonal antibodies directed against the MHC-encoded Hsp70s.
The top panel in Figure 3 shows that the monoclonal αHsc70 reacted preferentially with Hsc70 and also showed some cross-reactivity with Hsp70-Hom. The affinity-purified rabbit polyclonal, αHsp70-C (middle panel), reacted only with human Hsp70-1/2 (second lane) and the small amount of bovine Hsp70 (first lane, band of higher mobility than Hsc70) always present in preparations of Hsc70 from tissues. Thus, this antibody was highly specific for only the heat-inducible Hsp70 family member. The second polyclonal antibody, αHsp70–Hom-C (bottom panel), reacted with both Hsp70-1/2 and Hsp70-Hom but not with Hsc70. Thus, this antibody was equally specific for the 2 MHC-encoded Hsp70 proteins and is, to our knowledge, the only antibody that preferentially recognizes Hsp70-Hom over Hsc70.
Fig 3.
Specificity of antipeptide antibodies against MHC-encoded Hsp70s. Polyclonal rabbit antibodies αHsp70-C and αHsp70–Hom-C were raised against bovine serum albumin–conjugated peptides corresponding to sequences near the C-termini of Hsp70-1/2 and Hsp70-Hom. Equal quantities (0.5 μg) of purified bovine Hsc70, as well as Flag-tagged human Hsp70-1/2 and Hsp70-Hom, were subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted with the mouse monoclonal, MA3-014 (αHsc70), or the polyclonal antipeptide antibodies directed against the MHC-encoded Hsp70s. The results are from 1 of 2 similar experiments
Endogenous expression and inducibility of MHC-encoded Hsp70s in cell lines
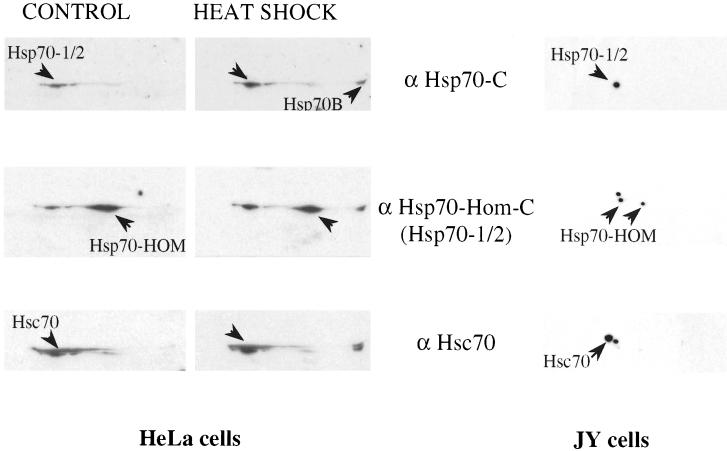
To examine endogenous expression of the MHC-encoded Hsp70s in human cell lines, lysates were prepared from control and heat-shocked HeLa cells and from the B-cell line, JY. The lysates were subjected to 2-dimensional electrophoresis and transferred to nitrocellulose, followed by sequential immunoblotting with the polyclonal antibodies described above, in addition to monoclonal αHsc70.
In control HeLa cells (top left panel in Fig 4), αHsp70-C recognized one major 70-kDa protein spot, of which the levels increased approximately 3-fold after heat shock, in addition to the appearance of another more basic protein, presumably the strictly heat-inducible Hsp70-B (Leung et al 1990). JY cells also exhibited detectable basal levels of Hsp70-1/2 (top right panel). Subsequent blotting of the same membranes with αHsp70–Hom-C (center panels) revealed an additional protein in HeLa cells, not induced by heat shock and of slightly higher mobility and more basic pI than Hsp70-1/2. From the specificity of this antibody, we concluded that the additional protein spot represented Hsp70-Hom. In JY cells, αHsp70–Hom-C recognized 2 spots in addition to Hsp70-1/2, one of similar mobility and isoelectric point (pI) to Hsp70-Hom in HeLa cells and another of similar mobility to Hsp70-Hom and a pI close to that of Hsp70-1/2. This may represent an additional isoform of either Hsp70-Hom or Hsp70-1/2, present in JY, but not HeLa cells. Subsequent blotting with αHsc70 detected a protein spot of the expected lower mobility and slightly more acidic pI than Hsp70-1/2 (Tavaria et al 1996) in both cell types (lower panels). Thus, Hsp70-Hom protein was endogenously expressed in 2 different human cell lines and was not induced by heat shock.
Fig 4.
Endogenous expression and inducibility of MHC-encoded Hsp70s. Cell lysates from HeLa cells (control and heat shocked) (left panels) and JY cells (right panels) were subjected to 2-dimensional electrophoresis (IEF and SDS-PAGE) and then transferred to nitrocellulose, followed by immunoblotting with the polyclonal antibodies directed against the MHC-encoded Hsp70s or the mouse monoclonal MA3-014 (αHsc70). Antigens recognized by the specific antibodies are indicated by arrowheads. The experiment was performed once on JY cells and 3 times, with similar results, for HeLa cells
Subcellular locations of Hsp70-1/2 and Hsp70-Hom in HeLa cells
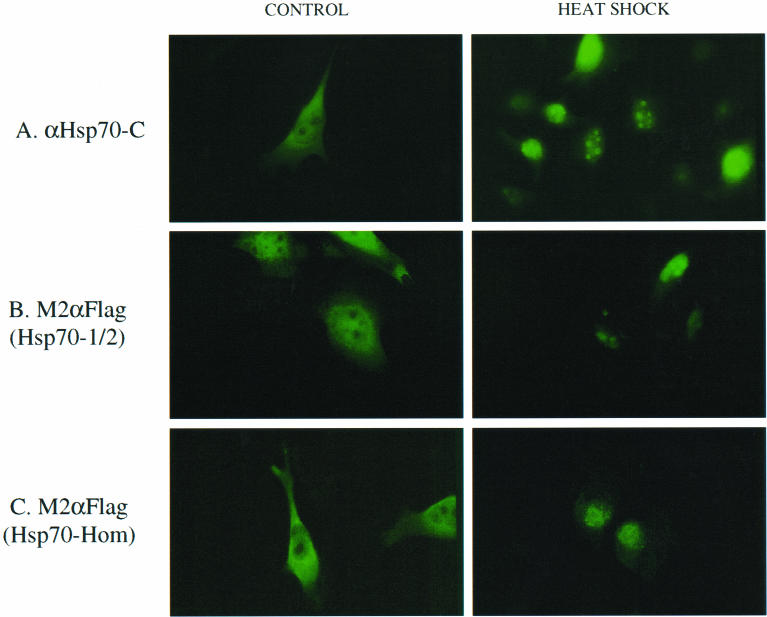
Previous studies have shown that Hsp70-1/2 is predominantly in the cytoplasm under basal conditions and is targeted to the nucleus and nucleolus after heat shock (Milarski and Morimoto 1989). Hsp70-Hom was predicted to be a cytoplasmic protein (Tavaria et al 1996), but no studies have been done to confirm this. To determine the subcellular location of Hsp70-Hom under basal and heat shock conditions, in comparison to Hsp70-1/2, immunofluorescence analysis was performed on HeLa cells expressing Flag-tagged MHC-encoded Hsp70s.
Under basal conditions at 37°C, immunostaining with anti–Hsp70-C antibodies of HeLa cells expressing endogenous and Flag-tagged Hsp70-1/2 showed mainly cytoplasmic but also nuclear staining, and the staining was clearly excluded from the nucleoli (control panel, Fig 5A). When the cells were heated to 43°C for 1.5 hours, the staining was concentrated to the nucleus and particularly to the nucleoli (heat shock panel, Fig 5A). The same results were obtained for endogenous Hsp70-1/2 in untransfected HeLa cells (not shown). When the cells were stained with the M2α-Flag antibody, similar staining patterns to the anti–Hsp70-C antibody were observed for both control and heat-shocked cells (Fig 5B), indicating that the transfected, Flag-tagged Hsp70-1/2 behaved similarly to the endogenous wild-type protein under both control and stress conditions.
Fig 5.
Subcellular locations of Hsp70-1/2 and Hsp70-Hom in HeLa cells. HeLa cells expressing Flag-tagged Hsp70-1/2 (A and B) or Hsp70-Hom (C) were grown on coverslips at 37°C (control) or shifted to 43°C for 1.5 hours (heat shock) before formaldehyde fixation, permeabilization, and immunostaining with polyclonal antipeptide anti–Hsp70-C (A) or monoclonal M2α-Flag (B and C) antibodies, followed by fluorescein isothiocyanate–labeled goat–anti-rabbit or goat–anti-mouse immunoglobulin G, respectively. Similar results were obtained in 3 experiments
When cells transfected with Flag-tagged Hsp70-Hom were stained with M2α-Flag antibody, the staining pattern was mainly cytoplasmic (Fig 5C), similar to that observed for endogenous and Flag-tagged Hsp70-1/2. This experimental evidence confirms the prediction (Tavaria et al 1996) of a cytoplasmic localization of Hsp70-Hom. On heat shock, there was clearly a concentration in the nucleus of Flag-tagged Hsp70-Hom but not in the nucleoli. A punctate-type staining was observed as opposed to the nucleolar concentration seen for Hsp70-1/2. Thus, although Hsp70-Hom protein appeared to move to the nucleus in response to the stress of heat shock, the distribution in the nucleus seemed to be different to that observed for Hsp70-1/2. Nevertheless, there was clearly a response of Hsp70-Hom subcellular localization to heat shock, even though no induction in the levels of Hsp70-Hom RNA (Milner and Campbell 1990) or protein (this article, Fig 4) took place. It was not possible to stain the endogenous Hsp70-Hom in HeLa cells, because the antipeptide antibody that recognized denatured Hsp70-Hom was not able to recognize the protein in immunofluorescence or immunoprecipitation assays (not shown). However, based on the results obtained for endogenous and Flag-tagged Hsp70-1/2, we would predict that the endogenous Hsp70-Hom would behave similarly to the transfected Flag-tagged protein.
Tissue-specific expression of Hsp70-Hom and Hsp70-1/2 RNA
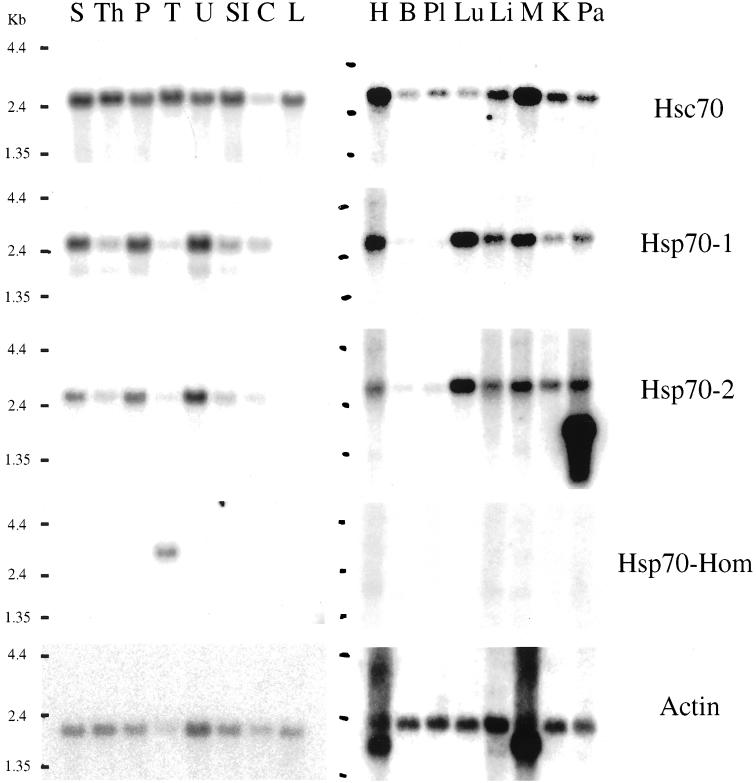
The results in Figure 4 showed endogenous expression of Hsp70-1/2 and Hsp70-Hom protein in lines of both antigen-presenting B cells (JY) and cervical carcinoma cells (HeLa). We next examined the relative expression of Hsp70-Hom, Hsp70-1/2, and Hsc70 RNA in various human tissues.
Both actin and Hsc70 RNA expression were observed in all the tissues (Fig 6), although the quantity varied somewhat for Hsc70. In contrast, both Hsp70-1 and Hsp70-2, and particularly Hsp70-Hom RNA, showed a more restricted and/or variable pattern of expression. Although expression of the hsp70-1 and hsp70-2 genes cannot be distinguished at the protein level, differences in their 3′-untranslated regions allowed analysis of their individual RNA expression profiles. Quantitative differences could be observed; for example, the relative expression of Hsp70-1 RNA in the spleen and heart was higher than that of Hsp70-2. High expression of both Hsp70-1 and -2 RNA was observed in the uterus, skeletal muscle, and particularly lung tissue. A second, lower band of RNA for Hsp70-2 was strongly detected in the pancreas. This may represent a degradation product or a shorter transcript. Significant RNA expression for Hsp70-1 and -2 was also observed in the spleen, prostate, heart, liver, and kidney. Low Hsp70-1 and -2 RNA expression was detected in the small intestine, thymus, and colon; very low levels in the testis, brain and placenta; and no expression in peripheral blood leukocytes. Hsp70-Hom RNA expression could only be detected in the testis, similarly to results obtained for the murine homologue, Hsc70t (Matsumoto and Fujimoto 1990). Thus, each individual hsp70 gene exhibited a unique pattern of tissue-specific RNA expression.
Fig 6.
Tissue-specific expression of MHC-encoded Hsp70-Hom and Hsp70-1/2 RNA. Northern blots of RNA from human tissues were hybridized with 32P-labeled oligonucleotides corresponding to 3′-untranslated regions of Hsc70, Hsp70-1, Hsp70-2, and Hsp70-Hom, respectively. As a control, hybridization to a probe for actin mRNA was performed. Tissues analyzed are spleen (S), thymus (Th), prostate (P), testis (T), uterus (U), small intestine (SI), colon (C), peripheral blood leukocyte (L), heart (H), brain (B), placenta (Pl), lung (Lu), liver (Li), skeletal muscle (M), kidney (K), and pancreas (Pa)
Effects of interferon gamma treatment of HeLa cells on endogenous expression of MHC-encoded Hsp70s and other proteins
The cytokine interferon gamma plays a critical role in the induction of an immune response (Fourie and Yang 1998). Therefore, to investigate the possible involvement of the MHC-encoded Hsp70s in antigen presentation, we studied their expression in response to interferon gamma treatment.
Analysis of Hsp70 expression at the RNA level showed that, before interferon treatment, only Hsc70 RNA was detectable (results not shown). Hsc70 RNA expression increased slightly up to 7 hours and then declined to below the starting level by 22 hours of interferon treatment. RNA for Hsp70-1 and -2 was undetectable except after 7 to 8 hours of interferon treatment, where some expression was observed, after which levels declined to become undetectable again after 22 hours. Hsp70-Hom RNA was undetectable under all conditions, similarly to previous results for HeLa cells (Milner and Campbell 1990). In contrast, significant Hsp70-Hom expression was observed at the protein level, as shown in 2-dimensional immunoblots of HeLa cells (Fig 4). Therefore, the effects of interferon gamma on MHC-encoded Hsp70 expression at the protein level was investigated.
Extracts were prepared from HeLa cells that had been treated with interferon gamma for 0, 12, 24, and 48 hours and analyzed by Western blotting for protein expression of MHC class I, the constitutively expressed and interferon-inducible proteasomal subunits, C3 and LMP7, respectively and the chaperones gp96, Hsc70, Hsp70-1/2, and Hsp70-Hom. The results in Figure 7 were normalized relative to the constitutive expression of C3 at each time point.
Fig 7.
Effects of interferon gamma treatment on expression of MHC-encoded Hsp70s and other proteins. HeLa cells were treated for different times as indicated up to 48 hours with interferon gamma (1000 U/mL), and samples were analyzed at 0, 12, 24, and 48 hours by Western blotting for expression of MHC class I, the constitutively expressed and interferon-inducible proteasomal subunits C3 and LMP7, and the chaperones gp96, Hsc70, Hsp70-1/2, and Hsp70-Hom. The results were normalized relative to the constitutive expression of C3 at each time point. A single-course experiment was performed, but similar results for single time points of Hsp70 levels plus and minus interferon gamma were obtained in 2 other experiments
As expected, LMP7 was induced several-fold by interferon gamma within 12 hours, reaching a maximum after 24 hours. MHC class I induction had a less rapid course, showing increasing levels up to 48 hours. Compared with these classic interferon-responsive, MHC-encoded proteins, changes in the levels of the MHC-encoded Hsp70s were relatively minor, and levels of the constitutive Hsc70 appeared to decrease, rather than increase, in response to interferon. The chaperone gp96 showed a several-fold induction that reached maximum protein levels at 24 hours, in agreement with previous results for mRNA levels (Anderson et al 1994).
Before interferon gamma treatment, Hsc70 levels were higher than Hsp70-1/2 and Hsp70-Hom. Hsc70 and, to a lesser extent, Hsp70-1/2 levels declined during interferon gamma treatment. Therefore, by 48 hours, Hsp70-Hom showed the predominant immunodetectable expression of the 3 Hsp70s. If one considers that the antibody used for the Western blotting cannot distinguish Hsp70-Hom from Hsp70-1/2, the actual relative increase in Hsp70-Hom levels would be more dramatic, particularly in the first 24 hours, than that shown for the combined expression of Hsp70-Hom and Hsp70-1/2. Thus, although the changes in Hsp70 protein levels during interferon treatment were small, there was a relative increase in levels of Hsp70-Hom protein compared with other Hsp70s.
Inducibility of MHC-encoded Hsp70s in vivo by LPS
Bacterial LPS is known to be a potent inducer of the inflammatory response. Therefore, to investigate the possibility that the MHC-encoded Hsp70s may have a role in this pathway, we examined their expression in blood cells during an in vivo inflammatory response. We analyzed data from a study conducted to study gene expression after systemic administration of LPS in 3 human subjects. At various times after LPS injection, white blood cell RNA was extracted and gene expression analyzed using DNA microarrays, as described in Materials and Methods. The results for a number of chaperones are shown in Figure 8. Three hours after LPS administration, mRNA levels for Hsp70-1/2 had increased 7-fold, whereas Hsp70-Hom expression increased 4-fold. The mRNA levels for the MHC-encoded Hsp70s remained at maximum levels between 3 and 6 hours and then decreased dramatically, although Hsp70-1/2 levels remained elevated above basal levels even after 24 hours. In contrast, mRNA levels for Hsc70 and gp96 showed no significant induction by LPS. After an initial decrease in expression at 2 hours, Hsp70-B mRNA reached a maximum induction of about 2.5-fold at 6 hours, decreasing to basal levels by 24 hours.
Similarly to the MHC-encoded Hsp70s, the mRNA expression for the DnaJ homologue, hdj2 was induced 9-fold by 3 hours, in contrast to another DnaJ homologue, hdj1, which showed only 2-fold induction. After reaching maximal levels 3 hours following LPS administration, the mRNA levels for hdj1 and hdj2 decreased, returning to basal levels by 6 and 10 hours, respectively. The specific induction, with similar kinetics to those of the heat shock response, of only the MHC-encoded chaperone proteins and a specific DnaJ homologue suggests that they may cooperate to regulate functions of specific substrate proteins during the inflammatory response.
DISCUSSION
Hsp70 molecular chaperones are a highly conserved family of proteins. However, there is evidence that they are not functionally interchangeable in vivo (Gao et al 1991; Terlecky et al 1992; Brodsky et al 1993; Wiech et al 1993). Furthermore, although there is considerable overlap in their overall substrate specificity, individual members of the family also exhibit unique sequence preferences (Fourie et al 1994; Gragerov and Gottesman 1994). We have investigated the possibility that the MHC class III–encoded Hsp70s, particularly Hsp70-Hom, may have evolved to perform specific functions, possibly through binding to unique substrates.
We demonstrate for the first time that Hsp70-Hom is capable of binding peptides, with substrate specificity similar to other mammalian Hsp70s. In additional experiments (A. M. Fourie, unpublished results), we have shown that Hsp70-Hom does not exclusively bind peptides but also full-length, unfolded proteins. In this study, we show that the overall substrate specificities of the MHC-encoded Hsp70s are similar to those of other Hsp70 proteins in that they have a preference for binding peptides with a central, large hydrophobic amino acid. Detailed sequence preference analysis showed that Hsp70-Hom and Hsp70-1/2 were closer in specificity to mammalian Hsc70 than to bacterial DnaK. Furthermore, we found that the peptide binding specificities of Hsp70 proteins, including the MHC-encoded members, were not similar to any particular MHC class I allele (results not shown), which makes sense on the basis of their very different peptide binding structures (Matsamura et al 1992; Zhu et al 1996).
We determined the subcellular localization of Hsp70-Hom protein to be cytoplasmic under basal conditions. In response to the stress of heat shock, targeting of the protein to the nucleus was observed, similar to Hsp70-1/2. However, the distribution of Hsp70-Hom in the nucleus was punctate, as opposed to the clearly nucleolar concentration seen for Hsp70-1/2. The lack of distinct nucleolar concentration of Hsp70-Hom may be related to divergence relative to Hsp70-1/2 in the region of the sequence (residues 480 to 641) previously shown to be required for the nucleolar localization of Hsp70 (Milarski and Morimoto 1989). This is the first report of this response of Hsp70-Hom subcellular localization to heat shock, even though no induction in the levels of Hsp70-Hom RNA (Milner and Campbell 1990) or protein (this article) was observed on heat shock.
Interestingly, we found unique RNA expression profiles in human tissues for each of the MHC-encoded Hsp70s, significantly different from that for the constitutive Hsc70, suggesting some functional specialization for the individual Hsp70s. Although previous studies of RNA expression (Milner and Campbell 1990) predicted that Hsp70-Hom would be expressed exclusively in testis, our results also showed significant expression of the protein in HeLa and JY cell lines. The human tissue distribution of Hsp70-Hom and Hsp70-1/2 RNA or protein (not shown) was apparently not specific for immunologically relevant tissues. In contrast, MHC class I, the interferon-inducible proteasomal subunits LMP2 and 7, and the transporters associated with antigen processing were preferentially expressed (at the protein level) in the spleen, lymph nodes, and thymus (results not shown). Calnexin and gp96, 2 chaperones implicated in class I assembly and/or peptide loading, showed significant expression in lymph nodes and spleen or thymus, respectively (not shown)
The functional diversity of Hsp70 proteins in general (Gao et al 1991; Terlecky et al 1992; Brodsky et al 1993; Wiech et al 1993; James et al 1997) and the linkage of particular Hsp70s with the MHC for 350 million years (Salter-Cid et al 1994) suggested that these proteins may have evolved to perform specific functions in antigen processing and/or presentation. Although previous evidence has implicated Hsp70s in antigen presentation (Pierce et al 1991; Pierce 1994; Wells et al 1998; Panjwani et al 1999), no clear correlation has been made. A number of studies have implicated Hsp70 chaperones as having a role in immune responses, particularly as adjuvants or cross-priming agents (reviewed in Srivastava et al 1998). We found that overexpression of either of the MHC-encoded Hsp70s in HeLa cells did not significantly affect the class I surface expression levels (A. M. Fourie, unpublished results). Similar results were recently obtained for overexpression of rat Hsp70-1 in a human melanoma cell line (Dressel et al 1999). However, the finding that interferon gamma treatment of HeLa cells apparently leads to a relative increase in Hsp70-Hom protein levels, compared with Hsp70-1/2 and Hsc70, suggests some preferential role for this particular Hsp70 protein in the immune response. Interestingly, a recent study (Multhoff et al 1999) showed that the C-terminal domains of Hsp70-1/2 or Hsp70-Hom, but not that of Hsc70, stimulate proliferation and cytolytic activity of natural killer cells..
It has been shown that Hsp70s and DnaJ homologues are required for ubiquitin-mediated degradation of some proteins (Lee et al 1996; Yaglom et al 1996; Bercovich et al 1997; Fisher et al 1997). Thus, although the MHC-encoded Hsp70s do not seem to be dedicated chaperones for antigen presentation, it is possible that they may affect the repertoire or amount of cytosolic peptides produced by the proteasome, particularly during interferon induction (Fourie and Yang 1998), and in this manner subtly alter the peptide content of surface class I molecules. The dramatic and specific induction of the MHC-encoded Hsp70s in vivo by LPS treatment suggests that they may have a regulatory role in the inflammatory stress response. Detailed analysis of gene expression patterns in response to a number of stimuli will shed light on which proteins are involved in MHC-encoded Hsp70-mediated pathways.
The central 18-kDa domain of Hsp70 proteins, defined by Wang et al (1993) as the minimal peptide binding domain, is highly conserved (60% identical) between different Hsp70s, whereas the most divergent region between different members of the Hsp70 family is the C-terminal 10-kDa domain. The function of this domain remains obscure, but it may be involved in interactions with co-chaperone DnaJ homologues (Freeman et al 1995; Demand et al 1998) or other unknown cellular factors. Specific DnaJ homologues could target Hsp70s to particular locations or substrates, and their functions may also be regulated at the level of tissue-specific expression. Our finding that the DnaJ homologue hdj2 was specifically induced by LPS, in a manner similar to the MHC-encoded Hsp70s, suggests that hdj2 may function together with these Hsp70s to modulate function of particular proteins during the inflammatory response. From the findings in this study, we propose that the MHC-encoded Hsp70s have evolved to play specialized roles in vivo, mediated by specific DnaJ homologues.
Fig 1.
Continued
Acknowledgments
We thank the members of the vivarium, peptide synthesis, DNA core, and DNA microarray groups at the R. W. Johnson Pharmaceutical Institute for their support in this study; Dr Jackson Wan for assistance in analysis of the DNA microarray data; Jesse Pang, Veronica Moreno, Stephen Ho, and Xiaodong Wu for technical assistance; and Dr Lars Karlsson for critical reading of the manuscript.
REFERENCES
- Anderson SL, Shen T, Lou J, Xing L, Blachere NE, Srivastava PK, Rubin BY. The endoplasmic reticular heat shock protein gp96 is transcriptionally upregulated in interferon-treated cells. J Exp Med. 1994;180:1565–1569. doi: 10.1084/jem.180.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell JCA, Craig EA. Major heat shock gene of Drosophila and the Escherichia coli heat-inducible dnaK gene are homologous. Proc Natl Acad Sci U S A. 1984;81:848–852. doi: 10.1073/pnas.81.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Craig EA. Heat-shock proteins as molecular chaperones. Eur J Biochem. 1994;219:11–23. doi: 10.1007/978-3-642-79502-2_2. [DOI] [PubMed] [Google Scholar]
- Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJH. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993a;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MJH. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993b;268:12730–12735. [PubMed] [Google Scholar]
- Brodsky JL, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. J Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J, Angulo A, and Amaratunga D. et al. . 1999 DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J Virol. 73:5757–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E, Ziegelhoffer T, Nelson J, Laloraya S, Halladay J. Complex multigene family of functionally distinct Hsp70s of yeast. Cold Spring Harb Symp Quant Biol. 1995;60:441–449. doi: 10.1101/sqb.1995.060.01.049. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homolog Ydj1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- Demand J, Luders J, Hohfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18:2023–2028. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressel R, Lubbers M, Walter L, Herr W, Gunther E. Enhanced susceptibility to cytotoxic T lymphocytes without increase of MHC class I antigen expression after conditional overexpression of heat shock protein 70 in target cells. Eur J Immunol. 1999;29:3925–3935. doi: 10.1002/(SICI)1521-4141(199912)29:12<3925::AID-IMMU3925>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, Goldberg AL, Ginsberg HN. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Fourie AM, Hupp TR, Lane DP, Sang BC, Barbosa MS, Sambrook JF, Gething MJH. Hsp70 binding sites in the tumor suppressor protein p53. J Biol Chem. 1997;272:19471–19479. doi: 10.1074/jbc.272.31.19471. [DOI] [PubMed] [Google Scholar]
- Fourie AM, Sambrook JF, Gething MJH. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J Biol Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- Fourie AM, Yang Y. Molecular requirements for assembly and intracellular transport of class I major histocompatibility complex molecules. Curr Topics Microbiol Immunol. 1998;232:49–74. doi: 10.1007/978-3-642-72045-1_3. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frueh K, Yang Y, Arnold D, Chambers J, Wu L, Waters JB, Spies T, Peterson PA. Alternative exon usage and processing of the major histocompatibility complex-encoded proteasome subunits. J Biol Chem. 1992;267:22131–22140. [PubMed] [Google Scholar]
- Gao B, Biosca J, Craig EA, Greene LE, Eisenberg E. Uncoating of coated vesicles by yeast hsp70 proteins. J Biol Chem. 1991;266:19565–19571. [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Gragerov A, Gottesman ME. Different peptide binding specificities of hsp70 family members. J Mol Biol. 1994;241:133–135. doi: 10.1006/jmbi.1994.1482. [DOI] [PubMed] [Google Scholar]
- Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Itoh T, Matsuda H, Mori H. Phylogenetic analysis of the third Hsp70 homolog in Escherichia coli; a novel member of the Hsc66 subfamily and its possible co-chaperone. DNA Res. 1999;6:299–305. doi: 10.1093/dnares/6.5.299. [DOI] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Konings AWT, Evers AJ, Brunsting JF, Misfud N, Anderson RL. Resistance to heat radiosensitization and protein damage in thermotolerant and thermoresistant cells. Int J Radiat Biol. 1997;71:315–326. doi: 10.1080/095530097144201. [DOI] [PubMed] [Google Scholar]
- Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelivelt MJ, Kawula TH. Hsc66, an Hsp70 homolog in Escherichia coli, is induced by cold shock but not by heat shock. J Bacteriol. 1995;177:4900–4907. doi: 10.1128/jb.177.17.4900-4907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung TKC, Rajendran MY, Monfries C, Hall C, Lim L. The human heat-shock protein family: expression of a novel heat-inducible Hsp70 (Hsp70B’) and isolation of its cDNA and genomic DNA. Biochem J. 1990;267:125–132. doi: 10.1042/bj2670125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GC, Li L, Liu YK, Mak JY, Chen L, Lee WMF. Thermal response of rat fibroblasts stably transfected with the human 70-kDa heat shock protein-encoding gene. Proc Natl Acad Sci U S A. 1991;88:1681–1685. doi: 10.1073/pnas.88.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsamura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Fujimoto H. Cloning of a hsp70-related gene expressed in mouse spermatids. Biochem Biophys Res Commun. 1990;166:43–49. doi: 10.1016/0006-291x(90)91909-c. [DOI] [PubMed] [Google Scholar]
- McKay DB. Structure and mechanism of 70-kDa heat-shock-related proteins. Adv Protein Chem. 1993;44:67–98. doi: 10.1016/s0065-3233(08)60564-1. [DOI] [PubMed] [Google Scholar]
- Milarski KL, Morimoto RI. Mutational analysis of the human Hsp70 protein: distinct domains for nucleolar localization and adenosine triphosphate binding. J Cell Biol. 1989;109:1947–1962. doi: 10.1083/jcb.109.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner C, Campbell R. Polymorphic analysis of the three MHC-linked Hsp70 genes. Immunogenetics. 1992;36:357–362. doi: 10.1007/BF00218042. [DOI] [PubMed] [Google Scholar]
- Milner C, Campbell RD. Structure and expression of the three MHC-linked Hsp70 genes. Immunogenetics. 1990;32:242–251. doi: 10.1007/BF00187095. [DOI] [PubMed] [Google Scholar]
- Multhoff G, Mizzen L, Winchester CC, Milner CM, Wenk S, Eissner G, Kampinga HH, Laumbacher B, Johnson J. Heat shock protein 70 (Hsp70) stimulates proliferation and cytolytic activity of natural killer cells. Exp Hematol. 1999;27:1627–1636. doi: 10.1016/s0301-472x(99)00104-6. [DOI] [PubMed] [Google Scholar]
- Panjwani N, Akbari O, Garcia S, Brazil M, Stockinger B. The HSC73 molecular chaperone: involvement in MHC class II antigen presentation. J Immunol. 1999;163:1936–1942. [PubMed] [Google Scholar]
- Pierce SK. Molecular chaperones in the processing and presentation of antigen to helper T cells. Experientia. 1994;50:1026–1030. doi: 10.1007/BF01923457. [DOI] [PubMed] [Google Scholar]
- Pierce SK, DeNagel DC, VanBuskirk AM. A role for heat shock proteins in antigen processing and presentation. Curr Top Microbiol Immunol. 1991;167:83–92. doi: 10.1007/978-3-642-75875-1_5. [DOI] [PubMed] [Google Scholar]
- Poirier GMC, Pyati J, Wan JS, Erlander MG. Screening differentially expressed cDNA clones obtained by differential display using amplified RNA. Nucleic Acid Res. 1997;25:913–914. doi: 10.1093/nar/25.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter-Cid L, Kasahara M, Flajnik MF. Hsp70 genes are linked to the Xenopus major histocompatibility complex. Immunogenetics. 1994;39:1–7. doi: 10.1007/BF00171790. [DOI] [PubMed] [Google Scholar]
- Srivastava PK, Menoret A, Basu S, Binder RJ, McQuade KL. Heat shock proteins come of age: primitive functions acquire new roles in an adaptive world. Immunity. 1998;8:657–665. doi: 10.1016/s1074-7613(00)80570-1. [DOI] [PubMed] [Google Scholar]
- Tavaria M, Gabriele T, Kola I, Anderson RL. A hitchhiker's guide to the human Hsp70 family. Cell Stress Chaperones. 1996;1:23–28. doi: 10.1379/1466-1268(1996)001<0023:ahsgtt>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlecky SR, Chiang HL, Olson TS, Dice JF. Protein and peptide binding and stimulation of in vitro lysosomal proteolysis by the 73-kDa heat shock cognate protein. J Biol Chem. 1992;267:9202–9209. [PubMed] [Google Scholar]
- Wang TF, Chang J, Wang C. Identification of the peptide binding domain of Hsc70. J Biol Chem. 1993;268:26049–26051. [PubMed] [Google Scholar]
- Wells AD, Rai SK, Salvato MS, Band H, Malkovsky M. Hsp72-mediated augmentation of MHC class I surface expression and endogenous antigen presentation. Int Immunol. 1998;10:609–617. doi: 10.1093/intimm/10.5.609. [DOI] [PubMed] [Google Scholar]
- Wiech H, Buchner J, Zimmermann M, Zimmermann R, Jakob U. Hsc70, immunoglobulin heavy chain binding protein, and Hsp90 differ in their ability to stimulate transport of precursor proteins into mammalian microsomes. J Biol Chem. 1993;268:7414–7421. [PubMed] [Google Scholar]
- Wu B, Hunt C, Morimoto R. Structure and expression of the human gene encoding major heat shock protein Hsp70. Mol Cell Biol. 1985;5:330–341. doi: 10.1128/mcb.5.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom JA, Goldberg AL, Finley D, Sherman MY. The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol Cell Biol. 1996;16:3679–3684. doi: 10.1128/mcb.16.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Frueh K, Ahn K, Peterson PA. In vivo assembly of the proteasomal complexes, implications for antigen processing. J Biol Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- Yang Y, Waters JB, Frueh K, Peterson PA. Proteasomes are regulated by interferon. gamma: implications for antigen processing. Proc Natl Acad Sci U S A. 1992;89:4928–4932. doi: 10.1073/pnas.89.11.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Furkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]