Abstract
Metallothionein (MT), a cysteine-rich metal binding protein, is considered to play an essential role in the regulation of intracellular metals. Induction of MT in mammalian and nonmammalian tissues following heavy metal exposure may serve as a defense mechanism and a biomarker of environmental exposure to chemical stressors such as toxic metals. In this study, MT messenger RNA (mRNA) expression was characterized in male and female nonspawning and spawning killifish (Fundulus heteroclitus) following an 8-day exposure to specific sublethal stressors, which included temperature perturbation (26°C or 10°C) and/or 6 ppb of waterborne cadmium chloride (CdCl2). Hepatic, gill, and intestinal MT mRNA, expressed as copy number per microgram of total RNA, was assessed by reverse transcriptase–polymerase chain reaction and electrochemiluminescence using winter flounder (Pleuronectes americanus) MT complementary DNA primers. Liver, gill, and intestine MT mRNA expression was significantly (P < 0.05) increased in nonspawning killifish exposed to 26°C compared with those exposed to 19°C (control). In addition, a significant (P < 0.05) increase in gill MT mRNA induction was observed in nonspawning killifish exposed to 6 ppb of waterborne CdCl2 compared with controls. The results of this study demonstrate significant MT mRNA induction in nonspawning killifish following short-term exposure to physiological and chemical stressors. Thus, further research may be necessary before the use of killifish MT mRNA induction as a biomarker of environmental chemical stress exposure alone.
INTRODUCTION
Metallothioneins (MTs) are a family of low-molecular-weight cytosolic proteins that contain highly conserved cysteinyl residues. These residues allow MT to bind, transport, and store various transition metals via thiolate bonding (Hamer 1986; Dunn et al 1987). MT genes have been cloned and sequenced in several species of teleosts, including rainbow trout (Oncorhynchus mykiss), plaice (Pleuronectes platessa), winter flounder (Pleuronectes americanus), and stone loach (Noemacheilus barbatulus) (Bonham et al 1987; Chan et al 1989; Zafarullah et al 1989; Kille et al 1991). Although the specific physiological role of MT remains unclear (Palmiter 1998), MT has been proposed to have a significant role in regulating the intracellular availability of essential metals, such as zinc (Zn) and copper (Cu), and detoxification of poisonous metals such as cadmium (Cd) and mercury (Hg) (George et al 1990). MT is not only capable of donating Cu and Zn to metalloenzymes (Karin 1985; Brouwer and Brouwer-Hoexum 1991), but also donating metals, such as Zn, to transcription factors (Zeng et al 1991). The existence of specific metal-activated transcription factors for MT gene expression supports the hypothesis that MT induction by metals may be a specific cellular response to cellular metal concentrations (Thiele 1992). Metal sequestration may be a mechanism whereby MT confers organismal protection against toxicity from metals such as Cd (Klaassen et al 1999). Therefore, increased cellular resistance to metal toxicity may be conferred by processes that result in an increased capacity for MT synthesis (Roesijadi 1994, 1996).
Inductions of MT and/or MT mRNA in aquatic organisms by heavy metals have been considered potential biomarkers for environmental metal exposure (Olafson and Thompson 1974; Pruell and Engelhardt 1980; Stegeman et al 1992; Linde et al 1999; Cajaraville et al 2000). In addition, field studies demonstrate that MT concentrations in fish and shellfish correlate well with environmental levels of exposure to Cd, Zn, and Cu (Roch et al 1982; Weidow et al 1982; Engel and Brouwer 1984, 1987; Roch and McCarter 1984; Brown et al 1987; Hogstrand et al 1991; Hamza-Chaffai et al 2000; Wong et al 2000).
Because MT gene transcription may also be induced by factors such as glucocorticoids, inflammatory and cytotoxic agents, and general stress-producing conditions (Kagi 1991; Palmiter 1998), MT may be involved in generalized cellular stress responses (Overnell et al 1987; Jacob et al 1999). Very little is known about the stress-induced MT gene expression in the killifish (Fundulus heteroclitus). Knowledge concerning MT gene expression in killifish is particularly important given the recent use of these fish in laboratory studies to assess the toxic effects of environmental pollutants (Weis 1984; Kaplan et al 1995).
The killifish is a small, bottom-feeding teleost that is common in estuaries along the US Atlantic coast from the Gulf of St Lawrence to Florida (Crawford and Powers 1992). It adapts well to laboratory conditions and is found in both contaminated and pristine habitats (Weis and Weis 1989). Killifish can tolerate relatively high temperature extremes (up to 34°C) and a wide range of salinities. Killifish spawn between April and August and are widely used as experimental animals because of their hardiness (Atz 1986). Effects of salinity and temperature on Cd toxicity and metal tolerance in killifish have been addressed previously (Eisler 1971; Weis and Weis 1989). Although considered to be a relatively metal-tolerant species, killifish may still potentially serve as sentinel species in many estuaries contaminated with xenobiotics due to their ubiquitous distribution along the Atlantic coast of North America.
MT mRNA levels, quantitated by Northern blot (Kaplan et al 1995), reverse transcriptase–polymerase chain reaction (RT-PCR) (Schlenk et al 1997, 2000), or RT-PCR and electrochemiluminescence (ECL) (Jessen-Eller et al 1994; Jessen and Crivello 1998; Van Cleef et al 2000; Van Cleef-Toedt et al 2000) have been useful in mechanistic studies on gene regulation of the stress response in several teleosts (Ryan and Hightower 1996). In this study, RT-PCR with ECL was used to characterize liver, gill, and intestine MT mRNA expression in spawning and nonspawning killifish exposed to temperature perturbation and in nonspawning killifish exposed to both temperature perturbation and waterborne Cd. The results of these studies may contribute to an overall understanding of the complex biochemical mechanisms that occur following exposure to physiological and toxic heavy metal stressors in aquatic organisms.
MATERIALS AND METHODS
Animals
Adult male and female killifish were collected with a box trap from the Great South Bay (Nassau County, Long Island, NY, USA) during the 1995–1996 nonspawning and 1996 spawning seasons. Nonspawning animals were collected in September and spawning animals were collected in May. After collection, approximately 200 killifish were housed in 200-L, oval, plastic holding tanks equipped with constant aeration and carbon filtration (Magnum filters, Marineland, Moorpark, CA, USA). Fish were maintained in clean water for 4 weeks. Filtered tap water (pH 7.5–8.0) was treated with 0.1% sodium thiosulfate (50 mL, 10 L−1) and Instant Ocean (Aquarium Systems Inc, Mentor, OH, USA) (28 ppt) to ensure proper salinity. Animals were fed ad libitum with Tetra Marine (Tetra, Blacksburg, VA, USA) fish flakes during the 4-week period. All tanks were kept in temperature-controlled rooms at 19°C with light-dark cycles as follows: 14:10 hour (May) and 13:11 hour (September). The temperature and light-dark cycles resembled natural environmental conditions during the trapping periods.
Experimental design
Temperature challenge experiments
Following the 4-week holding period, subset groups of killifish were removed from the oval tanks, separated by sex (as determined by coloration and external markings), and placed in standard 20-L glass tanks (maximum of 10 fish per tank) for an 8-day period. Nonspawning (n = 24) and spawning (n = 24) control animals were maintained at 19°C. For both the nonspawning and spawning seasons, temperature stress included 10°C (n = 50, nonspawning; n = 40, spawning) or 26°C (n = 24, nonspawning; n = 24, spawning). Approximately equal numbers of male and female animals were used for all temperature experiments. During each 8-day exposure period, animals were maintained in static water with constant water aeration. Fish were fed once daily with fish flakes and debris subsequently siphoned. Salinity and light-dark cycle conditions were maintained as described in the above section.
Temperature challenge with waterborne CdCl2 experiments
During the nonspawning season, temperature challenge in addition to waterborne CdCl2 exposures were performed. Following the 4-week holding period, subset groups of killifish were removed from the oval tanks, separated by sex (as determined by coloration and external markings), and placed in standard 20-L glass tanks (maximum of 10 fish per tank). Killifish were then exposed to 6 ppb of waterborne CdCl2 (Sigma, St Louis, MO, USA) while maintained at 19°C (control; n = 22), 10°C (n = 40), or 26°C (n = 20) for an 8-day period. Approximately equal numbers of male and female animals were used for each exposure. During the 8-day exposure period, animals were maintained in static water with constant aeration. Fish were fed once daily with fish flakes and debris subsequently siphoned. Salinity and light-dark cycle conditions were maintained as described in the above section.
Following all exposure regimens, all fish were pithed. Total length (in centimeters), weight (in grams), sex, spawning condition, and gross pathological condition (fin rot, ichthyophthiriasis, dropsy) were noted. Liver, gill, and intestine tissues were removed from each animal, snap frozen in liquid nitrogen, and stored at −70°C for subsequent analysis.
RNA analysis using quantitative RT-PCR
MT mRNA was quantified with RT-PCR and the PerkinElmer (Norwalk, CT, USA) QPCR 5000. This method, detailed by Jessen-Eller et al (1994) and Van Cleef (1998), is briefly described below. Tissues stored at −70°C were thawed on ice and homogenized with a variable-speed tissue tearer in RNAzol reagent (guanidium-thyocyanate and phenol). Total RNA was extracted as described by Kreamer et al (1991). Total RNA concentration was determined by absorbance at 260 nm, and purity was determined by the 260/280-nm absorption ratio. Approximately 1 μg of total RNA was used in RT reactions with thermostable rTth enzyme (PerkinElmer) to generate complementary DNA (cDNA).
An aliquot of each cDNA sample was amplified via PCR with winter flounder MT-specific primers (Jessen-Eller et al 1994). These primers were previously shown to target MT in killifish cDNA templates (Van Cleef 1998). Biotin was attached to the 5′ end of one primer, whereas a trivalent oxidized form of ruthenium (tributyl ruthenium) was attached to the 3′ end of the other primer. Optimal PCR conditions to yield the desired 75-bp product were as follows: 94°C (30 seconds), 56°C (30 seconds), and 72°C (45 seconds) using 2.5 mM MgCl2, 0.3 μM primers, and 1 U Taq polymerase in a final reaction volume of 100 μL.
At time zero (cycle 0) and between cycles 16 and 30, 5-μL aliquots of each 100-μL reaction were sampled. The product in each aliquot was captured by streptavidin beads and analyzed via the QPCR 5000 (PerkinElmer) for chemiluminescence (DiCesare et al 1993). The PCR product was expressed as ECL units. Each unit is equivalent to 7.4 × 10−13 moles MT mRNA (Jessen-Eller et al 1994).
cDNA samples derived from liver, gill, and intestine were randomized within each 96-well PCR plate. MT mRNA was standardized to a plasmid MT cDNA for winter flounder. Each PCR plate included serial dilutions of this plasmid MT cDNA (data used to construct a standard curve), 1–5 replicates of each sample, and at least 5 control (blank) wells. Percentage of recovery of product was determined by spiking at least one replicate with the plasmid MT cDNA. MT mRNA (normalized to the plasmid MT cDNA) was expressed as starting copy number of MT per microgram of total RNA.
To confirm that the cDNA PCR product obtained using winter flounder primers (WFRu: 5′-TGC ACC ACC TGC AAC AAG AGC−3′; bases 73–93 and WF2-: 5′-GCA CAC GCA GCC AGA GG-3′; bases 131–147) during quantitative RT-PCR was killifish MT, the PCR product was cloned using the TA Cloning System (Invitrogen, Groningen, The Netherlands), and sequenced using the Cy5 AutoRead Sequencing System (Pharmacia, Peapack, NJ, USA) with the ALF DNA Sequencer (Pharmacia) (Van Cleef 1998).
Metal analysis
Water samples were taken from the exposure tanks both before and after each experiment for metal analysis (Cd). Metal analysis (Cd) was also conducted on water taken from the Great South Bay (Nassau County, NY, USA) near the location of the killifish collection site. These samples were acid treated, prepared according to US Environmental Protection Agency (EPA) method 7131A, and analyzed with a graphite furnace (GFAA).
Fish food flakes were prepared for metal analysis according to a scaled-down version (10 mL total volume) of the nitric acid/hydrogen peroxide EPA method 3050. Arsenic (As), Cd, and Cu were analyzed using GFAA (EPA methods 7060A, 7131A, and 7211, respectively), and Zn and nickel (Ni) were analyzed with an inductively coupled plasma spectrometer (EPA method SW 846–6010). The respective detection limits for all these metals were 1.0, 0.5, 1.0, 5.0, and 20.0 μg/L. Standard metal analysis quality controls were used to assess percentage of recovery and repeatability within and across experiments and were calibrated with Trace Metal AA Standard 9954.
Statistical analysis
Nonparametric data were analyzed across and within tissue groups using the Kruskal-Wallis Test (P < 0.05) and Tukey's Studentized Range Test (SAS System, Cary, NC, USA). Statistical analysis was based on the ranking of each fish liver, gill, and intestine MT mRNA copy number values within a comparison group. Mortality was compared between exposure groups using χ2 analysis (P < 0.05).
RESULTS
MT cDNA
Sequencing of the killifish cDNA-nested PCR product yielded a 75-bp fragment (Van Cleef 1998) that was more than 90% similar to other teleost MT cDNA sequences such as winter flounder (Chan et al 1989), plaice (George et al 1990), stone loach, northern pike (Esox lucius), rainbow trout MT-A (Kille et al 1991), goldfish (Carassius auratus), and tilapia (Tilapia mossambica) (Chan 1994) between base positions 73–150. Based on the size of the cDNA PCR product, similarity with other teleost MT cDNA sequences, and high cysteine content, the cDNA PCR product obtained using the ruthenium (WFRu) and biotin-labeled (WF2-) heterologous primers during the quantitative RT-PCR procedure was assumed to be killifish MT (Van Cleef 1998).
For all tissue types, duplicate RT-PCR samples within the same PCR plate using the same master mix averaged a 10% error as demonstrated by quantitative PCR. This variability increased to approximately 15% when duplicate samples were compared between PCR plates where a different master mix was used.
Animals
The average length and weight of all fish used in this study were 7.12 (±1.23) cm and 5.65 (±0.78) g, respectively. No significant differences in length and weight between nonspawning and spawning killifish were observed within any experimental group. Mature female and male spawning killifish maintained reproductive season colorations and markings throughout the experimental period. Both nonspawning and spawning control animals maintained at 19°C exhibited no mortality or morbidity during the 4-week holding period and during all 8-day experiments. For other experimental groups that exhibited mortality during the 8-day exposure period, MT mRNA expression was only characterized in those killifish that survived the 8-day period. No gross pathological condition was observed at the time of pithing. No significant differences in tissue MT mRNA expression between male and female killifish were observed within or between any control or exposure group (data not shown). Unless otherwise noted, male and female MT mRNA expression data were pooled for each category.
Temperature experiments
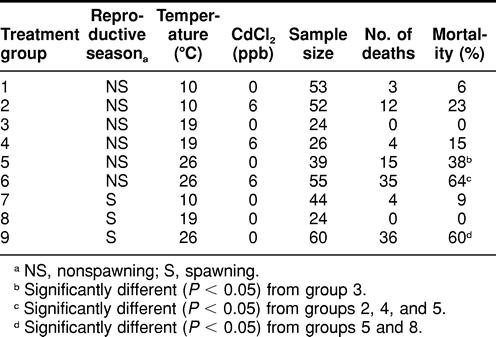
Spawning killifish exposed to 26°C exhibited significantly higher (P < 0.05) mortality following the 8-day exposure period compared with those maintained at 19°C (Table 1). Also, spawning animals exposed to 26°C demonstrated significantly higher (P < 0.05) mortality compared with nonspawning killifish housed at the same temperature. In addition, nonspawning killifish exposed to 26°C exhibited significantly higher (P < 0.05) mortality compared with those exposed to 19°C (control) (Table 1).
TABLE 1.
Mortality among spawning and nonspawning Fundulus heteroclitus during an 8-day simultaneous exposure to temperature perturbation and waterborne cadmium chloride (CdCl2)
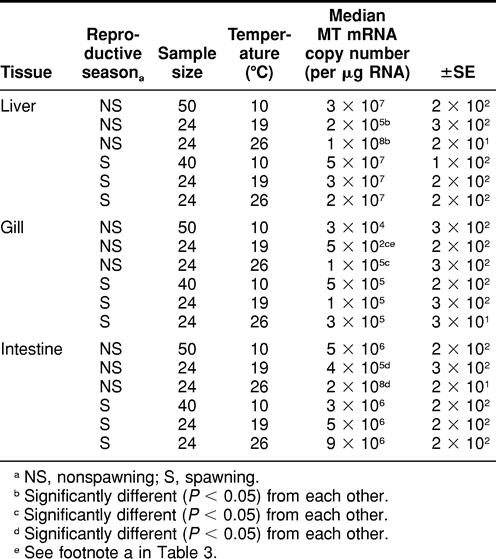
During the nonspawning season, killifish exposed to 26°C exhibited significantly higher (P < 0.05) MT mRNA expression in all 3 tissues analyzed compared with killifish exposed to 19°C (control) (Table 2). There were, however, no significant differences in liver, gill, or intestine MT mRNA expression between killifish exposed to 19°C (control) and those exposed to 10°C during the nonspawning and spawning seasons.
TABLE 2.
Spawning and nonspawning Fundulus heteroclitus metallothionein (MT) messenger RNA (mRNA) expression in liver, gill and intestine tissues among animals exposed to 8 days of temperature perturbation
Temperature challenge with waterborne CdCl2 experiments
Metal analysis of all 20-L exposure tanks confirmed the Cd exposure conditions to be 0 ppb of Cd (control tanks) and 6 ppb of Cd (waterborne CdCl2 tanks) before the addition of animals. Cd concentration in each tank was determined immediately following removal of animals after 8-day exposure; control tanks contained 0 ppb of Cd, and exposure tanks (previously containing 6 ppb of Cd) were found to contain 4 ppb of Cd. No Cd was detected following metal analysis of water samples from the killifish capture site (Great South Bay, Nassau County, NY, USA) or from food source.
Nonspawning killifish simultaneously exposed to 6 ppb of Cd and 26°C exhibited significantly higher (P < 0.05) mortality compared with those exposed to 26°C alone (Table 1). Also, nonspawning killifish exposed to 26°C in addition to 6 ppb of Cd exhibited significantly higher (P < 0.05) mortality compared with those exposed to 19°C with 6 ppb of Cd and to those exposed to 10°C with 6 ppb of Cd (Table 1).
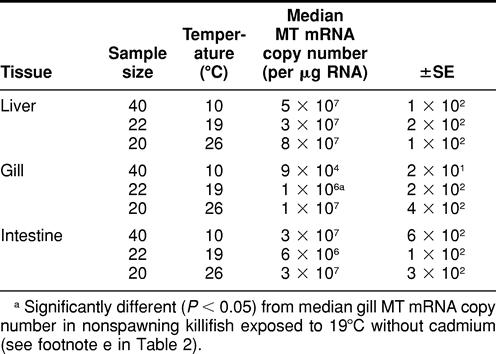
When killifish were maintained at 19°C, there was a significant difference observed in gill (not liver or intestine) MT mRNA expression between killifish exposed to 0 ppb of Cd (control) and those exposed to 6 ppb of Cd (Tables 2 and 3).
There were no significant differences in liver, gill, or intestine MT mRNA expression between animals exposed to heat stress (26°C) and those exposed to 26°C in addition to 6 ppb of Cd during the 8-day period (Tables 2 and 3). There were also no significant differences observed in liver, gill, or intestine MT mRNA expression between nonspawning killifish exposed to 10°C (cold stress) and those exposed to 10°C in addition to 6 ppb of waterborne Cd (Tables 2 and 3).
DISCUSSION
An understanding of the relationships between MT mRNA expression and exposure to exogenous stresses will undoubtedly contribute to improved strategies for use of MT in assessing aquatic organismal health. For this to be achieved, there needs to be a better understanding of MT function and the dynamics of MT mRNA induction (Roesijadi 1992). The presence of MT protein has been previously demonstrated in killifish (Pruell and Engelhardt 1980; Weis 1984). To the authors' knowledge, this is the first study to demonstrate MT mRNA expression in liver, gill, and intestine tissues of killifish following acute exposure to a physiological stress such as temperature perturbation.
The killifish has been found to be successful in surviving and reproducing in metal-polluted estuaries (Weis 1984). However, previous studies by Mitton and Koehn (1976) demonstrated specific morphological divergence in north shore populations of Long Island killifish exposed to thermal stress alone (compared with controls). Although synthesis of MT in these feral animals may be a response mechanism to heavy metal exposure within the aquatic environment, this study demonstrated that exposure to thermal stress alone results in a significant elevation in liver, gill, and intestine MT mRNA expression in killifish compared with controls during the nonspawning season.
Mammalian in vitro studies by Bauman et al (1993) using normal rat hepatocytes demonstrated that exposure to short-term heat shock (15–60 minutes at 43°C–44°C) did not increase MT protein. Although early in vitro studies with embryonic Chinook salmon (Oncorhynchus tshawytscha) and rainbow trout hepatoma cells also demonstrated that heat shock did not induce MT or MT mRNA (Heikkila et al 1982; Misra et al 1989), recent studies with yeast (Saccharomyces cerevisiae) have shown that MT gene expression may be activated through the action of heat shock factor and heat shock elements located within the yeast MT gene promoter upstream regulatory region following exposure to both heat and oxidative stress (Tamai et al 1994; Santoro et al 1998). Thus, MT biosynthesis is considered an important response to heat shock stress in yeast (Silar et al 1991). Bonneton et al (1996) recently demonstrated enhanced expression of 2 MT transgenes of Drosophila melanogaster (Mtn and Mto) after heat shock treatment. As suggested by Bauman et al (1993), there may be a relationship between conditions of heat shock and MT gene expression. The significant increase in MT mRNA expression observed in nonspawning killifish exposed to 8 days of heat stress (compared with controls) supports previous suggestions that MT may belong to the common group of heat shock or general stress proteins (Ryan and Hightower 1996). This proposed role of MT as a protein involved in the general heat shock response metabolic cascade is also supported by the observation that no difference in MT mRNA expression was observed in the same tissues when nonspawning killifish were exposed to cold stress (compared with controls).
The effect of heat stress was also demonstrated in this study by the significantly high mortality observed in nonspawning and spawning killifish exposed to heat stress compared with controls. The high mortality observed in spawning killifish exposed to heat stress compared with spawning controls may reflect various endogenous gonadal and hormonal fluctuations and adaptations occurring during teleost reproduction, such as sex steroid synthesis and increased plasma cortisol levels (Day and Taylor 1984). Previously, 100% mortality was observed in both nonspawning and spawning killifish exposed to 30°C for an 8-day period (Van Cleef 1998). These findings support past work that indicates that 26°C may be the highest temperature extreme for killifish maintained in aquaria (Hellner 1990).
An increase in murine MT mRNA has been shown to correlate well with a rise in physiological stress-induced serum corticosterone (Jacob et al 1999). Also, studies with largemouth bass (Micropterus salmoides) have confirmed that fish exposed to physiological stress display elevated serum cortisol, Cu, and Zn levels (Weber et al 1992). Following the observation that cortisol and corticosterone induced a 2-fold increase in MT levels of rainbow trout hepatocytes in primary culture, it was suggested that cortisol may be involved in the regulation of MT in rainbow trout (Hyllner et al 1989). In addition, Muto et al (1999) demonstrated that exposure to physical stress in crucian carp (Carassius cuvieri) may induce MT synthesis, in part, by the release of endogenous factors such as glucocorticoids. Although the MT gene has not been thoroughly characterized in killifish, recent structural and functional analysis of many teleost MT genes has demonstrated the presence of upstream metal responsive element (MRE) and glucocorticoid responsive element sequences that are similar to those observed for mammalian MT (Olsson et al 1995; Olsson and Kille 1997). In addition to mediating Zn induction of MT synthesis, MREs may make important contributions to nonmetal-induced promoter activity (Samson and Gedamu 1998).
MRE-binding transcription factor 1 (MTF-1) has been shown to play a central role in the transcriptional activation of specific mammalian and teleost MT genes through MRE binding in response to metals such as Zn and oxidative stress (Samson and Gedamu 1998; Andrews 2000). Expression of the pufferfish (Fugu rubripes) cDNA in mammalian cells by Auf der Maur et al (1999) showed that pufferfish MTF-1 had the same DNA-binding specificity as its mammalian counterpart. In addition, Dalton et al (2000) showed that both zebrafish (Danio rerio) and rainbow trout express MRE-specific binding activities immunologically similar to mammalian MTF-1. Interestingly, Zn reversibly modulated 1 of the 2 isoforms of MTF-1 in rainbow trout in a temperature-dependent manner (Dalton et al 2000). Future studies are needed to examine the 5′ upstream region(s) of the MT gene(s) in killifish to elucidate the events related to MT mRNA expression following exposure to physiological stress.
A significant increase in gill MT mRNA expression was observed in killifish exposed to Cd compared with controls. This result supports previous studies with teleosts that have demonstrated a significant increase in gill MT and MT mRNA expression following Cd exposure (Cosson 1994; George et al 1996). Interestingly, Cd, which has been shown to have little effect on the DNA-binding activity of mammalian and teleost MTF-1 in vivo or in vitro, is often considered a more potent inducer of MT gene expression than Zn (Chu et al 1999; Andrews 2000; Dalton et al 2000).
The effects of multiple environmental stressors on MT expression in fish have been unclear (Thomas and Wofford 1984; Olsson et al 1987; Overnell et al 1987). Previous studies have demonstrated a temperature dependence of hepatic MT synthesis in fish exposed to Cd, such that Cd sequestration by MT increased in fish maintained at 20°C compared with those reared at 10°C (Carpene et al 1992). Also, the accumulation of Cd has been shown to be a temperature-dependent process (Failla et al 1979). However, the results of the current study demonstrated that nonspawning killifish exposed to thermal stress alone exhibited significantly elevated liver, gill, and intestine MT mRNA expression compared with controls. Thus, exposure to a low dose of an additional stress (6 ppb of Cd) did not result in further significant MT mRNA expression when comparing nonspawning killifish exposed to heat stress. The results of the current study also showed no significant increase in liver, gill, or intestine MT mRNA expression in nonspawning killifish exposed to cold stress compared with those exposed to cold stress and Cd. As the temperature is lowered, the fluidity of plasma membranes has been shown to decrease (Krasne et al 1971). This process could then affect ion channel permeability of Cd into tissues and subsequent MT mRNA induction at lower temperatures.
To identify relationships between physiological stressors and MT mRNA expression, characterization of a model system using a sentinel species must be performed with an evaluation of specific inducers. For utilization in potential environmental monitoring projects, RT-PCR has been shown to provide the ability to sensitively and accurately quantitate levels of MT mRNA expression in target tissues of killifish and various other aquatic species (Jessen-Eller et al 1994; Kaplan et al 1995; Schlenk et al 1997, 2000; Jessen-Eller and Crivello 1998; Van Cleef et al 2000; Van Cleef-Toedt et al 2000). Because the effects of heat stress on MT mRNA expression in this aquatic organism may possibly serve to confound the analysis of MT mRNA expression in killifish sampled from contaminated aquatic ecosystems, further analyses are required before the use of MT mRNA induction in killifish as a biomarker of heavy metal exposure alone.
TABLE 3.
Metallothionein (MT) messenger RNA (mRNA) expression in liver, gill, and intestine tissues in nonspawning Fundulus heteroclitus exposed to 8 days of temperature perturbation and 6 ppb of waterborne cadmium chloride (CdCl2)
Acknowledgments
This study was supported by the 1997 Gladys Mateyko Award (New York University, Department of Biology) and grants from Connecticut Innovations Inc, University of Connecticut Biotechnology Center, and the Environmental Research Institute at the University of Connecticut. K. Chan of the Chinese University, Hong Kong, kindly donated the MT cDNA used to optimize the RT-PCR procedure. The authors wish to thank C. Schlichting, L. Fumal, A. Moiseff, E. Lechowicz, K. Jessen-Eller, and S. Gerdes for all their assistance and advice.
REFERENCES
- Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- Atz JW. Fundulus heteroclitus in the laboratory: a history. Am Zool. 1986;26:111–120. [Google Scholar]
- Auf der Maur A, Belser T, Elgar G, Georgiev O, Schaffner W. Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol Chem. 1999;380:175–185. doi: 10.1515/BC.1999.026. [DOI] [PubMed] [Google Scholar]
- Bauman JW, Liu J, Klaassen CD. Production of metallothionein and heat-shock proteins in response to metals. Fundam Appl Toxicol. 1993;21:15–22. doi: 10.1006/faat.1993.1066. [DOI] [PubMed] [Google Scholar]
- Bonham K, Zafarullah M, Gedamu L. The rainbow trout metallothioneins: molecular cloning and characterization of two distinct cDNA sequences. DNA. 1987;6:519–528. doi: 10.1089/dna.1987.6.519. [DOI] [PubMed] [Google Scholar]
- Bonneton F, Theodore L, Silar P, Maroni G, Wegnez M. Response of Drosophila metallothionein promoters to metallic, heat shock and oxidative stress. FEBS Lett. 1996;380:33–38. doi: 10.1016/0014-5793(95)01544-2. [DOI] [PubMed] [Google Scholar]
- Brouwer M, Brouwer-Hoexum T. Interaction of copper—metallothionein from the American lobster, Homarus americanus, with glutathione. Arch Biochem Biophys. 1991;290:207–213. doi: 10.1016/0003-9861(91)90610-u. [DOI] [PubMed] [Google Scholar]
- Brown MW, Shurben D, Solbe JF, Cryer A, Kay J. Sequestration of environmental cadmium by metallothionein in the roach (Rutilus rutilus) and the stone loach (Noemacheilus barbatulus) Comp Biochem Physiol C. 1987;87:65–69. doi: 10.1016/0742-8413(87)90182-4. [DOI] [PubMed] [Google Scholar]
- Cajaraville MP, Bebianno MJ, Blasco J, Porte C, Sarasquete C, Viarengo A. The use of biomarkers to assess the impact of pollution in coastal environments of the Iberian Peninsula: a practical approach. Sci Total Environ. 2000;247:295–311. doi: 10.1016/s0048-9697(99)00499-4. [DOI] [PubMed] [Google Scholar]
- Carpene E, Camatti A, Isani G, Cattani O, Cortesi P. Cd-metallothionein in liver and kidney of goldfish (Carassius auratus): effects of temperature and salinity. Ital J Biochem. 1992;41:273–282. [PubMed] [Google Scholar]
- Chan KM. PCR-cloning of goldfish and tilapia metallothionein complementary DNAs. Biochem Biophys Res Comm. 1994;205:368–374. doi: 10.1006/bbrc.1994.2674. [DOI] [PubMed] [Google Scholar]
- Chan KM, Davidson WS, Hew CL, Fletcher GL. Metallothionein cDNA production of a cRNA probe to detect metallothionein gene expression in winter flounder. Can J Zool. 1989;67:2520–2527. [Google Scholar]
- Chu WA, Moehlenkamp JD, Bittel D, Andrews GK, Johnson JA. Cadmium-mediated activation of the metal response element in human neuroblastoma cells lacking functional metal response element-binding transcription factor-1. J Biol Chem. 1999;274:5279–5284. doi: 10.1074/jbc.274.9.5279. [DOI] [PubMed] [Google Scholar]
- Cosson RP. Heavy metal intracellular balance and relationship with metallothionein induction in the gills of carp: after contamination by Ag, Cd, and Hg following pretreatment with Zn or not. Biol Trace Elem Res. 1994;46:229–245. doi: 10.1007/BF02789299. [DOI] [PubMed] [Google Scholar]
- Crawford DL, Powers DA. Evolutionary adaptation to different thermal environments via transcriptional regulation. Mol Biol Evol. 1992;9:806–813. doi: 10.1093/oxfordjournals.molbev.a040762. [DOI] [PubMed] [Google Scholar]
- Dalton TP, Solis WA, Nebert DW, Carvan MJ III.. Characterization of the MTF-1 transcription factor from zebrafish and trout cells. Comp Biochem Physiol B. 2000;126:325–335. doi: 10.1016/s0305-0491(00)00182-6. [DOI] [PubMed] [Google Scholar]
- Day JR, Taylor MH. Photoperiod and temperature interaction in the seasonal reproduction of female mummichogs. Trans Am Fish Soc. 1984;113:452–457. [Google Scholar]
- DiCesare J, Grossman B, Katz E, Picozza E, Ragusa R, Woudenberg T. A high-sensitivity electrochemiluminescence-based detection system for automated PCR production quantitation. Biotechniques. 1993;15:152–157. [PubMed] [Google Scholar]
- Dunn MA, Blalock TL, Cousins RJ. Metallothionein. Proc Soc Exp Biol Med. 1987;185:107–119. doi: 10.3181/00379727-185-42525a. [DOI] [PubMed] [Google Scholar]
- Eisler R. Cadmium poisoning in Fundulus heteroclitus (Cyprinodontidae) and other marine organisms. J Fish Res Board Can. 1971;28:1225–1234. [Google Scholar]
- Engel DW, Brouwer M. Cadmium-binding proteins in the blue crab, Callinectes sapidus: laboratory and field comparison. Mar Environ Res. 1984;14:139–151. [Google Scholar]
- Engel DW, Brouwer M. Metal regulation and molting in the blue crab, Callinectes sapidus, metallothionein function in metal metabolism. Biol Bull. 1987;173:239–251. doi: 10.2307/1541876. [DOI] [PubMed] [Google Scholar]
- Failla ML, Cousins RJ, Mascenik MJ. Cadmium accumulation and metabolism by rat liver parenchyma cells in primary monolayer culture. Biochim Biophys Acta. 1979;583:63–72. doi: 10.1016/0304-4165(79)90310-6. [DOI] [PubMed] [Google Scholar]
- George S, Leaver M, Frerichs N, Burgess D. Fish metallothioneins: molecular cloning studies and induction in cultured cells. Mar Environ Res. 1990;28:173–177. [Google Scholar]
- George SG, Todd K, Wright J. Regulation of metallothionein in teleosts: induction of MT mRNA and protein by cadmium in hepatic and extrahepatic tissues of a marine flatfish, the turbot (Scophthalmus maximus) Comp Biochem Physiol C. 1996;113:109–115. doi: 10.1016/0742-8413(95)02076-4. [DOI] [PubMed] [Google Scholar]
- Hamer DH. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- Hamza-Chaffai A, Amiard JC, Pellerin J, Joux L, Berthet B. The potential use of metallothionein in the clam Ruditapes decussatus as a biomarker of in situ metal exposure. Comp Biochem Physiol C. 2000;127:185–197. doi: 10.1016/s0742-8413(00)00147-x. [DOI] [PubMed] [Google Scholar]
- Heikkila JJ, Schultz GA, Iatrou K, Gedamu L. Expression of a set of fish genes following heat or metal ion exposure. J Biol Chem. 1982;257:12000–12005. [PubMed] [Google Scholar]
- Hellner S 1990 Killifish. Barron's Educational Series Inc, New York, 10–20. [Google Scholar]
- Hogstrand C, Lithner G, Haux C. The importance of metallothionein for the accumulation of copper, zinc and cadmium in environmentally exposed perch, Perca fluviatilis. Pharmacol Toxicol. 1991;68:492–501. doi: 10.1111/j.1600-0773.1991.tb01275.x. [DOI] [PubMed] [Google Scholar]
- Hyllner SJ, Andersson T, Haux C, Olsson P-E. Cortisol induction of metallothionein in primary culture of rainbow trout hepatocytes. J Cell Physiol. 1989;139:24–38. doi: 10.1002/jcp.1041390105. [DOI] [PubMed] [Google Scholar]
- Jacob ST, Ghoshal K, Sheridan JF. Induction of metallothionein by stress and its molecular mechanisms. Gene Expr. 1999;7:301–10. [PMC free article] [PubMed] [Google Scholar]
- Jessen-Eller K, Crivello JF. Subcutaneous NaAs3+ exposure increases metallothionein mRNA and protein expression in juvenile winter flounder. Aquat Toxicol. 1998;42:301–320. [Google Scholar]
- Jessen-Eller K, Picozza E, Crivello JF. Quantitation of metallothionein mRNA by RT-PCR and chemiluminescence. Biotechniques. 1994;17:962–973. [PubMed] [Google Scholar]
- Kagi JHR 1991 Overview of metallothionein. In: Methods in Enzymology: Metallobiochemistry (B), ed Riordan JF, Vallee BL. Academic Press, San Diego, 613–626. [DOI] [PubMed] [Google Scholar]
- Kaplan LAE, Van Cleef K, Wirgin I, Crivello JF. A comparison of RT-PCR and Northern blot analysis in quantifying metallothionein mRNA levels in killifish exposed to waterborne cadmium. Mar Environ Res. 1995;39:137–141. [Google Scholar]
- Karin M. Metallothioneins: proteins in search of function. Cell. 1985;41:9–10. doi: 10.1016/0092-8674(85)90051-0. [DOI] [PubMed] [Google Scholar]
- Kille P, Stephens PE, Kay J. Elucidation of cDNA sequences for metallothioneins from rainbow trout, stone loach and pike liver using the polymerase chain reaction. Biophys Acta. 1991;1089:407–410. doi: 10.1016/0167-4781(91)90187-q. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Liu J, Choudhuri S. Metallothionein: an intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol. 1999;39:267–294. doi: 10.1146/annurev.pharmtox.39.1.267. [DOI] [PubMed] [Google Scholar]
- Krasne S, Eisenman G, Szabo G. Freezing and melting of lipid bilayers and the mode of action of nonactin, valinomycin, and gramicidin. Science. 1971;174:412–415. doi: 10.1126/science.174.4007.412. [DOI] [PubMed] [Google Scholar]
- Kreamer GL, Squibb K, Gioeli D, Garte SJ, Wirgin I. Cytochrome P450IA mRNA expression in feral Hudson River tomcod. Environ Res. 1991;55:64–78. doi: 10.1016/s0013-9351(05)80141-0. [DOI] [PubMed] [Google Scholar]
- Linde AR, Sanchez-Galan S, Klein D, Garcia-Vazquez E, Summer KH. Metallothionein and heavy metals in brown trout (Salmo trutta) and European eel (Anguilla anguilla): a comparative study. Ecotoxicol Environ Saf. 1999;44:168–73. doi: 10.1006/eesa.1999.1819. [DOI] [PubMed] [Google Scholar]
- Misra S, Zafarullah M, Price-Haughey J, Gedamu L. Analysis of stress-induced gene expression in fish cell lines exposed to heavy metals and heat shock. Biochim Biophys Acta. 1989;1007:325–333. doi: 10.1016/0167-4781(89)90155-3. [DOI] [PubMed] [Google Scholar]
- Mitton JB, Koehn RK. Morphological adaptation to thermal stress in a marine fish, Fundulus heteroclitus. Biol Bull. 1976;151:548–549. doi: 10.2307/1540505. [DOI] [PubMed] [Google Scholar]
- Muto N, Ren HW, Hwang GS, Tominaga S, Itoh N, Tanaka K. Induction of two major isoforms of metallothionein in crucian carp (Carassius cuvieri) by air-pumping stress, dexamethasone, and metals. Comp Biochem Physiol C. 1999;122:75–82. doi: 10.1016/s0742-8413(98)10081-6. [DOI] [PubMed] [Google Scholar]
- Olafson RW, Thompson JAJ. Isolation of heavy metal binding proteins from marine vertebrates. Mar Biol. 1974;28:83–86. [Google Scholar]
- Olsson P-E, Kille P. Functional comparison of the metal-regulated transcriptional control regions of metallothionein genes from cadmium-sensitive and tolerant fish species. Biochim Biophys Acta. 1997;1350:325–334. doi: 10.1016/s0167-4781(96)00173-x. [DOI] [PubMed] [Google Scholar]
- Olsson P-E, Haux C, Forlin L. Variations in hepatic metallothionein, zinc and copper levels during an annual reproductive cycle in rainbow trout, Salmo gairdneri. Fish Physiol Biochem. 1987;3:39–47. doi: 10.1007/BF02183992. [DOI] [PubMed] [Google Scholar]
- Olsson P-E, Kling P, Erkell LJ, Kille P. Structural and functional analysis of the rainbow trout (Oncorhynchus mykiss) metallothionein-A gene. Eur J Biochem. 1995;230:344–349. doi: 10.1111/j.1432-1033.1995.tb20569.x. [DOI] [PubMed] [Google Scholar]
- Overnell J, McIntosh R, Fletcher TC. The enhanced induction of metallothionein by zinc, its half-life in the marine fish Pleuronectes platessa, and the influence of stress factors on metallothionein levels. Experimentia. 1987;43:178–181. doi: 10.1007/BF01942842. [DOI] [PubMed] [Google Scholar]
- Palmiter RD. The elusive function of metallothionein. Proc Natl Acad Sci U S A. 1998;95:8428–8430. doi: 10.1073/pnas.95.15.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruell RJ, Engelhardt FR. Liver cadmium uptake, catalase inhibition and cadmium thionein production in the killifish (Fundulus heteroclitus) induced by experimental exposure. Mar Environ Res. 1980;3:101–111. [Google Scholar]
- Roch M, McCarter JA. Hepatic metallothionein production and resistance to heavy metals by rainbow trout (Salmo gairdneri) II: held in a series of contaminated lakes. Comp Biochem Physiol. 1984;77:77–82. doi: 10.1016/0742-8413(84)90133-6. [DOI] [PubMed] [Google Scholar]
- Roch M, McCarter JA, Matheson M, Clark M, Olafson RW. Hepatic metallothionein in rainbow trout (Salmo gairdneri) as an indicator of metal pollution in the Campell River system. Can J Fish Aquat Sci. 1982;39:1596–1601. [Google Scholar]
- Roesijadi G. Metallothioneins in metal regulation and toxicity in aquatic animals. Aquat Toxicol. 1992;22:81–113. [Google Scholar]
- Roesijadi G. Metallothionein induction as a measure of response to metal exposure in aquatic animals. Environ Health Perspect. 1994;102:91–95. doi: 10.1289/ehp.94102s1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesijadi G. Metallothionein and its role in toxic metal regulation. Comp Biochem Physiol. 1996;113:117–123. [Google Scholar]
- Ryan JA, Hightower LE 1996 Stress proteins as molecular biomarkers for environmental toxicology. In: Stress-Inducible Cellular Responses, ed Friege U, Morimoto RI, Yahara I, Polla B. Birkhauser-Verlag, Basel, Switzerland, 411–424. [DOI] [PubMed] [Google Scholar]
- Samson SL, Gedamu L. Molecular analyses of metallothionein gene regulation. Prog Nucleic Acids Res Mol Biol. 1998;59:257–288. doi: 10.1016/s0079-6603(08)61034-x. [DOI] [PubMed] [Google Scholar]
- Santoro N, Johansson N, Thiele DJ. Heat shock architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol. 1998;18:6340–6352. doi: 10.1128/mcb.18.11.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenk D, Chelius M, Wolford L, Khan S, Chan KM. Characterization of hepatic metallothionein expression in channel catfish (Ictalurus punctatus) by reverse-transcriptase polymerase chain reaction. Biomarkers. 1997;2:161–167. doi: 10.1080/135475097231698. [DOI] [PubMed] [Google Scholar]
- Schlenk D, Colley WC, El-Alfy A, Kirby R, Griffin BR. Effects of the oxidant potassium permaganate on the expression of gill metallothionein mRNA and its relationship to sublethal whole animal endpoints in channel catfish. Toxicol Sci. 2000;54:177–182. doi: 10.1093/toxsci/54.1.177. [DOI] [PubMed] [Google Scholar]
- Silar P, Butler G, Thiele DJ. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Mol Cell Biol. 1991;11:1232–1238. doi: 10.1128/mcb.11.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman JJ, Brouwer M, DiGiulio RT, Forlin L, Fowler B, Sanders BM, and Van Veld PA 1992 Molecular responses to environmental contamination: enzyme and protein systems as indicators of chemical exposure and effect. In: Biomarkers: Biochemical, Physiological, and Histological Markers of Anthropogenic Stress, ed Huggett RJ, Kimerle RA, Mehrle PM, Bergman HL. Lewis Publishers, Chelsea, NC, 235–335. [Google Scholar]
- Tamai KT, Liu X, Silar P, Sosinowski T, Thiele DJ. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signaling pathways. Mol Cell Biol. 1994;14:8155–8165. doi: 10.1128/mcb.14.12.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele DJ. Metal-regulated transcription in eukaryotes. Nucleic Acids Res. 1992;20:1183–1191. doi: 10.1093/nar/20.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Wofford HW. Effects of metals and organic compounds on hepatic glutathione, cysteine, and acid-soluble thiol levels in mullet (Mugil cephalus) Toxicol Appl Pharmacol. 1984;76:172–182. doi: 10.1016/0041-008x(84)90040-1. [DOI] [PubMed] [Google Scholar]
- Van Cleef K 1998 Induction and modulation of metallothionein mRNA in Fundulus heteroclitus. PhD Thesis, New York University, Department of Biology, New York, 25–65. [Google Scholar]
- Van Cleef K, Kaplan LAE, Crivello JF. The relationship between reproductive status and metallothionein mRNA expression in the common killifish, Fundulus heteroclitus. Environ Biol Fishes. 2000;57:97–105. [Google Scholar]
- Van Cleef-Toedt K, Kaplan LAE, Crivello JF. Metallothionein mRNA expression in spawning and non-spawning Fundulus heteroclitus following acute exposure to starvation and waterborne cadmium. Fish Physiol Biochem. 2000;22:319–327. [Google Scholar]
- Weber DN, Eisch S, Spieler RE, Petering DH. Metal redistribution in largemouth bass (Micropterus salmoides) in response to restrainment stress and dietary cadmium: role of metallothionein and other metal-binding proteins. Comp Biochem Physiol C. 1992;101:255–262. doi: 10.1016/0742-8413(92)90270-h. [DOI] [PubMed] [Google Scholar]
- Weidow MA, Kneip TJ, Garte SJ. Cadmium-binding protein from blue crabs (Callinectes sapidus) environmentally exposed to cadmium. Environ Res. 1982;28:164–170. doi: 10.1016/0013-9351(82)90165-7. [DOI] [PubMed] [Google Scholar]
- Weis P. Metallothionein and mercury tolerance in the killifish, Fundulus heteroclitus. Mar Environ Res. 1984;14:153–166. [Google Scholar]
- Weis JS, Weis P. Tolerance and stress in a polluted environment. BioScience. 1989;39:89–95. [Google Scholar]
- Wong CK, Yeung HY, Cheung RY, Yung KK, Wong MH. Ecotoxicological assessment of persistent organic and heavy metal contamination in Hong Kong coastal sediment. Arch Environ Contam Toxicol. 2000;38:486–493. doi: 10.1007/s002449910064. [DOI] [PubMed] [Google Scholar]
- Zafarullah M, Olsson P, and Gedamu L 1989 Rainbow trout metallothionein gene structure and regulation. In: Oxford Survey of Eukaryotic Genes, ed Maclean N. Oxford University Press, Oxford, 112–143. [PubMed] [Google Scholar]
- Zeng J, Vallee BL, Kagi JHR. Zinc transfer from transcription factor IIIA fingers to thionein clusters. Proc Natl Acad Sci U S A. 1991;88:9984–9988. doi: 10.1073/pnas.88.22.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]


