Abstract
Patterns of metabolic rate variation have been documented extensively in animals, but their functional basis remains elusive. The membrane pacemaker hypothesis proposes that the relative abundance of polyunsaturated fatty acids in membrane phospholipids sets the metabolic rate of organisms. Using species of tropical orchid bees spanning a 16-fold range in body size, we show that the flight muscles of smaller bees have more linoleate (%18 : 3) and stearate (%18 : 0), but less oleate (%18 : 1). More importantly, flight metabolic rate (FlightMR) varies with the relative abundance of 18 : 3 according to the predictions of the membrane pacemaker hypothesis. Although this relationship was found across large differences in metabolic rate, a direct association could not be detected when taking phylogeny and body mass into account. Higher FlightMR, however, was related to lower %16 : 0, independent of phylogeny and body mass. Therefore, this study shows that flight muscle membrane composition plays a significant role in explaining diversity in FlightMR, but that body mass and phylogeny are other factors contributing to their variation. Multiple factors are at play to modulate metabolic capacity, and changing membrane composition can have gradual and stepwise effects to achieve a new range of metabolic rates. Orchid bees illustrate the correlated evolution between membrane composition and metabolic rate, supporting the functional link proposed in the membrane pacemaker hypothesis.
Keywords: membrane composition, metabolic rate, membrane pacemaker theory, flight, bees, phospholipid
1. Introduction
Membranes play key roles in catalysis, transport and reception because they anchor essential proteins and outline every cell compartment. The composition of the phospholipid bilayer can vary greatly, and this affects the functional properties of membrane proteins by altering their local molecular environment [1–4]. Changing the composition of membrane phospholipids modulates the activity of many oxidative enzymes, ATPases, hormone receptors and ion channels [5–11]. Hulbert & Else [12–14] formulated the membrane pacemaker theory of metabolism by relating the separate observations that mass-specific metabolic rate of mammals scales with body size [15], fatty acid (FA) composition of membrane phospholipids also changes allometrically [16], and most cell processes that consume energy depend on membrane function [17]. They postulate that relative abundance of polyunsaturated FAs sets the molecular activity of membrane proteins, and consequently, the metabolic rate of cells, tissues and organisms [18]. Analyses of endotherm classes varying in body mass by four orders of magnitude provide support for the theory. They show that birds [19] and mammals [16] of increasing body mass reduce membrane unsaturation by substituting docosahexaenoate (DHA or 22 : 6: a FA with 22 carbons and six double bonds) with oleate (18 : 1, a monounsaturated FA). Yet, the correlated evolution between membrane composition and metabolic rate has not been demonstrated.
More direct support for the hypothesis is provided by the experimental manipulation of membrane phospholipids, which shows that various mitochondrial properties, as well as the activity of Na+–K+–ATPase, are altered predictably through specific changes in FA composition [4,20,21]. The mechanisms responsible for activation of the membrane proteins remain unknown, but double bonds constrain rotation between carbons and cause kinks in the acyl chains. Therefore, the relative abundance of polyunsaturated FAs likely affects protein activity by modulating biophysical membrane properties such as fluidity and permeability [22,23]. However, the observed relationships between basal metabolic rate (BMR) and membrane composition in mammals and birds do not demonstrate a functional link between those variables. Their association may simply result from the fact that metabolic rate and membrane composition are both correlated with body mass [24]. In addition, phylogenetic background links species and can impose statistical dependence among phenotypic trait values. The resultant phylogenetic signal can be detected and should be accounted for when comparing species [25]. To address such issues, Valencak & Ruf [26] re-examined this relationship using 30 species of mammals and found no support for the hypothesis. After correcting for the effects of body mass and phylogeny, no significant correlation remains between BMR and any membrane parameter, including unsaturation, % polyunsaturated FAs and % DHA. Similarly, several studies that report intraspecific experiments fail to support the theory [27–29]. Mice selected for high BMR over 20 generations decrease their relative abundance of membrane DHA [28], and natural variation in the BMR of mice is positively correlated with % palmitate (16 : 0, a saturated FA) [27]. Therefore, both studies contradict predictions from the theory. More recently, mice selected for high maximal metabolic rate (MMR) over seven generations did not change membrane unsaturation, but showed an increase in polyunsaturated FAs consistent with the hypothesis [30]. Overall, the membrane pacemaker hypothesis may help explain natural variation in metabolic rate, particularly if observations made at different scales of genetic diversity (interspecific versus intraspecific) are reconciled.
The goal of this study was to test the membrane pacemaker hypothesis using natural variation in metabolic rates during flight in a lineage of tropical bees [31,32]. This model was selected because (i) phylogenetic relationships among orchid bee species have been established and the importance of phylogenetic signals for membrane composition can be investigated and accounted for, (ii) this group of closely related species spanning a 20-fold range in body size covers an intermediate evolutionary timescale (species within a tribe) that conveniently fills the gap between the intraspecific studies and studies comparing species within classes, (iii) the model allows analysis of a cohesive dataset where active metabolism and membrane composition of flight muscle, the tissue accounting for more than 90% of hovering metabolic rate, can both be measured in the same individuals, and (iv) the hypothesis has never been tested for invertebrates. We hypothesized that membrane composition of orchid bees would vary with body size and with mass-specific metabolic rate according to the theory. To improve current understanding of metabolic rate diversity and evolution, our study addresses key unresolved issues of the membrane pacemaker hypothesis.
2. Material and methods
(a). Sampling of orchid bees and flight measurements
Two to 14 males from 22 species of orchid bees were captured using chemical baits. Fifteen species were found on Barro Colorado Island, Smithsonian Tropical Research Institute, whereas the remaining seven species were captured at Cerro Campana and Santa Rita Ridge in Panama. Bees from Barro Colorado Island were immediately transferred to a respirometry chamber consisting of a 1 l glass flask with a sidearm. Rates of CO2 production during hovering flight were measured using a FOXBOX flow-through field respirometry system (Sable Systems International, Las Vegas, NV). Flight trials lasted approximately 5 min, and only periods of successful hovering flight were considered. The criteria used were a stable CO2 level lasting over 30 s, corresponding to a steady-state CO2 emission while aloft and away from chamber walls [33]. Ambient air was dried and pushed through a flow controller and a flow meter at a rate of 800 ml min−1, before entering the respirometry chamber, being dried again and finally passed through the gas analyser. Atmospheric CO2 levels were monitored before and after each flight measurement to obtain incurrent gas content used for metabolic rate calculation and to ensure no drift in the signal was present. Orchid bees fuel flight exclusively with carbohydrates, yielding a respiratory quotient of 1 [34]; hence, 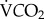 values are equivalent to rates of O2 consumption and were used to estimate metabolic rate [35]. The gas analyser was calibrated daily. Thoracic surface temperature of each bee was also measured immediately after flight and upon landing using an infrared camera (EX300, FLIR, Danderyd, Sweden) and using an empirically determined emissivity of 0.97. Ambient temperature at our field site was fairly constant during our measurement period, with a mean value of 27.3°C and a standard deviation of 1.3°C. Thoracic temperature did not vary with species body mass (p = 0.564; electronic supplementary material, table S1 and figure S1). A total of 188 individuals were caught, measured and transported to the laboratory in 50 ml tubes maintained on ice. Body masses of the bees ranged from 50 to 1081 mg. Animals were then identified, dissected, and body parts were weighed to the nearest milligram, freeze-dried and placed in 1.5 ml CryoTube vials for transport back to Canada where they were stored at −80°C.
values are equivalent to rates of O2 consumption and were used to estimate metabolic rate [35]. The gas analyser was calibrated daily. Thoracic surface temperature of each bee was also measured immediately after flight and upon landing using an infrared camera (EX300, FLIR, Danderyd, Sweden) and using an empirically determined emissivity of 0.97. Ambient temperature at our field site was fairly constant during our measurement period, with a mean value of 27.3°C and a standard deviation of 1.3°C. Thoracic temperature did not vary with species body mass (p = 0.564; electronic supplementary material, table S1 and figure S1). A total of 188 individuals were caught, measured and transported to the laboratory in 50 ml tubes maintained on ice. Body masses of the bees ranged from 50 to 1081 mg. Animals were then identified, dissected, and body parts were weighed to the nearest milligram, freeze-dried and placed in 1.5 ml CryoTube vials for transport back to Canada where they were stored at −80°C.
(b). Fatty acid composition of membrane phospholipids
Total lipids from flight muscle were extracted by homogenizing the thorax (Polytron, Kinematica, Luzern, Switzerland) in 2 : 1 chloroform : methanol (v/v) [36] and completing three cycles of shaking, centrifugation (10 min, 2000g) and filtration. Before the last cycle, 0.25% KCl was added to help remove aqueous contaminants. The aqueous phase was discarded, and the organic phase was dried on a rotating evaporator (Büchi Rotavapor, Flawil, Switzerland). Phospholipids were separated by resuspending total lipids in chloroform before loading on solid-phase extraction columns (Supelclean 1 ml, 100 mg LC-NH2; Sigma-Aldrich, St. Louis, MO). Neutral lipids, non-esterified FAs and phospholipids were separated by sequential elution using solvents of increasing polarity: isopropyl ether : acetic acid (98 : 2 v/v), chloroform : isopropanol (3 : 2 v/v) and methanol [37]. The FA composition of membrane phospholipids was measured after acid transesterification in 1 M acetyl chloride and methanol (90°C for 2 h). FA methyl esters were analysed on an Agilent Technologies 6890N gas chromatograph (Mississauga, Ontario, Canada) equipped with a flame-ionization detector and a fused silica capillary column (Supelco DB-23, 60 m, 0.25 mm i.d., 0.25 μm film thickness; Sigma-Aldrich) using published procedures [38]. Individual FAs were identified by determining exact retention time with authentic standards (Supelco, Bellefonte, PA). Only the FAs accounting for more than 1% of total FAs in membrane phospholipids are reported.
(c). Statistical and phylogenetic analyses
All statistical analyses were performed using Systat v. 13 software. The relative abundance of each FA (expressed in weight%) in phospholipids is presented as species mean ± s.e. Variables were first tested for normality using the Shapiro–Wilk test. Relationships between body mass and the different variables were expressed as power functions, where Y = aXb, as such relationships have been well documented for physiological variables in animals in general [39] and orchid bees in particular [32]. Moreover, relationships between body mass and FA relative abundance in birds and mammals also follow power functions [40]. Therefore, variables were log-transformed to linearize the relationships. Normality of residuals was also verified using the Shapiro–Wilk test. We used linear regressions to test for relationships between body mass and flight metabolic rate (FlightMR) or FA relative abundance in each fraction. An ANCOVA test was used to evaluate differences among genera, and the interaction term was shown only when significant. Kolmogorov–Smirnov and Levene tests were used to check for normality and homoscedasticity.
The relationship between FlightMR and each FA relative abundance was first tested using simple linear regression. In order to test how differences in muscle tissue metabolic intensity relate to membrane composition, we expressed FlightMR on a mass-specific basis, but also performed analyses using whole-animal values (see description below and §3). We further investigated the fine association between FlightMR and FA relative abundance by accounting for the effects of body mass and thoracic temperature. Initial models included genus and interaction terms but only final models with significant variables are presented. We then used backward stepwise multiple regression to test the effect of body mass, thoracic temperature and the relative abundance of all FAs on FlightMR. The same analysis was conducted on whole-animal values. We report these findings graphically using residuals obtained from a multiple regression including body mass and thoracic temperature effects on FlightMR. All analyses were also conducted using the standardized contrasts obtained from a phylogenetically independent contrasts (PICs) analysis described below.
To assess and correct for the degree of relatedness among species, we analysed the data using phylogenetic comparative methods in two steps. We first evaluated the presence and strength of a phylogenetic signal, the tendency of related species to resemble each other, using Pagel's λ and Blomberg's K indices. Then we conducted PIC analysis. Both analyses were done using the phylogenetic tree obtained from RNA polymerase II (RNAP II), cytochrome oxidase subunit 1 (CO1), arginine kinase (argK) and elongation factor 1-alpha (EF-1 alpha) gene sequences. To generate the tree, we first compiled the sequences for our 22 species using GenBank based on the analysis found in Ramírez et al. [41]. We included two outgroup species in the analysis (Apis cercana and Apis dorsata), as well as Aglae caerulea, the putative ancestral genus at the base of the orchid bee tribe. We then aligned the sequences obtained using the muscle software [42], and we determined the best-fit model using the MrModel software [43]. The aligned sequences were concatenated into a singular long sequence with the help of a Perl script, replacing the missing data with a ‘question mark’. We used MrBayes v. 3.1.2 [44] to reconstruct the phylogeny through a partitioned dataset (four independent genes) and an averaged generalized time-reversible gamma model. Through a Markov chain Monte Carlo method, six million trees were generated, keeping every thousandth generation, giving a total of 6000 trees. To measure the phylogenetic signal, we used the phylosig function (v0.2) included in the phytools package [45], in the R environment [46]. We then conducted a PIC analysis using the phenotypic diversity analysis program (PDAP) module [47] in Mesquite [48]. We obtained standardized independent contrasts from the log-transformed character data and tested relationships using linear or multiple regressions fitted through the origin [49]. To assess the influence of the phylogeny, topology and branch length on the PIC results, we conducted all analyses using the last 2041 trees obtained from the Bayesian analysis; the same analysis was performed on a set of 10 000 simulated trees and essentially yielded the same results. Results are reported as Pearson correlation coefficient (two-tailed p ≤ 0.05).
3. Results
(a). Flight metabolic rate diversity in orchid bees
Whole-animal FlightMR, measured as CO2 production rate or 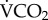 , was strongly dependent on species body mass, following the allometric relationship
, was strongly dependent on species body mass, following the allometric relationship 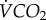 = 51.29 Mb0.800 (95% CI:
±
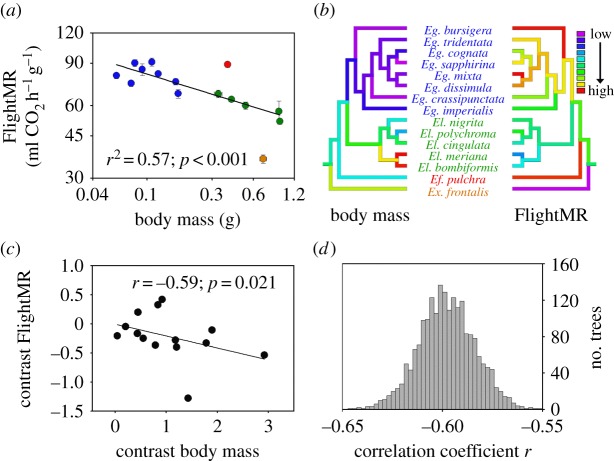
0.104) (r2 = 0.96; p < 0.001). Owing to the allometric exponent below 1, FlightMR expressed on a mass-specific basis decreased with increasing size (figure 1a; FlightMR = 51.64 Mb−0.198 (±0.104), r2 = 0.57, p < 0.001), such that orchid bee species sampled varied by approximately 2.5-fold, from 90.8 in Euglossa mixta to 36.0 ml CO2 h−1 g−1 in Exaerete frontalis. To reflect species differences in flight muscle metabolic intensity, mass-specific FlightMR was used unless otherwise mentioned. Two species representing the genera Exaerete and Eufriesea deviated from the body mass regression with high Studentized residual values (−4.00 and 2.60, respectively). An ANCOVA showed no difference between Euglossa and Eulaema after accounting for body mass (mass: F1,10 = 7.21, p = 0.023; genus: F1,10 = 0.02, p = 0.900). A multiple regression including both body mass and thoracic temperature explained a larger proportion of the FlightMR variance (r2 = 0.77; mass: β = −0.178, p < 0.001; temp.: β = 5.29, p = 0.006). Body mass had a significant phylogenetic signal (λ = 1.25, p < 0.001 and K = 1.54, p = 0.001), as did FlightMR (λ = 1.17, p < 0.001 and K = 1.16, p = 0.024). Groupings of trait values associated with phylogeny, where closely related species have similar trait values, are shown for body mass and FlightMR in figure 1b. Using PIC analysis, the relationship between body mass and FlightMR remained significant (r = −0.59, p = 0.021; figure 1c). The same analysis was performed using 2041 trees obtained from the Bayesian analysis and showed that the average correlation coefficient of the distribution (r) was −0.60, and all absolute values of correlation coefficients obtained were higher than the critical r value of 0.532 (two-tailed p ≤ 0.05; d.f. = 12; figure 1d).
= 51.29 Mb0.800 (95% CI:
±
0.104) (r2 = 0.96; p < 0.001). Owing to the allometric exponent below 1, FlightMR expressed on a mass-specific basis decreased with increasing size (figure 1a; FlightMR = 51.64 Mb−0.198 (±0.104), r2 = 0.57, p < 0.001), such that orchid bee species sampled varied by approximately 2.5-fold, from 90.8 in Euglossa mixta to 36.0 ml CO2 h−1 g−1 in Exaerete frontalis. To reflect species differences in flight muscle metabolic intensity, mass-specific FlightMR was used unless otherwise mentioned. Two species representing the genera Exaerete and Eufriesea deviated from the body mass regression with high Studentized residual values (−4.00 and 2.60, respectively). An ANCOVA showed no difference between Euglossa and Eulaema after accounting for body mass (mass: F1,10 = 7.21, p = 0.023; genus: F1,10 = 0.02, p = 0.900). A multiple regression including both body mass and thoracic temperature explained a larger proportion of the FlightMR variance (r2 = 0.77; mass: β = −0.178, p < 0.001; temp.: β = 5.29, p = 0.006). Body mass had a significant phylogenetic signal (λ = 1.25, p < 0.001 and K = 1.54, p = 0.001), as did FlightMR (λ = 1.17, p < 0.001 and K = 1.16, p = 0.024). Groupings of trait values associated with phylogeny, where closely related species have similar trait values, are shown for body mass and FlightMR in figure 1b. Using PIC analysis, the relationship between body mass and FlightMR remained significant (r = −0.59, p = 0.021; figure 1c). The same analysis was performed using 2041 trees obtained from the Bayesian analysis and showed that the average correlation coefficient of the distribution (r) was −0.60, and all absolute values of correlation coefficients obtained were higher than the critical r value of 0.532 (two-tailed p ≤ 0.05; d.f. = 12; figure 1d).
Figure 1.
(a) Relationship between body mass and flight metabolic rate (FlightMR) in 15 orchid bee species. Values are means (±s.e.) and colours represent the four genera sampled (blue, Euglossa; red, Eufriesea; green, Eulaema; orange, Exaerete). (b) Trait values were mapped on the phylogeny using colour coding such that low values are represented by cold colours and high values by warm colours. Grouping of traits associated with the phylogeny, where closely related species share similar values, was found for both body mass and FlightMR (see §3 for phylogenetic signal results). (c) Relationship between phylogenetically independent contrasts for body mass and FlightMR. (d) Distribution of correlation coefficients from independent contrasts analyses performed with 2041 trees and testing for different tree topologies and branch lengths (see §2). All correlations were higher than the critical r-values for significance at the p = 0.05 level (two-tailed; r = 0.532; d.f. = 12).
(b). Correlated evolution between membrane composition and body mass in orchid bees
The thoracic membranes were composed of five major FAs: 16 : 0 and 18 : 0 were the main saturated FAs, 18 : 1 (n − 9) was the main monounsaturated FA, 18 : 2 (n − 6) and 18 : 3 (n − 3) were the main polyunsaturated FAs. In all species, these five FAs accounted for at least 95% of the total. Other FAs were found in low abundances ( < 1%) and their presence varied among individuals within species: 14 : 0, 16 : 1, 20 : 0, 20 : 1, 21 : 0, 22 : 0, 22 : 1, 20 : 5 (see the electronic supplementary material, tables S2 and S3).
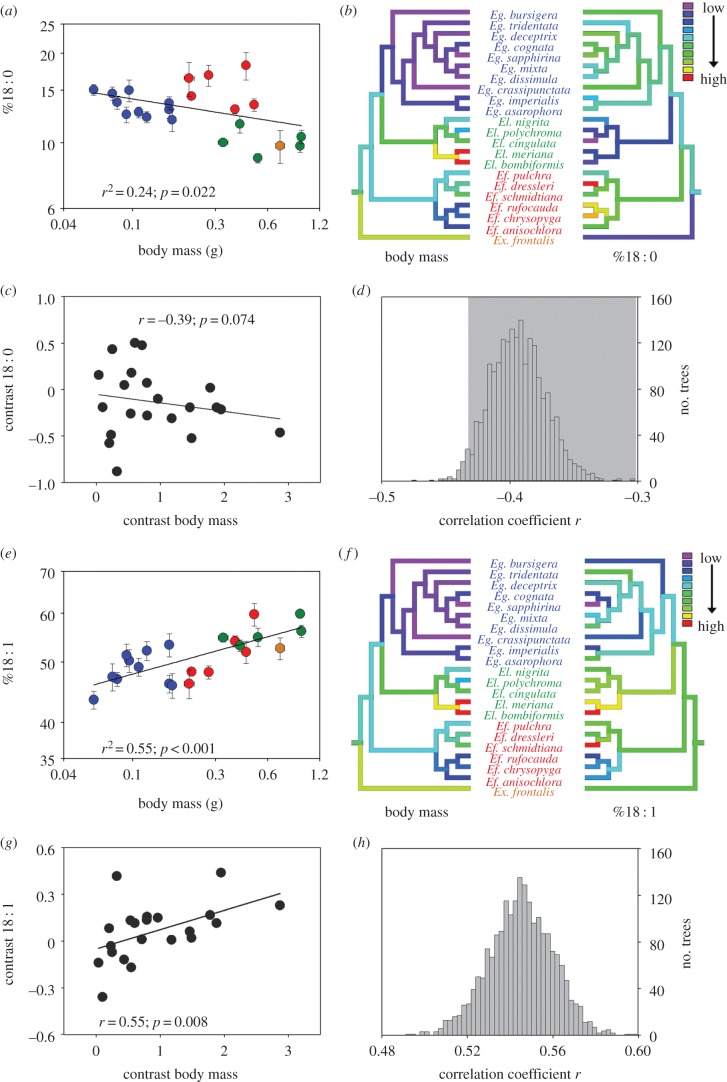
The relative abundance of palmitate, 16 : 0, was not associated with variation in species body mass (figure 2a; p = 0.638). Stearate, 18 : 0, in membranes decreased significantly with body mass (figure 3a; 18 : 0 = 10.89 Mb−0.110(±0.091), r2 = 0.24, p = 0.022), and ranged from 8.9% (Eulaema cingulata) to 18.2% (Eufriesea dressleri). Using an ANCOVA accounting for body mass variation showed that the mean of species belonging to the Eufriesea genus had greater 18 : 0 content than other genera (mass: F1,17 = 2.38, p = 0.142; genus: F2,17 = 18.97, p < 0.001). 18 : 0 had a significant phylogenetic signal (λ = 1, p < 0.001 and K = 0.92, p = 0.001), and trait values mapped on the phylogeny show groupings associated with phylogenetic relatedness (figure 3b). The relationship between body mass and %18 : 0 became non-significant when applying the PIC analysis (r = − 0.39, p = 0.074; figure 3c); the mean correlation coefficient of analyses conducted with Bayesian trees was − 0.39, and the vast majority of trees (1974 of 2041 trees) showed absolute correlation coefficient below the critical value of 0.433 (two-tailed p ≤ 0.05; d.f. = 19; figure 3d).
Figure 2.
Relationship between body mass and the relative abundance of (a) 16 : 0, (b) 18 : 2 and (c) 18 : 3 in the thorax membrane phospholipids of 22 orchid bee species. Values are means (±s.e.), and colours represent the four genera sampled (blue, Euglossa; red, Eufriesea; green, Eulaema; orange, Exaerete).
Figure 3.
Relationship between body mass and the relative abundance of (a) 18 : 0, (e) 18 : 1 in the thorax membrane phospholipids of 22 orchid bee species. Values are means (±s.e.), and colours represent the four genera sampled (blue, Euglossa; red, Eufriesea; green, Eulaema; orange, Exaerete). Trait values were mapped on the phylogeny using colour coding such that low values are represented by cold colours and high values by warm colours. Grouping of traits associated with the phylogeny was found for (b) body mass and 18 : 0, as well as (f) body mass and 18 : 1 (see text for phylogenetic signal results). Relationship between phylogenetically independent contrasts for (c) body mass and 18 : 0 (r = −0.39, p = 0.074), and for (g) body mass and 18 : 1 (r = 0.55, p = 0.008). (d,h) Distribution of correlation coefficients from independent contrasts analyses performed with 2041 trees and testing for different tree topologies and branch lengths (see §2). Shaded areas represent correlation coefficients below the critical r-value of 0.433 (two-tailed p ≤ 0.05; d.f. = 19).
The strongest relationship with body mass was found for oleate (18 : 1; figure 3e; 18 : 1 = 57.02 Mb0.076(±0.031), r2 = 0.55, p < 0.001). This FA was the most abundant in the membrane, ranging from 43.4% in Euglossa sapphirina (smallest species) to 59.8% in Eulaema bombiformis (second-biggest species). An ANCOVA accounting for body mass did not detect any differences among genera (mass: F1,17 = 9.08, p = 0.008; genus: F2,17 = 1.10, p = 0.355). A significant phylogenetic signal was found for 18 : 1 (λ = 0.98, p = 0.03 and K = 0.69, p = 0.02), and groupings of trait values associated with phylogeny are shown in figure 3f. PIC analysis showed that the effect of body mass on %18 : 1 remained significant when correcting for phylogenetic relationships (r = 0.55, p = 0.008; figure 3g). Using Bayesian trees showed an average correlation coefficient of 0.55, where all trees showed significant correlations (figure 3h).
The percentage of 18 : 2, linoleate, did not vary significantly with the orchid bees' mass (figure 2b; r2 = 0.03, p = 0.434). This was owing to different relationships across genera shown by the ANCOVA (mass: F1,15 = 1.54, p = 0.233; genus: F2,15 = 18.06, p < 0.001; genus × mass: F2,15 = 21.68, p < 0.001). Regressions within genera showed a significant increase in %18 : 2 with mass among Euglossa species (r2 = 0.80, p < 0.001), whereas the opposite relationship could be observed among Eufriesea species (r2 = 0.82, p = 0.013). The composition in Eulaema species did not vary significantly with mass (r2 = 0.22, p = 0.424). These opposing relationships between genera explained the overall bell-shaped distribution for this polyunsaturated FA.
Finally, alpha-linolenate, 18 : 3, decreased significantly (figure 2c; 18 : 3 = 10.19 Mb−0.186(±0.0181), r2 = 0.19, p = 0.045) with body mass. The abundance varied from 25.7% in the smallest species (Euglossa sapphirina) to 5.4% (Eufriesea dressleri). An ANCOVA accounting for body mass did not detect any differences among genera (mass: F1,17 = 1.86, p = 0.191; genus: F2,17 = 1.81, p = 0.193). No significant phylogenetic signal was detected for 18 : 2 (λ = 0, p = 1 and K = 0.39, p = 0.4) and 18 : 3 (λ = 0.16, p = 0.51 and K = 0.41, p = 0.3).
(c). Flight metabolic rate varies with membrane composition
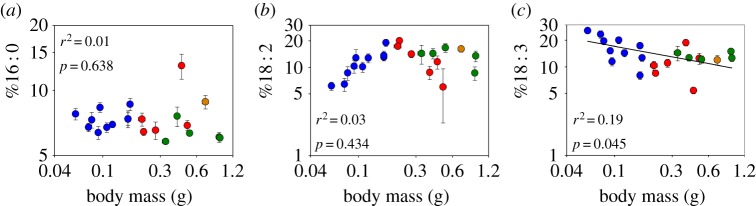
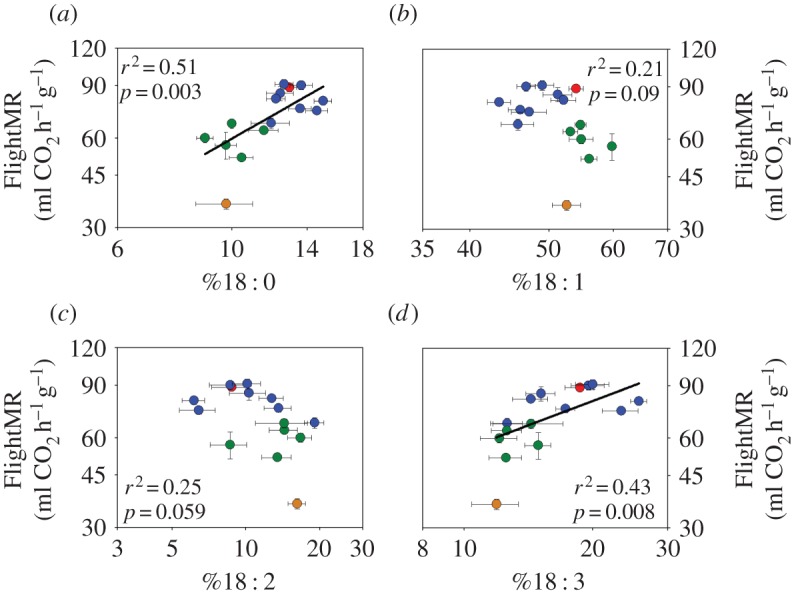
FlightMR increased significantly with the relative abundance of the saturated FA stearate, 18 : 0 (figure 4a; r2 = 0.51, p = 0.003), and alpha-linolenate, 18 : 3 (figure 4d; r2 = 0.43, p = 0.008). Such a relationship could not be detected for %18 : 1 (figure 4b; r2 = 0.21, p = 0.09), %18 : 2 (figure 4c; r2 = 0.25, p = 0.059) or %16 : 0 (figure 5a; r2 = 0.10, p = 0.252). Genera with multiple species representative formed clusters that differed in FlightMR (F1,11 = 25.73, p < 0.001) and the abundance of all FAs (p < 0.05) except for 18 : 2 (p = 0.213). Further analysis showed that the relationship between FlightMR and FA content was dependent on body mass for both 18 : 0 (mass: β = −0.21, p < 0.001; temp.: β = 2.49, p = 0.018; 18 : 0: β = −0.28, p = 0.286) and 18 : 3 (mass: β = −0.16, p < 0.001; temp.: β = 2.69, p = 0.023; 18 : 3: β = 0.05, p = 0.686).
Figure 4.
Relationship between flight metabolic rate (FlightMR) and the relative abundance of fatty acids in the thorax membrane phospholipids of 15 orchid bee species. Metabolic rate as a function of relative abundance is presented for (a) 18 : 0, (b) 18 : 1, (c) 18 : 2 and (d) 18 : 3. Values are means (±s.e.), and colours represent the four genera sampled (blue, Euglossa; red, Eufriesea; green, Eulaema; orange, Exaerete).
Figure 5.
Relationship between (a) flight metabolic rate (FlightMR) and the relative abundance of 16 : 0 in the thorax membrane phospholipids of 15 orchid bee species, (b) residual variation of FlightMR and %16 : 0 and (c) residual variation of FlightMR independent contrasts and %16 : 0 independent contrasts. Colours represent the four genera sampled (blue, Euglossa; red, Eufriesea; green, Eulaema; orange, Exaerete).
In order to assess the influence of multiple variables on FlightMR, a backward stepwise multiple regression, including body mass, thoracic temperature, and the relative abundance of all FAs was performed. The final model (r2 = 0.94; mass: β = −0.214, p < 0.001; temp.: β = 3.743, p = 0.002; %16 : 0: β = −0.645, p < 0.001) showed that variation in FlightMR, independent of body mass or thoracic temperature, was negatively correlated with the relative abundance of 16 : 0. This relationship was represented using the residual variation in FlightMR from the relationship with body mass and thoracic temperature, and plotted against %16 : 0 (figure 5b, r = −0.79, p < 0.001). The same analysis performed using whole-animal FlightMR yields essentially the same results and correlation coefficient. To address the potential that two extreme species may drive the observed correlation (Ex. frontalis and Eufriesea pulchra), we further analysed this relationship using PIC, which strengthened the correlation even more (figure 5c, r = −0.82, p < 0.001). The same results are obtained when using whole-animal FlightMR.
4. Discussion
This study provides direct support for the membrane pacemaker hypothesis in a group of closely related insect species. We show that species variation in flight muscle membrane composition plays a significant role in explaining diversity of FlightMR, but that body mass and phylogeny are other factors also involved in driving variation in membrane composition. Consistent with mammals and birds, the relative abundance of key FAs varies with body mass in orchid bees (figures 2 and 3). Smaller bees have less oleate (18 : 1) in their membranes than larger species, even after accounting for phylogenetic relatedness. Whereas DHA levels in tissue membranes decrease with size in mammals and birds, in orchid bees another omega-3 FA varies the same way—smaller species have membranes with more linolenate (18 : 3) than larger bees. Furthermore, a saturated FA, stearate (18 : 0), shows a significant decrease with body mass, although this relationship appears to be driven by phylogenetic association among species. Moreover, we find a twofold increase in FlightMR and %18 : 3, where bees with high metabolic activity have higher 18 : 3 content. When considering only fine variation in FlightMR after accounting for body mass, thoracic temperature and phylogeny, we show that species with higher FlightMR have lower 16 : 0 content. Overall, our results show the correlated evolution of membrane composition and metabolic rate in flying orchid bees, supporting the proposed functional link between these variables [12].
The original studies that led to the pacemaker hypothesis of metabolism were done on mammals [16] and birds [19]. Here, insects were used to examine the relationship between membrane composition and metabolic rate, and we report evidence of such a pattern outside the chordates. The resulting implications for the pacemaker hypothesis could be important if strong support can be demonstrated across phyla, because recent research questions its validity [26,28–30]. Furthermore, we test the pacemaker hypothesis using an intermediate phylogenetic scale (within the orchid bee tribe, Euglossini). Previous evidence comes from comparisons within classes of vertebrates (mammals or birds) or intraspecific studies (artificially selected mice). Our results show that relationships apply on a narrower range in body mass than in the original studies, a range of 16-fold, versus 9000- and 11 000-fold ranges in mammals and birds, respectively.
Previous research on the membrane pacemaker hypothesis of metabolism often overlooks the influence of phylogenetic relatedness among the species studied. Such lack of independence in the comparison of various traits was noted in early comparative work [25], and recently revisited in a context of membrane composition and metabolic rate [26], or ageing [24]. The important issue of relatedness among species selected for the study of membrane composition–body mass relationships is absent from the original work that led to the pacemaker hypothesis. Here, the presence of a phylogenetic signal for FlightMR and body mass in bees indicates that relatedness among species clearly affects metabolic rate (figure 1b). After correcting for the phylogenetic associations among species, the relationship remains significant (figure 1c,d), further supporting previous findings [31]. Phylogeny evidently also affects membrane composition for 18 : 0 and 18 : 1 according to body mass (figure 3b,f). After the correction for phylogenetic relatedness, this relationship persists for 18 : 1 (figure 3g,h), but not for 18 : 0 (figure 3c,d). Thus, variation in 18 : 0 and 18 : 1 content is associated with body mass, but phylogenetic association also drives part (18 : 1) or most (18 : 0) of the variation in this cellular trait.
Tropical orchid bees prove to be a convenient model to assess the membrane pacemaker hypothesis of metabolism. Flying insects sustain some of the highest mass-specific metabolic rates of any animals, and it is estimated that the thoracic flight muscle accounts for more than 90% of the metabolic rate [50]. Indeed, our previous work on orchid bees finds that wing-loading (body mass per unit wing area) drives wing-beat frequency, whereas wing-beat frequency, a major determinant of power output [51], drives hovering metabolic rate [32], including deviations observed for species such as Ex. frontalis and Ef. pulchra in figure 1a. Carbohydrate oxidation fuels the stretch-activated wing muscles of these flying insects [34]. We show that the membrane lipid composition of tropical orchid bee flight muscles comprises only five main FAs: two saturates, one monounsaturate and two polyunsaturates. Here, 18 : 1 accounts for 40–60% of membrane FAs. That is in clear contrast with vertebrate models, in which up to nine different acids account for more than 5% of total FAs in the skeletal muscle of mammals [16], birds [19] or fish [52]. In orchid bees, other acyl chains are found in trace quantities (less than 1%) and very long-chain FAs (22 carbons or more) are virtually absent from the thoracic membranes. Orchid bees are thus a simpler model that offers many advantages for the study of covariation between membrane lipid composition and metabolism.
This work represents the first test of the pacemaker hypothesis on metabolic rate during locomotion among different species. Previous investigations focus primarily on BMR and FA composition in organs that are the main contributors to BMR. In a recent study, mice selected for increased MMR were analysed for FA composition of gastrocnemius muscle and liver membranes, and compared with control lines. Selection for increased MMR did not result in direct changes in FA desaturation. However, individual variation in the percentages of some FAs correlated with MMR and BMR. In the liver, %18 : 0 and %22 : 6, among others, were predictors of MMR, but no FAs were predictors of BMR. In muscle, %18 : 2 and %22 : 6 were predictors of MMR, whereas %18 : 1 was one of the predictors of BMR [30]. In our study, hovering FlightMR (analogous to MMR) is correlated with the abundance of key FAs (figures 4 and 5), as predicted by the pacemaker hypothesis.
Variation in composition of the most polyunsaturated FA found in orchid bees, 18 : 3, follows the predictions of the membrane pacemaker hypothesis. Its abundance varies with body mass, following the same allometric exponent as FlightMR (−0.19 and −0.20, respectively), such that a twofold difference in FlightMR corresponds to a similar difference in %18 : 3. This parallel association with body mass supports their functional link. The relationship between FlightMR and the abundance of FAs, however, shows strong species groupings (figure 4). Essentially, genera differ in %18 : 0, %18 : 1, %18 : 3 and FlightMR, but the association between these FAs and metabolic rate is not found within genus and not independent of body mass, thus questioning direct support of the pacemaker hypothesis. Our results illustrate a recurring conundrum in similar studies, such as in experimental evolution studies [28] or studies controlling for body mass and phylogeny [26] that seldom find strong support for the pacemaker hypothesis. Nonetheless, we did find a direct relationship between FlightMR and the abundance of the saturated FA 16 : 0, after controlling for body mass and phylogeny (figure 5). Species with high FlightMR have lower %16 : 0, according to the pacemaker hypothesis. Our interpretation of such findings is twofold: (i) a given membrane composition can accommodate species ranging in metabolic intensity, but transitions in membrane composition are necessary to achieve larger changes in metabolic intensity, such as differences observed between orchid bee genera; (ii) adjustments in the composition of some FAs are associated with finer gradual changes in metabolic activity.
Hovering FlightMR of orchid bees is driven by variation in wing morphology and kinematics [32], and muscle tissue metabolic capacity evolved according to metabolic rate variation. Darveau et al. [31] report that the activity of enzymes involved in ATP production also differs across species in conjunction with species FlightMR. Suarez et al. [53] quantify the relative importance of level of enzyme expression and metabolic regulation to account for interspecific variation in metabolic rates. These studies emphasize that differences in metabolic capacity are explained by multiple cellular properties. Membrane composition can have a major influence on the activity of membrane-bound proteins affecting metabolic capacities, such as cellular transport and oxidative phosphorylation. These effects on cellular activity could be owing to bulk changes in membrane composition affecting multiple functions, combined with specific changes targeting certain properties. Still, the activity of non-membrane-bound proteins can be modulated and affect metabolic capacity. The evolution of metabolic phenotype is therefore not necessarily gradual and direct, but can involve stepwise changes in properties that allow new metabolic capacities.
Using orchid bees as a model system, this study demonstrates that membrane composition evolves with metabolic rate, in accordance with the membrane pacemaker hypothesis. However, results also show that body mass and phylogeny play an important role in explaining variation in membrane composition across species. It is our view that multiple factors are at play that provide the means to alter metabolic capacity, such that some changes in membrane composition would be analogous to switching gears and achieving a new range of metabolic rates, whereas others have more gradual effects. Quantifying the relative contributions of membrane composition and other factors in setting metabolic capacity strikes us as a major challenge for future work.
Supplementary Material
Acknowledgements
The authors acknowledge the work of Emmanuelle Godbout-Gauthier, who generated preliminary results that guided this study. We also thank the staff from Barro Colorado Island in Panama for their help in making part of this work possible. The comments of anonymous referees greatly helped to improve the paper.
Funding statement
This study was supported by an NSF grant award to R.K.S., and NSERC Discovery Grants to J.-M.W. and C.-A.D.
References
- 1.Gerson AR, Brown JCL, Thomas R, Bernards MA, Staples JF. 2008. Effects of dietary polyunsaturated fatty acids on mitochondrial metabolism in mammalian hibernation. J. Exp. Biol. 211, 2689–2699. ( 10.1242/jeb.013714) [DOI] [PubMed] [Google Scholar]
- 2.Guderley H, Kraffe E, Bureau W, Bureau DP. 2008. Dietary fatty acid composition changes mitochondrial phospholipids and oxidative capacities in rainbow trout red muscle. J. Comp. Physiol. B, Biochem. Syst. Environ. Physiol. 178, 385–399. ( 10.1007/s00360-007-0231-y) [DOI] [PubMed] [Google Scholar]
- 3.Murphy MG. 1990. Dietary fatty acids and membrane protein function. J. Nutr. Biochem. 1, 68–79. ( 10.1016/0955-2863(90)90052-m) [DOI] [PubMed] [Google Scholar]
- 4.Stillwell W, Jenski LJ, Crump FT, Ehringer W. 1997. Effect of docosahexaenoic acid on mouse mitochondrial membrane properties. Lipids 32, 497–506. ( 10.1007/s11745-997-0064-6) [DOI] [PubMed] [Google Scholar]
- 5.Guo W, Xie W, Lei T, Hamilton JA. 2005. Eicosapentaenoic acid, but not oleic acid, stimulates β-oxidation in adipocytes. Lipids 40, 815–821. ( 10.1007/s11745-005-1443-8) [DOI] [PubMed] [Google Scholar]
- 6.Power GW, Newsholme EA. 1997. Dietary fatty acids influence the activity and metabolic control of mitochondrial carnitine palmitoyltransferase I in rat heart and skeletal muscle. J. Nutr. 127, 2142–2150. [DOI] [PubMed] [Google Scholar]
- 7.Miyasaka CK, Azevedo RB, Curi R, Filho JM, Lajolo FM. 1996. Administration of fish oil by gavage increases the activities of hexokinase, glucose-6-phosphate dehydrogenase, and citrate synthase in rat lymphoid organs. Gen. Pharmacol. 27, 991–994. ( 10.1016/0306-3623(96)00041-9) [DOI] [PubMed] [Google Scholar]
- 8.Corcoran MP, Lamon-Fava S, Fielding RA. 2007. Skeletal muscle lipid deposition and insulin resistance: effect of dietary fatty acids and exercise. Am. J. Clin. Nutr. 85, 662–677. [DOI] [PubMed] [Google Scholar]
- 9.Leaf A, Xiao YF, Kang JX, Billman GE. 2005. Membrane effects of the n-3 fish oil fatty acids, which prevent fatal ventricular arrhythmias. J. Membr. Biol. 206, 129–139. ( 10.1007/s00232-005-0789-9) [DOI] [PubMed] [Google Scholar]
- 10.Swanson JE, Lokesh BR, Kinsella JE. 1989. Ca2+-Mg2+ ATPase of mouse cardiac sarcoplasmic reticulum is affected by membrane n-6 and n-3 polyunsaturated fatty acid content. J. Nutr. 119, 364–372. [DOI] [PubMed] [Google Scholar]
- 11.Turner N, Haga KL, Hulbert AJ, Else PL. 2005. Relationship between body size, Na+–K+–ATPase activity, and membrane lipid composition in mammal and bird kidney. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R301–R310. ( 10.1152/ajpregu.00297.2004) [DOI] [PubMed] [Google Scholar]
- 12.Hulbert AJ, Else PL. 1999. Membranes as possible pacemakers of metabolism. J. Theor. Biol. 199, 257–274. ( 10.1006/jtbi.1999.0955) [DOI] [PubMed] [Google Scholar]
- 13.Hulbert AJ, Else PL. 2000. Mechanisms underlying the cost of living in animals. Annu. Rev. Physiol. 62, 207–235. ( 10.1146/annurev.physiol.62.1.207) [DOI] [PubMed] [Google Scholar]
- 14.Hulbert AJ. 2003. Life, death and membrane bilayers. J. Exp. Biol. 206, 2303–2311. ( 10.1242/jeb.00399) [DOI] [PubMed] [Google Scholar]
- 15.Kleiber M. 1961. The fire of life: an introduction to animal energetics, p. 454 New York, NY: Wiley. [Google Scholar]
- 16.Couture P, Hulbert AJ. 1995. Membrane fatty acid composition of tissues is related to body mass of mammals. J. Membr. Biol. 148, 27–39. ( 10.1007/bf00234153) [DOI] [PubMed] [Google Scholar]
- 17.Rolfe DFS, Brown GC. 1997. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 77, 731–758. [DOI] [PubMed] [Google Scholar]
- 18.Hulbert AJ, Else PL. 2005. Membranes and the setting of energy demand. J. Exp. Biol. 208, 1593–1599. ( 10.1242/jeb.01482) [DOI] [PubMed] [Google Scholar]
- 19.Hulbert AJ, Faulks S, Buttemer WA, Else PL. 2002. Acyl composition of muscle membranes varies with body size in birds. J. Exp. Biol. 205, 3561–3569. [DOI] [PubMed] [Google Scholar]
- 20.Wu BJ, Hulbert AJ, Storlien LH, Else PL. 2004. Membrane lipids and sodium pumps of cattle and crocodiles: an experimental test of the membrane pacemaker theory of metabolism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 287, R633–R641. ( 10.1152/ajpregu.00549.2003) [DOI] [PubMed] [Google Scholar]
- 21.Else PL, Wu BJ. 1999. What role for membranes in determining the higher sodium pump molecular activity of mammals compared to ectotherms? J. Comp. Physiol. B 169, 296–302. ( 10.1007/s003600050224) [DOI] [PubMed] [Google Scholar]
- 22.Hazel JR. 1995. Thermal adaptation in biological membranes: is homeoviscous adaptation the explanation? Annu. Rev. Physiol. 57, 19–42. ( 10.1146/annurev.ph.57.030195.000315) [DOI] [PubMed] [Google Scholar]
- 23.Yehuda S, Rabinovitz S, Carasso R, Mostofsky D. 2002. The role of polyunsaturated fatty acids in restoring the aging neuronal membrane. Neurobiol. Aging 23, 843–853. ( 10.1016/s0197-4580(02)00074-x) [DOI] [PubMed] [Google Scholar]
- 24.Speakman JR. 2005. Correlations between physiology and lifespan: two widely ignored problems with comparative studies. Aging Cell 4, 167–175. ( 10.1111/j.1474-9726.2005.00162.x) [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15. ( 10.1086/284325) [DOI] [Google Scholar]
- 26.Valencak TG, Ruf T. 2007. N−3 polyunsaturated fatty acids impair lifespan but have no role for metabolism. Aging Cell 6, 15–25. ( 10.1111/j.1474-9726.2006.00257.x) [DOI] [PubMed] [Google Scholar]
- 27.Haggerty C, Hoggard N, Brown DS, Clapham JC, Speakman JR. 2008. Intra-specific variation in resting metabolic rate in MF1 mice is not associated with membrane lipid desaturation in the liver. Mech. Ageing Dev. 129, 129–137. ( 10.1016/j.mad.2007.11.001) [DOI] [PubMed] [Google Scholar]
- 28.Brzęk P, Bielawska K, Książek A, Konarzewski M. 2007. Anatomic and molecular correlates of divergent selection for basal metabolic rate in laboratory mice. Physiol. Biochem. Zool. 80, 491–499. ( 10.1086/520617) [DOI] [PubMed] [Google Scholar]
- 29.Pannorfi R, Zee BM, Vatnick I, Berner N, Hiebert SM. 2012. Dietary lipid saturation influences environmental temperature preference but not resting metabolic rate in the Djungarian hamster (Phodopus sungorus). Physiol. Biochem. Zool. 85, 405–414. ( 10.1086/666473) [DOI] [PubMed] [Google Scholar]
- 30.Wone BWM, Donovan ER, Cushman JC, Hayes JP. 2013. Metabolic rates associated with membrane fatty acids in mice selected for increased maximal metabolic rate. Comp. Biochem. Physiol. A, Comp. Physiol. 165, 70–78. ( 10.1016/j.cbpa.2013.02.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darveau C-A, Hochachka PW, Roubik DW, Suarez RK. 2005. Allometric scaling of flight energetics in orchid bees: evolution of flux capacities and flux rates. J. Exp. Biol. 208, 3593–3602. ( 10.1242/jeb.01777) [DOI] [PubMed] [Google Scholar]
- 32.Darveau C-A, Hochachka PW, Welch KC, Roubik DW, Suarez RK. 2005. Allometric scaling of flight energetics in Panamanian orchid bees: a comparative phylogenetic approach. J. Exp. Biol. 208, 3581–3591. ( 10.1242/jeb.01776) [DOI] [PubMed] [Google Scholar]
- 33.Skandalis DA, Darveau C-A. 2012. Morphological and physiological idiosyncrasies lead to interindividual variation in flight metabolic rate in worker bumblebees (Bombus impatiens). Physiol. Biochem. Zool. 85, 656–670. ( 10.1086/665568) [DOI] [PubMed] [Google Scholar]
- 34.Suarez RK, Darveau C-A, Welch KC, O'Brien DM, Roubik DW, Hochachka PW. 2005. Energy metabolism in orchid bee flight muscles: carbohydrate fuels all. J. Exp. Biol. 208, 3573–3579. ( 10.1242/jeb.01775) [DOI] [PubMed] [Google Scholar]
- 35.Lighton JRB. 2008. Measuring metabolic rates: a manual for scientists. New York, NY: Oxford University Press. [Google Scholar]
- 36.Folch J, Lees M, Stanley GHS. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509. [PubMed] [Google Scholar]
- 37.Maillet D, Weber J-M. 2006. Performance-enhancing role of dietary fatty acids in a long-distance migrant shorebird: the semipalmated sandpiper. J. Exp. Biol. 209, 2686–2695. ( 10.1242/jeb.02299) [DOI] [PubMed] [Google Scholar]
- 38.Magnoni L, Weber J-M. 2007. Endurance swimming activates trout lipoprotein lipase: plasma lipids as a fuel for muscle. J. Exp. Biol. 210, 4016–4023. ( 10.1242/jeb.007708) [DOI] [PubMed] [Google Scholar]
- 39.Schmidt-Nielsen K. 1984. Scaling, why is animal size so important? 1st edn Cambridge, UK: Cambridge University Press. [Google Scholar]
- 40.Hulbert A, Pamplona R, Buffenstein R, Buttemer WA. 2007. Life and death: metabolic rate, membrane composition and life span of animals. Physiol. Rev. 87, 1175–1213. ( 10.1152/physrev.00047.2006) [DOI] [PubMed] [Google Scholar]
- 41.Ramírez SR, Roubik DW, Skov CE, Pierce NE. 2010. Phylogeny, diversification patterns and historical biogeography of euglossine orchid bees (Hymenoptera: Apidae). Biol. J. Linn. Soc. 100, 552–572. ( 10.1111/j.1095-8312.2010.01440.x) [DOI] [Google Scholar]
- 42.Edgar R. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5, 113 ( 10.1186/1471-2105-5-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nylander JAA. 2004. MrModeltest. (ed. author P.d.b.t.), V2 ed. Uppsala University, Evolutionary Biology Centre. [Google Scholar]
- 44.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. ( 10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 45.Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 46.R Development Core Team. 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 47.Midford PE, Garland T, Jr, Maddison WP. 2010. PDAP package of MESQUITE. Version 1.16.
- 48.Maddison WP, Maddison DR. 2011. Mesquite: a modular system for evolutionary analysis. Version V2.75. See http://mesquiteproject.org.
- 49.Garland T, Harvey PH, Ives AR. 1992. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 41, 18–32. ( 10.2307/2992503) [DOI] [Google Scholar]
- 50.Suarez RK. 1998. Oxygen and the upper limits to animal design and performance. J. Exp. Biol. 201, 1065–1072. [DOI] [PubMed] [Google Scholar]
- 51.Pennycuick CJ, Rezende MA. 1984. The specific power output of aerobic muscle, related to the power density of mitochondria. J. Exp. Biol. 108, 377–392. [Google Scholar]
- 52.Martin N, Bureau DP, Marty Y, Kraffe E, Guderley H. 2013. Dietary lipid quality and mitochondrial membrane composition in trout: responses of membrane enzymes and oxidative capacities. J. Comp. Physiol. B, Biochem. Syst. Environ. Physiol. 183, 393–408. ( 10.1007/s00360-012-0712-5) [DOI] [PubMed] [Google Scholar]
- 53.Suarez RK, Darveau C-A, Hochachka PW. 2005. Roles of hierarchical and metabolic regulation in the allometric scaling of metabolism in Panamanian orchid bees. J. Exp. Biol. 208, 3603–3607. ( 10.1242/jeb.01778) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.