Abstract
While introductions and supplementations using non-native and potentially domesticated individuals may have dramatic evolutionary effects on wild populations, few studies documented the evolution of genetic diversity and life-history traits in supplemented populations. Here, we investigated year-to-year changes from 1989 to 2009 in genetic admixture at 15 microsatellite loci and in phenotypic traits in an Atlantic salmon (Salmo salar) population stocked during the first decade of this period with two genetically and phenotypically distinct source populations. We detected a pattern of temporally increasing introgressive hybridization between the stocked population and both source populations. The proportion of fish returning to the river after a single winter at sea (versus several ones) was higher in fish assigned to the main source population than in local individuals. Moreover, during the first decade of the study, both single-sea-winter and multi-sea-winter (MSW) fish assigned to the main source population were smaller than local fish. During the second decade of the study, MSW fish defined as hybrids were lighter and smaller than fish from parental populations, suggesting outbreeding depression. Overall, this study suggests that supplementation with non-local individuals may alter not only the genetic diversity of wild populations but also life-history traits of adaptive significance.
Keywords: genetic admixture, introgressive hybridization, life-history traits, outbreeding depression, Salmo salar, stocking
1. Introduction
The evolutionary impact of deliberate or unintentional introductions into wild populations of non-native and/or domesticated individuals is a growing concern for the management of endangered species [1–3]. The success of non-native species has been related to multiple introductions leading to high levels of genetic diversity in the invaded range [4–6]. Alternatively, the translocation of non-local individuals may threaten the local adaptation of native individuals [7–9], notably via introgressive hybridization leading possibly to hybrid vigour (i.e. heterosis) but also often to lower performances of hybrids relative to parental lineages (i.e. outbreeding depression, e.g. [10,11]) (hereafter we refer to non-local individuals as individuals from the species' native range which do not originate from the population where they were released). Moreover, captive breeding may select, after only one generation, for maladaptive traits in the wild hence potentially affecting the fitness of wild populations [12–14]. Therefore, it is crucial to document and understand the consequences of supplementation on the genetic makeup of populations but also on life-history traits of individuals to mitigate threats to wild populations and maximize their evolutionary potential over the long term [10,15,16].
Numerous studies have recently used pre- and post-stocking samples from supplemented populations together with samples from source populations or captivity to examine admixture at individual and population levels, especially in fish species for which archived samples are often available [16–18]. While the analysis of historical samples allowed assessment of the ‘natural’ genetic structure of wild populations prior to stocking operations, the analysis of post-introduction samples revealed variable loss of genetic integrity of target populations in terms of increased admixture [18–20], reduced differentiation with donor stocks [21] or disruption of relationships between genetic structure and geographical distance [17]. However, even if such comparisons between pre- and post-stocking situations inform on the degree of admixture in stocked populations, they do not provide any information on the temporal dynamics of admixture or on the phenotypic consequences of stocking in recipient populations.
Many animal traits are partially under genetic control and morphological traits like length and weight have high heritability values [22]. In salmonid fishes, morphological variations including body length, weight or life-history differences have been potentially linked to local adaptation [8,23,24]. Moreover, selective pressures in a captive environment can differ from those encountered in the wild and may lead to differences in life-history traits among wild-born and captive-reared individuals [13,15,25,26]. Therefore, stocked and wild individuals may harbour different fitness-related traits values, and consequently population admixture may alter phenotypic variation within the stocked population, although this has rarely been investigated ([27–29], but see [30]). For example, anadromous fishes (i.e. species that reproduce in freshwater and grow to adult size at sea) reared in a hatchery may have different migration tactics [14] and body length [31–33]. Fine long-term monitoring of traits and population assignment of individuals could help examine such phenotypic effects of supplementation. This is particularly important in the case of migratory species like Atlantic salmon (Salmo salar) in which the phenology of migration can be affected by stocking and for which consequences of supplementations will be observed only when individuals return from their potentially long migration [31–33]. The life history of Atlantic salmon is also extremely variable among populations: individuals spend 1–4 four years in freshwater as juveniles and then migrate to sea where they stay for 1–5 years (single-sea-winter (SSW) and multi-sea-winter (MSW) fish, respectively) before returning to their natal river for spawning.
Here, we examine the dynamics of population admixture and changes in phenotypic traits over 21 years in a small wild Atlantic salmon population (Sélune River, France) stocked using juveniles from two genetically [18,34] and phenotypically distinct populations. The main source population has been used from 1989 to 1994 and from 1996 to 1997, while the second one was used only in 1995. This unique configuration of stocking and record of individual phenotypic data allowed an analysis on a yearly basis of the evolution of genetically and phenotypically different groups of individuals sharing a common environment. We specifically tested whether: (i) a significant proportion of introduced fish survived and returned to the river, resulting in notable population admixture and in an erosion of genetic differences between sources and target populations few generations after the beginning of stocking, i.e. during the first decade of the study; (ii) introduced fish successfully reproduced together and with local fish, leading to notable proportions of fish assigned to stocking sources and defined as hybrids in generations following the end of stocking, i.e. during the second decade of the study; (iii) potential phenological or morphological differences among stocked and local individuals may persist beyond the first decade of the study; and (iv) individuals defined as hybrids between stocked and local individuals may present better or worse performances compared with parental populations, suggesting heterosis or outbreeding depression, respectively.
2. Material and methods
(a). Study populations, stocking and sample collection
The Sélune River is among the four Atlantic salmon rivers opening out into the Mont Saint Michel Bay, in the northwest of France (electronic supplementary material, figure S1; [18]). The salmon population from the Sélune River has been declining since the 1950s, partly as a result of local habitat degradation and the construction of two dams. In order to increase the population size and to support local recreational fishing activities, juvenile Atlantic salmons (young of the year) produced at the Favot hatchery (Brittany, France) were annually stocked in the Sélune from 1989 to 1997. Adult salmon originating from two rivers, the Aulne and Gave d'Oloron (Gave), were used as sources of progenitors. In total, 333 700 juveniles produced from Aulne progenitors were stocked in the Sélune from 1989 to 1994 and in 1996–1997, while 30 000 juveniles produced from Gave progenitors were released in 1995 only (figure 1a). Stocking was stopped in the Sélune after 1997 but it has been continued in the nearby Couesnon River (electronic supplementary material, table S1). Juveniles originating from Aulne were F1s (produced from wild parents), while those from Gave were F2s (produced from F1 hatchery reared parents). These two populations are genetically differentiated from the Sélune, hence hatchery reared individuals and wild x hatchery hybrids can be identified and population genetic admixture can be estimated [35].
Figure 1.
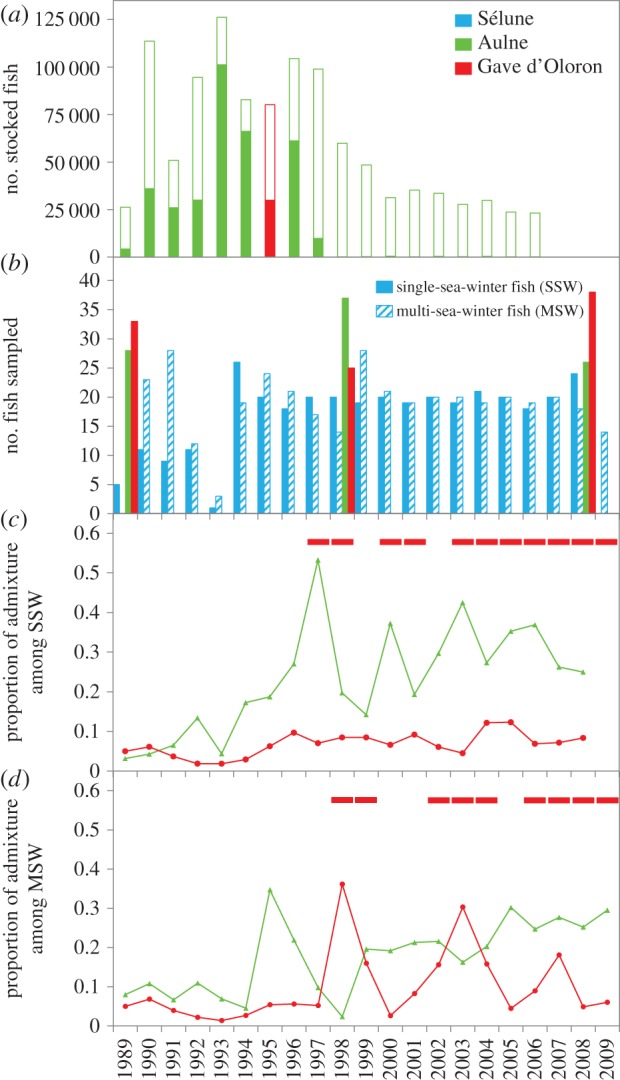
(a) Number of salmon from Aulne and Gave d'Oloron stocked in the Sélune River (filled bars) and in other proximal rivers from the Bay of Mont-Saint-Michel (open bars). (b) Number of SSW (coloured) and MSW (striped) fish sampled per year in the Sélune River, and number of individuals sampled in the Aulne and Gave d'Oloron rivers. Average admixture proportions (±s.d.) with Gave d'Oloron (circles) and Aulne (triangles) in the Sélune population for SSW (c) and MSW (d), estimated with Structure for k = 3. The squares in (c,d) represent the expected years of return for adults originating from Gave individuals released in 1995. Only main life histories are represented: 1-1+ (1 year in freshwater and 1 year at sea) and 2-1+ for SSW individuals and 1-2+ and 2-2+ for MSW fish. (Online version in colour.)
A total of 907 fish caught by anglers were sampled from 1989 to 2009 (figure 1b; see also electronic supplementary material, table S2). The body length and weight of each fish was recorded by anglers who also sampled scales on their catches. Scales stored in paper envelops at INRA were used: (i) to determine the age of individuals and the number of winters at sea (i.e. sea age), and (ii) for genetic analyses. In the Sélune River, 720 fish were selected between 1989 and 2009 to include in the analysis a similar proportion of SSW (n = 341) and MSW (n = 379) individuals in each cohort. Fish were selected randomly within each sea age category. We chose this balanced sampling strategy regardless of the natural proportion of these life histories in the population in order to obtain sufficient sample sizes to accurately estimate the evolution of admixture and phenotypic traits for both life strategies. In addition, over the years 1989, 1998 and 2008, 91 fish from the Aulne River and 96 fish from Gave d'Oloron River were genotyped as a baseline for admixture estimates (figure 1b).
(b). Genetic analyses
Genomic DNA was extracted in a solution with 10 µl of proteinase K (10 mg ml−1), 10 µl of TE (TrisEDTA 1x) and 100 µl of Chelex 100 sodium form (5%) incubated overnight at 56°C, and then 15 min at 100°C [36]. The DNA solution (10x) was used to genotype 15 microsatellite loci selected for the European SALSEA-Merge project [37]. A QIAGEN's Multiplex PCR Kit was used for PCR amplification according to manufacturer's recommendations, using an annealing temperature of 56°C. Forward primers were labelled with one of the three different Applied Biosystems (ABI) fluorescent dyes of set D (6-FAM, HEX or NED), and primer pairs were distributed in three multiplex sets: (FAM-SSsp2210, FAM-Ssa202, HEX-SSspG7, HEX-SSsp2201, NED-SsaD144, NED-SsaD157), (FAM-SSsp2216, HEX-Ssa289, HEX-SSsp1605, NED-Ssa14, NED-Ssa171) and (FAM-SsaF43, HEX-Ssa197, NED-SsaD486, NED-SSsp3016). The loci were analysed on an ABI 3130 Genetic Analyzer (ABI) and scored with Genemapper v. 3.7 software (ABI). The SsaD486 locus was used to identify and remove from the dataset putative brown trout (Salmo trutta) or F1 hybrids among brown trout and Atlantic salmon, which could have been sampled unintentionally [38]. Statistical analyses were thereafter performed with the 14 microsatellites left.
(c). Statistical analyses of molecular data
Micro-Checker was used to detect possible null alleles and allelic drop-outs [39]. The null hypothesis of independence between loci was tested from statistical genotypic disequilibrium analysis with the software Genepop v. 3.4 [40]. All linkage disequilibrium tests were corrected for multiple comparison by applying a false discovery rate (FDR) [41] (p.adjust of the statistical package R [42]). Allele frequencies, the total and average number of alleles, observed (Ho) and expected (He) heterozygosities were estimated at the population level with Genetix 4.05.2 [43]. Estimates of allelic richness (rarefaction size of 13 individuals), gene diversity, FIS (tested by permutation) and pairwise Fst were computed using Fstat, v. 2.9.3 [44].
(d). Admixture analyses
To assess the potential admixture at both population and individual levels, Bayesian clustering analyses were performed assuming an admixture model without using prior information on samples' origin as implemented in the software Structure 2.3.1 [45]. We tested from 1 to 6 genetic clusters (k) with 10 replicates for each k (burn-in period = 50 000 steps, Markov chain Monte Carlo replicates = 300 000 and estimation of 90% credible intervals). As previously found in [18], k = 3 was the best clustering solution based on L(K) and ΔK criteria [45,46] delineating Sélune, Aulne and Gave populations. The average admixture of temporal samples was estimated over the 10 Structure replicates to investigate the impact of stocking at the population level. Individual membership values to each cluster (q) were averaged over the 10 replicates to: (i) visually inspect trends in individual admixture; (ii) classify individuals as belonging to one of four origins: Sélune, Gave, Aulne (q-value ≥ 0.75) and a ‘admixed’ category (q-values < 0.75); and (iii) compare admixture proportions between SSW and MSW individuals.
We examined temporal trends in admixture of the Aulne and Gave populations into the Sélune samples from 1989 to 2009 using β regressions with a log–log transformation and β distribution error. The evolution of admixture proportions was analysed for overall fish and separately among SSW and MSW individuals for each stocking source from the beginning of stocking until the end of the survey, that is from 1989 to 2009 for the Aulne population and from 1995 to 2009 for the Gave population. These analyses were performed with the betareg function of the betareg package in R ([47]).
(e). Statistical analyses of phenotypic data
The weight, body length and the proportion of MSW individuals were compared among the four origins (Aulne, Gave, Sélune and admixed) over the whole period studied but also during the periods 1992–2000 and 2001–2009 to detect a potential erosion of differences among origins through time. Linear mixed models with a Gaussian error distribution were used to analyse log-transformed weight and body length data, and a logistic mixed model was used to test for differences in proportions of MSW individuals using the lme4 package [48] in R. The condition factor of fish was also considered with similar linear mixed models performed using residuals from the regression: log(weight) ∼ log(body length (estimated coefficient = 2.97, F625 = 3658.9, adjusted-R2 = 0.85, p < 0.001). A random cohort effect was included in all models and the significance of independent variables was tested with likelihood ratio tests. Multiple comparisons among groups were realized using the function pairwise.t.test for linear models and Fisher's exact tests for logistic models, with a FDR adjustment [41] as implemented in R. For each genetic origin and separately for SSW and MSW individuals, ANOVAs were performed on weight and body length data to test for differences between the two time periods (1992–2000 and 2001–2009). For the sake of comparison, weight and body length of SSW and MSW fish captured in their river of origin were also compared among populations using data from fish caught by anglers between 1989 and 2009 (SSW fish: n = 2037, 569, 997 and MSW fish: n = 1192, 933, 948 in Aulne, Gave and Sélune populations, respectively). These analyses were performed with linear models including population, age at sea and period (1992–2000 and 2001–2009) effects as independent variables. Multiple comparisons among populations were then carried out as explained above.
3. Results
(a). Genetic diversity within samples
Overall, 720 salmon from the Sélune River and 187 from Gave and Aulne rivers (n = 96 and 91, respectively) were successfully genotyped at a minimum of 10 microsatellite loci. No genotypic disequilibrium was detected over all populations. Micro-Checker analyses suggested the occurrence of null alleles in 18 out of 378 tests, yet they were not associated to specific samples or markers, and no evidence of large allelic drop-out or stuttering was found. Also, no significant departure from Hardy-Weinberg equilibrium was observed (electronic supplementary material, table S3).
The average gene diversity was similar in the three populations: 0.78 in Aulne, 0.77 in Gave and 0.76 in Sélune including all temporal samples. A tendency of increasing gene diversity was observed over time in the Sélune (He ranging from 0.76 to 0.80; electronic supplementary material, table S3). Allelic richness averaged 8.3 in Aulne and Gave rivers and ranged from 7.5 to 8.8 in Sélune temporal samples.
(b). Genetic differentiation among samples
The genetic differentiation among temporal samples from each source population was low: from 0.006 (p > 0.05) to 0.013 (p < 0.05) among samples from Gave and from 0.001 to 0.010 (p > 0.05) among samples from Aulne (electronic supplementary material, table S4). The differentiation was also low and not significant among pre-stocking samples from Sélune (1989, 1990 and 1991), ranging from 0.000 to 0.005 (p > 0.05). However, a significant differentiation (p < 0.05) among Gave, Aulne and Sélune pre-stocking samples was found with FSTs of 0.056 between Gave and Aulne, from 0.054 to 0.072 between Aulne and Sélune and from 0.078 to 0.083 between Gave and Sélune. A decrease of the genetic differentiation among Sélune and source populations was already noticed during the first decade of the study: FST between Aulne and Sélune was 0.035 and 0.065 between Gave and Sélune in 1998. This lower differentiation was still observed at the end of the study after the end of stocking events: FST between Aulne and Sélune was 0.032 and 0.064 between Gave and Sélune in 2008. On the other hand, the genetic differentiation between source populations only slightly declined during the study period: FST between Aulne and Gave was 0.049 and 0.045 in 1998 and 2008, respectively.
(c). Admixture
Individual admixture proportions are shown in figure 1 and electronic supplementary material, figure S2, for every year and separately for SSW and MSW fish (electronic supplementary material, table S5). Salmon from the Sélune population were sorted into four categories: 471 were assigned to Sélune, 21 to Gave, 103 to Aulne and 125 were classified as admixed individuals (‘admixed’) (electronic supplementary material, table S6). The occurrence of individuals assigned to the Gave and Aulne populations was consistent with the timing of supplementation operations (figure 1 and electronic supplementary material, figure S2). Overall, the admixture with the Aulne population increased significantly over time (average over the time series ±s.d = 0.26 ± 0.06; electronic supplementary material, table S7) and this pattern held for both SSW and MSW individuals. However, SSW individuals were more admixed than MSW individuals (mean ± s.d. = 0.23 ± 0.14 and mean ± s.d. = 0.18 ± 0.09, respectively, estimated coefficient = −0.17, log-likelihood ratio = 496.6, d.f. = 3, pseudo-R2 = 0.01, p = 0.021; figure 1c,d). The admixture with the Gave population also increased during the survey (estimated coefficient = 0.04, log-likelihood ratio = 39.47, d.f. = 3, pseudo-R2 = 0.31, p = 0.013) but this increase was no longer significant when considering only the 1995–2009 period following the stocking event. Analyses highlighted a cyclic increase following the unique release of the Gave salmon in 1995 with following peaks of admixture in 1998, 2003 and 2007 in MSW individuals (figure 1d). These individuals seemed more admixed with Gave than SSW individuals but this trend was not significant (mean ± s.d. = 0.12 ± 0.09 and mean ± s.d. = 0.08 ± 0.02, respectively, figure 1d,c). Interestingly, individuals assigned with high confidence to the Gave population were detected from 2002 to 2006 (electronic supplementary material, table S6) although no stocking operation using this population occurred after 1995.
(d). Life-history traits
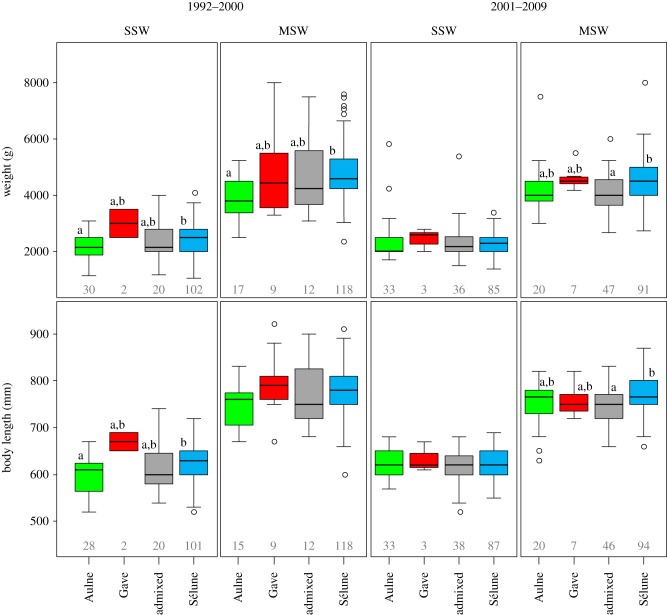
Phenotypic differences among genetic origins are presented in figures 2 and 3 (electronic supplementary material, table S8). We initially sampled similar proportions of SSW and MSW individuals in each cohort of the Sélune population (47.4% and 52.6%, respectively) but genetic assignment analyses revealed that proportions of MSW individuals significantly differed among the four origins (table 1 and figure 2). The lowest and highest proportions of MSW fish were observed in the Aulne population (37%, significantly lower than Gave and Sélune populations) and in the Gave population (76%, significantly higher than the Aulne population), respectively, whereas fish assigned as admixed individuals had a proportion (50%) similar to that observed for the pure Sélune population. Body length and weight were significantly different among SSW and MSW individuals and among the four origins (figure 3), yet no interaction between the age at sea and origins was detected (table 1). On the other hand, a significant interaction was found between origins and periods for these two traits. During the 1992–2000 period, SSW Aulne salmon were smaller and lighter than the SSW Sélune fish (figure 3). However, between 2001 and 2009, no significant difference was observed. Regarding MSW fish during the 1992–2000 period, Aulne salmon were also lighter than the Sélune salmon but no difference appeared for body length. MSW individuals sampled from 2001 to 2009 presented a different pattern with only one significant difference detected: admixed individuals were lighter and smaller than pure Sélune fish (figure 3). Therefore, we tested for a pattern of outbreeding depression by comparing the mean size and body length of these admixed individuals with the mean parental value (average value of the three genetic origins). We observed that both weight and body length of MSW admixed salmon sampled from 2001 to 2009 were significantly lower than the mean parental values (body length: t92.071 =−3.121, p = 0.002). Differences of body length and weight between the two periods were also detected within genetic origins: SSW Aulne salmon sampled in the 1992–2000 period were smaller than those from the 2001–2009 period (F1 = 7.48, p = 0.008) and MSW Sélune salmon were heavier in the first period than in the second one (F1 = 5.16, p = 0.024). Regarding the condition factor, only one significant effect was detected: MSW individuals had a better condition factor than SSW salmon (likelihood ratio = 10.97, d.f. = 1, p < 0.001).
Figure 2.
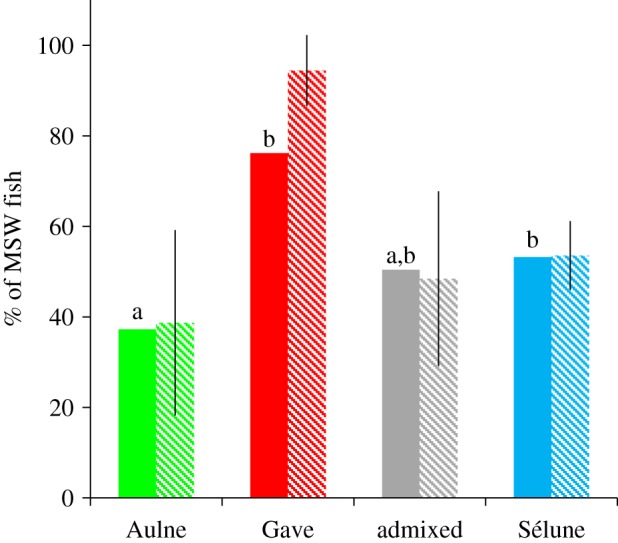
Percentages of MSW fish among individuals assigned to Sélune, Aulne and Gave d'Oloron clusters and to a admixed category from 1992 to 2009. Percentages are given for all years pooled (coloured bars) or as averages over the years (striped bars, ±s.d.). (Online version in colour.)
Figure 3.
Box plots of the body weight (g) and body length (mm) of fish assigned to Sélune, Aulne, Gave d'Oloron clusters and to a admixed category for two age at sea categories (SSW: single-sea-winter and MSW: multi-sea-winter) and for two periods: 1992–2000 and 2001–2009. Sample sizes are indicated for each box and different letters indicate significant pairwise differences. (Online version in colour.)
Table 1.
Results of logistic and linear mixed models testing the effect of origin (Aulne, Gave, Sélune and a admixed category) and period (1992–2000 and 2001–2009) on age at sea (SSW or MSW individuals) (a) and the effects of the genetic origin, period and age at sea on the weight (b) and body length of fish (c). (Significant p-values are in italic. χ d.f.: degree of freedom of χ2 distribution, L. ratio: likelihood ratio, P: p-value.)
| source of variation | parameters | χ d.f. | L. ratio | P |
|---|---|---|---|---|
| (a) number of winters at sea | ||||
| full model | origin × period | 3 | 3.6056 | 0.3073 |
| origin | 3 | 14.313 | <0.001 | |
| period | 1 | 0.1147 | 0.7349 | |
| (b) LOG weight (mm) | ||||
| full model | origin × age at sea × period | 3 | 1.92 | 0.589 |
| age at sea × period | 1 | 0.58 | 0.446 | |
| origin × age at sea | 3 | 4.02 | 0.259 | |
| origin × period | 3 | 8.24 | 0.041 | |
| period | 1 | 2.27 | 0.132 | |
| origin | 3 | 19.65 | <0.001 | |
| age at sea | 1 | 757.86 | <0.001 | |
| (c) LOG body length (mm) | ||||
| full model | origin × age at sea × period | 3 | 2.39 | 0.495 |
| age at sea × period | 1 | 0.60 | 0.440 | |
| origin × age at sea | 3 | 2.51 | 0.473 | |
| origin × period | 3 | 10.91 | 0.012 | |
| period | 1 | 0.84 | 0.359 | |
| origin | 3 | 20.57 | <0.001 | |
| age at sea | 1 | 800.04 | <0.001 | |
Comparing phenotypic traits on fish captured in their native river (electronic supplementary material, figure S3) revealed that SSW salmon of the Aulne and Sélune rivers were of similar body length (mean (mm) ± s.e.: 628.81 ± 43.20 and 626.60 ± 41.27, respectively) but both smaller than the Gave River salmon (668.46 ± 42.63). Regarding MSW individuals, Aulne River salmon (751.98 ± 48.13) were smaller than those from the Sélune River (778.81 ± 49.53), which were smaller than the Gave River salmon (798.51 ± 50.85; electronic supplementary material, figure S3 and table S9). Similar differences were observed for the weight of both SSW and MSW fish: Aulne individuals (SSW, mean (g) ± s.e.: 2270.14 ± 532.15, MSW: 3950.81 ± 865.46) being lighter than Sélune individuals (SSW: 2383.43 ± 579.79, MSW: 4709.47.81 ± 993.71) and Gave fish being the heaviest (SSW: 2809.21 ± 558.52, MSW: 5048.07.81 ± 967.48); electronic supplementary material, figure S3). The differences observed between populations were noted during the two periods (1992–2000 and 2001–2009; electronic supplementary material, figure S3).
4. Discussion
This study based on a longitudinal time series of Atlantic salmon sampled over 21 years revealed several major impacts of supplementation with non-local individuals into a wild population. First, genetic admixture rose rapidly in the Sélune population during the first decade of the study and was accompanied by a diminution of the differentiation with both source populations. Second, stocked fish reproduced with local fish producing hybrids but they also mated together as revealed by the detection of individuals assigned to the Gave population up to 11 years after the introduction of this source population in the Sélune River. Third, phenotypic differences detected among the three populations using individuals collected in their native river were also (for most of them) found in fish caught in the Sélune River during the first decade of the study. Specifically, Aulne salmon showed a higher proportion of SSW fish and were smaller and lighter than the Sélune and Gave salmon. Interestingly, body length and weight differences disappeared in the second decade of the monitoring, which could be explained by an adaptive or plastic response to local environmental conditions. Admixed MSW salmon sampled in the second decade of the study were also lighter and smaller than values averaged over parental populations, which may suggest a pattern of outbreeding depression.
To our knowledge, this study represents the first continuous temporal survey showing the evolution of admixture patterns before, during and after the end of stocking operations. The admixture of Sélune samples with the two stocking sources was overall consistent with the timing and intensity of supplementation: it increased steeply over time with highest values following the first significant stocking events and then persisted beyond the end of supplementations. We also detected an increase of the proportion of hybrids after the end of stocking, suggesting a potentially high reproductive success and/or survival of stocked fish as well as hybridization events among stocked and native fish. However, given the potentially reduced reproductive success of stocked fish in the wild [13,14,49], an effect of stocking was expected only in the short term. Immigration of fish stocked in nearby rivers may explain the persistence of admixture in the Sélune population as these rivers were stocked by similar source populations (electronic supplementary material, table S1). Importantly, a recent simulation study of the Sélune population suggested that stocked salmon in this system may have a 10–25 times lower survival than local individuals, hence the high levels of admixture observed are probably more linked to the high numbers of fish stocked than to a high survival of these individuals [18].
Interestingly, the patterns of admixture turned out to be different depending on the source population and/or stocking practice. The Aulne River salmon that were repeatedly stocked in the Sélune River from 1989 to 1997 resulted in a sharp increase of admixture that subsequently stabilized around roughly 30%, whereas the source population used once (Gave) produced a cyclic pattern of admixture in MSW individuals. Despite the relatively low number of Gave juveniles released in 1995, the proportion of fish from this origin was high among returning MSW adults in the Sélune in 1998: 0.36 (five of 14). This high proportion may explain why some Gave individuals mated together in 1998 as reflected by the moderate proportion of Gave fish among returning adults in 2003: 0.20 (four out of 20). This reproduction among stocked fish in the wild may just be due to the relatively high number of Gave individuals on the Sélune spawning grounds in 1998 but it may also suggest some degree of premating reproductive isolation between the two populations. Various mechanisms may explain the preferential reproduction of Gave individuals together including behavioural or genetic cues (e.g. genes of the major histocompatibility complex, [50]) as well as spatial or temporal isolation of spawners [20]. Post-zygotic isolation may have also occurred but it was probably weak as several admixed genotypes were detected (electronic supplementary material, figure S2 and table S6). Post-zygotic reproductive isolation has been described in various salmonid species through lower survival or body length of interpopulation hybrids (e.g. [28,51,52]).
The two source populations were phenotypically distinct from each other and also from the Sélune population. Only one of these differences persisted throughout the study period: the proportion of MSW individuals in fish assigned to the Gave population was higher than in individuals originating from the Aulne River Interestingly, the proportion of MSW salmon in the Gave River is also slightly higher than in the Aulne River: 0.19 and 0.13, respectively (and 0.17 for Sélune) [35]. The different proportions of MSW individuals observed for two decades in these populations introduced in a new environment may thus reflect intrinsic differences but dissimilarities in stocking practices may have also played a role. Fish released from the Aulne in the Sélune were F1s produced from a wild broodstock changed every year (for 8 years), while individuals from Gave were F2s from a captive F1 broodstock. Several studies showed a dramatic decline of the MSW proportion [14,31,53,54] in supplemented populations potentially owing to increased growth in the hatchery. However, the high number of MSW fish among Gave individuals is difficult to explain and may also be owing to a preferential (but not documented) use of MSW progenitors in the hatchery as the number of winter at sea could be partially genetically determined in Atlantic salmon [33,55].
Regarding other traits such as weight and body length, differences were also detected among genetic origins but only during the first decade of study: SSW Aulne fish captured in the Sélune were smaller and lighter than local individuals. Interestingly, Aulne salmon from the second period were larger than those measured during the first period, while no difference in weight or body length of Sélune individuals was observed between the two periods. Analyses of fish caught in their river of origin showed that SSW Aulne individuals were lighter and of similar body length than those from the Sélune for the two periods considered. Altogether these observations suggest that the increase in body length of only SSW Aulne individuals during the whole period in the Sélune may be due to selection for a body length similar to local individuals or to different reactions norms between genetic origins following some environmental changes (i.e. gene x environment interactions). A pure environmental effect would have probably altered the body length of fish of each genetic origin but further investigations would be necessary to demonstrate whether this change was adaptive, plastic or due to environmental variations.
Aulne MSW individuals caught in the Sélune during the 1992–2000 period were also lighter than local individuals but this difference did not persist during the 2001–2009 period. This pattern seems linked to a decrease in weight of MSW Sélune but a lack of significant difference between Aulne temporal samples may also be due to low sample sizes. Interestingly, a diminution of weight has also been observed in MSW salmon of several Scottish rivers from 1995 to 2006 [56], hence environmental changes at sea may be involved in the changes in weight of MSW Sélune individuals.
Importantly, the only significant phenotypic differences among genetic origins during the 2001–2009 period were the smaller body length and weight of MSW admixed individuals compared with local Sélune fish. These admixed individuals were also shown to be lighter and smaller than the mean values calculated over parental populations. These results may suggest some outbreeding depression, a hypothesis that would require an experimental approach to be validated. Outbreeding depression has been frequently described in numerous taxa [10] including salmonids ([27,28,51,57,58], review in [29]).
Overall, this temporal study highlights an important and rapid loss of local genetic integrity resulting from introgressive hybridization with non-local stocked individuals. This pattern of neutral genetic admixture persisted far beyond the end of supplementation operations. It is also possible that non-neutral genetic variation was introgressed into the native gene pool [7,59,60] which may alter local adaptation within an already small and threatened wild population and ultimately fail to contain its demographic decline [12,61]. Alternatively, this genetic admixture may have potential (and unnoticed) beneficial effects for the fitness of wild populations as often reported in non-native species for which multiple introductions of different lineages strongly contribute to their success in the invaded range [4–6,62,63]. The differences observed in life traits also suggested that supplementation programmes should carefully consider potential mechanisms leading to deviations from the life traits observed in the wild stocked populations. In particular, non-random sampling or artificial breeding can cause unintentional selection on particular traits. Accordingly, phenological differences (i.e. age at sea) of stocked adults depending on source populations observed in this study could have important consequences on the fitness of wild populations [64]. Differences in body length and weight between these individuals and local fish were also detected suggesting potential important impacts on the phenotypic variation of the local population. Lower body length and weight of fish originating from interbreeding between native and stocked individuals further indicate potential negative effects on the long-term viability of the local population. These results thus contrast with the prevailing view that translocations mainly bring positive effects to endangered populations including higher genetic diversity and possible ‘genetic rescue’ in small populations [9,61]. These potential negative impacts thus suggest that translocations of individuals should be cautiously considered for the conservation management of endangered populations and their consequences should be monitored in the long term to be fully appreciated.
Supplementary Material
Acknowledgements
We thank P. Provost, J.-P. Porcher (ONEMA), J. L. Baglinière and F. Marchand (INRA, experimental unit U3E) for their help in collecting scale samples, and A. Richard, F. Goulmi and M. A. Arago who helped us to collect demograhic data. We are grateful to two anonymous referees for their valuable comments on the manuscript.
Data accessibility
Molecular and phenotypic data has been deposited in the Dryad repository (doi:10.5061/dryad.044g3).
Funding statement
This study was supported by a grant from the Réseau Aquaculture Québec to C.P. and by the Plan Loire Grandeur Nature (ERDF funding, Agence de l'Eau Loire Bretagne and Etablissement Public Loire) and the European Interreg IV project ‘Atlantic Aquatic Resource Conservation’.
References
- 1.Allendorf FW, Leary RF, Spruell P, Wenburg JK. 2001. The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 16, 613–622. ( 10.1016/S0169-5347(01)02290-X) [DOI] [Google Scholar]
- 2.Randi E. 2008. Detecting hybridization between wild species and their domesticated relatives. Mol. Ecol. 17, 285–293. ( 10.1111/j.1365-294X.2007.03417.x) [DOI] [PubMed] [Google Scholar]
- 3.Facon B, Genton BJ, Shykoff J, Jarne P, Estoup A, David P. 2006. A general eco-evolutionary framework for understanding bioinvasions. Trends Ecol. Evol. 21, 130–135. ( 10.1016/j.tree.2005.10.012) [DOI] [PubMed] [Google Scholar]
- 4.Ellstrand NC, Schierenbeck KA. 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proc. Natl Acad. Sci. USA 97, 7043–7050. ( 10.1073/pnas.97.13.7043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molofsky J, Keller SR, Lavergne S, Kaproth MA, Eppinga MB. 2014. Human-aided admixture may fuel ecosystem transformation during biological invasions: theoretical and experimental evidence. Ecol. Evol. 4, 899–910. ( 10.1002/ece3.966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roman J, Darling JA. 2007. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 22, 454–464. ( 10.1016/j.tree.2007.07.002) [DOI] [PubMed] [Google Scholar]
- 7.Fitzpatrick BM, Johnson JR, Kump DK, Smith JJ, Voss SR, Shaffer HB. 2010. Rapid spread of invasive genes into a threatened native species. Proc. Natl Acad. Sci. USA 107, 3606–3610. ( 10.1073/pnas.0911802107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia de Leaniz C, et al. 2007. A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol. Rev. 82, 173–211. ( 10.1111/j.1469-185X.2006.00004.x) [DOI] [PubMed] [Google Scholar]
- 9.Laikre L, Schwartz MK, Waples RS, Ryman N. 2010. Compromising genetic diversity in the wild: unmonitored large-scale release of plants and animals. Trends Ecol. Evol. 25, 520–529. ( 10.1016/j.tree.2010.06.013) [DOI] [PubMed] [Google Scholar]
- 10.Edmands S. 2007. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 16, 463–475. ( 10.1111/j.1365-294X.2006.03148.x) [DOI] [PubMed] [Google Scholar]
- 11.Fraser DJ, Cook AM, Eddington JD, Bentzen P, Hutchings JA. 2008. Mixed evidence for reduced local adaptation in wild salmon resulting from interbreeding with escaped farmed salmon: complexities in hybrid fitness. Evol. Appl. 1, 501–512. ( 10.1111/j.1752-4571.2008.00037.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araki H, Cooper B, Blouin MS. 2007. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science 318, 100–103. ( 10.1126/science.1145621) [DOI] [PubMed] [Google Scholar]
- 13.Christie MR, Marine ML, French RA, Blouin MS. 2012. Genetic adaptation to captivity can occur in a single generation. Proc. Natl Acad. Sci. USA 109, 238–242. ( 10.1073/pnas.1111073109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milot E, Perrier C, Papillon L, Dodson JJ, Bernatchez L. 2013. Reduced fitness of Atlantic salmon released in the wild after one generation of captive breeding. Evol. Appl. 6, 472–485. ( 10.1111/eva.12028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankham R. 2008. Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 17, 325–333. ( 10.1111/j.1365-294X.2007.03399.x) [DOI] [PubMed] [Google Scholar]
- 16.Schwartz MK, Luikart G, Waples RS. 2007. Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 22, 25–33. ( 10.1016/j.tree.2006.08.009) [DOI] [PubMed] [Google Scholar]
- 17.Pearse DE, Martinez E, Garza JC. 2011. Disruption of historical patterns of isolation by distance in coastal steelhead. Conserv. Genet. 12, 691–700. ( 10.1007/s10592-010-0175-8) [DOI] [Google Scholar]
- 18.Perrier C, Baglinière JL, Evanno G. 2013. Understanding admixture patterns in supplemented populations: a case study combining molecular analyses and temporally explicit simulations in Atlantic salmon. Evol. Appl. 6, 218–230. ( 10.1111/j.1752-4571.2012.00280.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finnengan AK, Stevens JR. 2008. Assessing the long-term genetic impact of historical stocking events on contemporary populations of Atlantic salmon, Salmo salar. Fish. Manage. Ecol. 15, 315–326. ( 10.1111/j.1365-2400.2008.00616.x) [DOI] [Google Scholar]
- 20.Hansen MM, Mensberg KLD. 2009. Admixture analysis of stocked brown trout populations using mapped microsatellite DNA markers: indigenous trout persist in introgressed populations. Biol. Lett. 5, 656–659. ( 10.1098/rsbl.2009.0214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eldridge WH, Naish KA. 2007. Long-term effects of translocation and release numbers on fine-scale population structure among coho salmon (Oncorhynchus kisutch). Mol. Ecol. 16, 2407–2421. ( 10.1111/j.1365-294X.2007.03271.x) [DOI] [PubMed] [Google Scholar]
- 22.Visscher PM, Hill WG, Wray NR. 2008. Heritability in the genomics era: concepts and misconceptions. Nat. Rev. Genet. 9, 255–266. ( 10.1038/nrg2322) [DOI] [PubMed] [Google Scholar]
- 23.Oleksyk TK, Smith MW, O'Brien SJ. 2010. Genome-wide scans for footprints of natural selection. Phil. Trans. R. Soc. B 365, 185–205. ( 10.1098/rstb.2009.0219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor EB. 1991. A review of local adaptation in salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture 98, 185–207. ( 10.1016/0044-8486(91)90383-I) [DOI] [Google Scholar]
- 25.Ford MJ. 2002. Selection in captivity during supportive breeding may reduce fitness in the wild. Conserv. Biol. 16, 815–825. ( 10.1046/j.1523-1739.2002.00257.x) [DOI] [Google Scholar]
- 26.Mignon-Grasteau S, et al. 2005. Genetics of adaptation and domestication in livestock. Livestock Prod. Sci. 93, 3–14. ( 10.1016/j.livprodsci.2004.11.001) [DOI] [Google Scholar]
- 27.Fraser DJ, Houde ALS, Debes PV, O'Reilly P, Eddington JD, Hutchings JA. 2010. Consequences of farmed-wild hybridization across divergent wild populations and multiple traits in salmon. Ecol. Appl. 20, 935–953. ( 10.1890/09-0694.1) [DOI] [PubMed] [Google Scholar]
- 28.Gharrett AJ, Smoker WW, Reisenbichler RR, Taylor SG. 1999. Outbreeding depression in hybrids between odd-and even-broodyear pink salmon. Aquaculture 173, 117–129. ( 10.1016/S0044-8486(98)00480-3) [DOI] [Google Scholar]
- 29.McClelland EK, Naish KA. 2007. What is the fitness outcome of crossing unrelated fish populations? A meta-analysis and an evaluation of future research directions. Conserv. Genet. 8, 397–416. ( 10.1007/s10592-006-9178-x) [DOI] [Google Scholar]
- 30.Lamaze FC, Garant D, Bernatchez L. 2013. Stocking impacts the expression of candidate genes and physiological condition in introgressed brook charr (Salvelinus fontinalis) populations. Evol. Appl. 6, 393–407. ( 10.1111/eva.12022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonsson B, Jonsson N. 2006. Cultured Atlantic salmon in nature: a review of their ecology and interaction with wild fish. ICES J. Mar. Sci.: J du Conseil 63, 1162–1181. ( 10.1016/j.icesjms.2006.03.0) [DOI] [Google Scholar]
- 32.KallioNyberg I, Koljonen ML. 1997. The genetic consequence of hatchery-rearing on life-history traits of the Atlantic salmon (Salmo salar L): a comparative analysis of sea-ranched salmon with wild and reared parents. Aquaculture 153, 207–224. ( 10.1016/S0044-8486(97)00023-9) [DOI] [Google Scholar]
- 33.Naevdal G, Holm M, Ingebrigtsen O, Moller D. 1978. Variation in age at 1st spawning in Atlantic salmon (Salmo salar). J. Fish. Res. Board Can. 35, 145–147. ( 10.1139/f78-021) [DOI] [Google Scholar]
- 34.Perrier C, Daverat F, Evanno G, Pécheyran C, Bagliniere J-L, Roussel J-M. 2011. Coupling genetic and otolith trace element analyses to identify river-born fish with hatchery pedigrees in stocked Atlantic salmon (Salmo salar) populations. Can. J. Fish. Aquat. Sci. 68, 977–987. ( 10.1139/f2011-040) [DOI] [Google Scholar]
- 35.Perrier C, Guyomard R, Bagliniere J-L, Evanno G. 2011. Determinants of hierarchical genetic structure in Atlantic salmon populations: environmental factors vs. anthropogenic influences. Mol. Ecol. 20, 4231–4245. ( 10.1111/j.1365-294X.2011.05266.x) [DOI] [PubMed] [Google Scholar]
- 36.Estoup A, Largiader CR, Perrot E, Chourrout D. 1996. Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Mol. Mar. Biol. Biotechnol. 5, 295–298. [Google Scholar]
- 37.Ellis JS, et al. 2011. Microsatellite standardization and evaluation of genotyping error in a large multi-partner research programme for conservation of Atlantic salmon (Salmo salar L.). Genetica 139, 353–367. ( 10.1007/s10709-011-9554-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrier C, Grandjean F, Le Gentil J, Cherbonnel C, Evanno G. 2011. A species-specific microsatellite marker to discriminate European Atlantic salmon, brown trout, and their hybrids. Conserv. Genet. Resour. 3, 131–133. ( 10.1007/s12686-010-9307-1) [DOI] [Google Scholar]
- 39.Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P. 2004. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. ( 10.1111/j.1471-8286.2004.00684.x) [DOI] [Google Scholar]
- 40.Raymond M, Rousset F. 1995. Genepop (version-1.2) - Population-genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249. [Google Scholar]
- 41.Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. ( 10.1214/aos/1013699998) [DOI] [Google Scholar]
- 42.R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org/) [Google Scholar]
- 43.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. 2004. Genetix 4.05, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Université de Montpellier II; See http://kimura.univ-montp2.fr/genetix/. [Google Scholar]
- 44.Goudet J. 1995. Fstat (version 1.2): a computer program to calculate F-statistics. J. Hered. 86, 485–486. [Google Scholar]
- 45.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 14, 2611–2620. ( 10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 47.Cribari-Neto F, Zeileis A. 2010. Beta regression in R. J. Stat. Softw. 34, 1–24. [Google Scholar]
- 48.Bates D, Maechler M, Bolker B. 2012. lme4: linear mixed-effects models using S4 classes. (doi: citeulike-article-id: 12173300) [Google Scholar]
- 49.Thériault V, Moyer GR, Jackson LS, Blouin MS, Banks MA. 2011. Reduced reproductive success of hatchery coho salmon in the wild: insights into most likely mechanisms. Mol. Ecol. 20, 1860–1869. ( 10.1111/j.1365-294X.2011.05058.x) [DOI] [PubMed] [Google Scholar]
- 50.Evans ML, Dionne M, Miller KM, Bernatchez L. 2011. Mate choice for major histocompatibility complex genetic divergence as a bet-hedging strategy in the Atlantic salmon (Salmo salar). Proc. R. Soc. B 279, 379 ( 10.1098/rspb.2011.0909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Côte J, Roussel J-M, Le Cam S, Evanno G. 2014. Outbreeding depression in Atlantic salmon revealed by hypoxic stress during embryonic development. Evol. Biol. 41, 561–571. ( 10.1007/s11692-014-9289-0) [DOI] [Google Scholar]
- 52.Granier S, Audet C, Bernatchez L. 2011. Heterosis and outbreeding depression between strains of young-of-the-year brook trout (Salvelinus fontinalis). Can. J. Zool. 89, 190–198. ( 10.1139/Z10-108) [DOI] [Google Scholar]
- 53.Kallio-Nyberg I, Saloniemi I, Jutila E, Jokikokko E. 2011. Effect of hatchery rearing and environmental factors on the survival, growth and migration of Atlantic salmon in the Baltic Sea. Fish. Res. 109, 285–294. ( 10.1016/j.fishres.2011.02.015) [DOI] [Google Scholar]
- 54.Kostow KE. 2004. Differences in juvenile phenotypes and survival between hatchery stocks and a natural population provide evidence for modified selection due to captive breeding. Can. J. Fish. Aquat. Sci. 61, 577–589. ( 10.1139/f04-019) [DOI] [Google Scholar]
- 55.Gjerde B, Gjedrem T. 1984. Estimates of phenotypic and genetic parameters for carcass traits in Atlantic salmon and rainbow trout. Aquaculture 36, 97–110. ( 10.1016/0044-8486(84)90057-7) [DOI] [Google Scholar]
- 56.Bacon PJ, Palmer SCF, MacLean JC, Smith GW, Whyte BDM, Gurney WSC, Youngson AF. 2009. Empirical analyses of the length, weight, and condition of adult Atlantic salmon on return to the Scottish coast between 1963 and 2006. ICES J. Mar. Sci. 66, 844–859. ( 10.1093/icesjms/fsp096) [DOI] [Google Scholar]
- 57.Fraser DJ, Weir LK, Bernatchez L, Hansen MM, Taylor EB. 2011. Extent and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity 106, 404–420. ( 10.1038/hdy.2010.167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Houde ALS, Fraser DJ, O'Reilly P, Hutchings JA. 2011. Relative risks of inbreeding and outbreeding depression in the wild in endangered salmon. Evol. Appl. 4, 634–647. ( 10.1111/j.1752-4571.2011.00186.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bourret V, O'Reilly PT, Carr JW, Berg PR, Bernatchez L. 2011. Temporal change in genetic integrity suggests loss of local adaptation in a wild Atlantic salmon (Salmo salar) population following introgression by farmed escapees. Heredity 106, 500–510. ( 10.1038/hdy.2010.165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamaze FC, Sauvage C, Marie A, Garant D, Bernatchez L. 2012. Dynamics of introgressive hybridization assessed by SNP population genomics of coding genes in stocked brook charr (Salvelinus fontinalis). Mol. Ecol. 21, 2877–2895. ( 10.1111/j.1365-294X.2012.05579.x) [DOI] [PubMed] [Google Scholar]
- 61.Weeks AR, et al. 2011. Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol. Appl. 4, 709–725. ( 10.1111/j.1752-4571.2011.00192.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Facon B, Pointier J-P, Jarne P, Sarda V, David P. 2008. High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Curr. Biol. 18, 363–367. ( 10.1016/j.cub.2008.01.063) [DOI] [PubMed] [Google Scholar]
- 63.Lombaert E, Estoup A, Facon B, Joubard B, Grégoire JC, Jannin A, Blin A, Guillemaud T. 2014. Rapid increase in dispersal during range expansion in the invasive ladybird Harmonia axyridis. J. Evol. Biol. 27, 508–517. ( 10.1111/jeb.12316) [DOI] [PubMed] [Google Scholar]
- 64.Frankham R, Briscoe DA, Ballou JD. 2002. Introduction to conservation genetics. Cambridge, UK: Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Molecular and phenotypic data has been deposited in the Dryad repository (doi:10.5061/dryad.044g3).