Abstract
Cells have evolved exquisite mechanisms to fine-tune the rate of protein synthesis in response to stress. Systemic mapping of start codon positions and precise measurement of the corresponding initiation rate would transform our understanding of translational control. Here we present quantitative translation initiation sequencing (QTI-seq), where the initiating ribosomes can be profiled in real time at single nucleotide resolution. The resultant initiation map not only delineates variations of start codon selection, but also highlights a dynamic range of initiation rates in response to nutrient starvation. The integrated data set provides unique insights into principles of alternative translation and mechanisms controlling different aspects of translation initiation. Using RiboTag mice, QTI-seq permits tissue-specific profiling of initiating ribosomes in vivo. Liver cell-specific ribosome profiling uncovers a robust translational reprogramming of the proteasome system in fasted mice. Our findings illuminate the prevalence and dynamic nature of translational regulation pivotal to physiological adaptation in vivo.
Introduction
Translational regulation permits cells to respond swiftly to stress conditions via immediate and selective changes in protein levels1-3. Much of this translational control occurs at the initiation stage4-7. Despite long appreciation of the importance of translation initiation, direct measurement of the initiation rate on individual mRNAs has proven to be difficult. Recent development of ribosome profiling technology, based on deep sequencing of ribosome-protected mRNA fragments (RPFs), enables monitoring of ribosome dynamics with unprecedented resolution at the genome-wide scale8,9. The obtained snapshot of ribosome occupancy on the coding region has often been used to estimate relative changes of translation efficiency under different growth conditions. A caveat, however, exists because the average ribosome density on mRNAs is negatively influenced by the elongation speed10,11. Dynamic changes of initiation rates are thus masked by the varied elongation speed under different growth conditions. In addition, the average density of ribosome occupancy is not suitable to evaluate alternative translation events occurring on the same mRNA12. These constraints point to a need for a method capable of mapping start codon selection and quantifying the rate of 80S ribosome assembly at individual translation initiation sites.
Much of our understanding of the basic mechanisms of translation initiation has limited to cells in culture. Investigating translational control at the organismal level remains to be a pressing and formidable challenge. The cellular heterogeneity of tissues and organs confounds our efforts to achieve cell-type-specific genomic and proteomic interrogation. To monitor protein synthesis at the organismal level, one approach relies on tagged ribosomal proteins, such as HA-tagged Rpl2213 or EGFP-fused Rpl10a14. Following genetic targeting to specific cell populations, affinity purification allows isolation of polysome-bound mRNAs from specific cell types suitable for profiling assays. However, current methods do not allow tissue-specific profiling of initiating ribosomes, which bear more reliable information in evaluating translational control in vivo.
Here we present an approach called quantitative translation initiation sequencing (QTI-seq) that allows comprehensive monitoring of translation initiation in cells and solid tissues. Together with the RNA-seq and Ribo-seq data sets acquired in parallel, the integrated ribosome profiling data afford a holistic view of the translational landscape under different growth conditions.
Results
QTI-seq captures initiating ribosomes in an unbiased manner
We previously developed global translation initiation sequencing (GTI-seq)15, which capitalizes on the unique feature of lactimidomycin (LTM) that preferentially acts on the first 80S ribosome when its E-site is empty16. However, LTM-mediated enrichment of initiating ribosomes requires an incubation of the cell at 37°C to allow elongating ribosomes to run off. This procedure, albeit essential for high resolution mapping of translation initiation sites (TIS), may generate a number of artifacts. First, free ribosomes could start new rounds of initiation during the incubation period, resulting in an amplification of ribosome occupancy at the start codon (Supplementary Fig. 1a). Second, the amplified ribosome density is biased towards the upstream initiators, causing an unwanted 5′ end inflation of ribosome protected mRNA fragments (RPFs). When GTI-seq was applied to HEK293 cells with amino acid deprivation, the LTM peaks at the annotated start codons (aTIS) showed little changes despite the pervasive reduction of ribosome occupancy on many transcripts (Supplementary Fig. 1b). As a result, virtually no correlation was found between the differences of LTM-captured aTIS density and the ribosome occupancy on the coding region (CDS) (Supplementary Fig. 1c). The lack of quantitative features in GTI-seq largely hampers comparative interrogation of translational regulation.
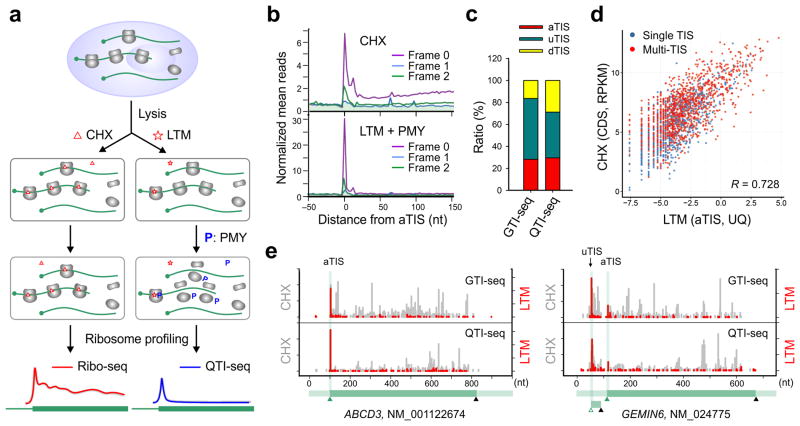
To circumvent this limitation, we set out to devise a new approach in order to preserve the small population of initiating ribosomes with minimal perturbation. Following extensive optimization, we implemented three steps to achieve this goal: (1) rapidly break down cells using Matrix-D that has a minimal effect on ribosome stability; (2) specifically freeze the initiating ribosomes by treating cell lysates with LTM; and (3) effectively deplete the elongating ribosomes by introducing another translation inhibitor puromycin (PMY) (Fig. 1a). PMY does not affect translation initiation and has been used previously to enrich the initiating ribosomes17. Acting as a tRNA analog, PMY releases the nascent chain and dissociates the ribosome into separate subunits18. In the presence of elongation inhibitors like CHX, however, addition of PMY catalyzes puromycylation of the nascent chain without triggering ribosome dissociation19. The same feature holds true for cell-free samples as the presence of CHX completely prevented PMY-induced polysome disassembly (Supplementary Fig. 2). Since LTM uses the similar mechanism as CHX but selectively acts on the first 80S ribosome16, we reasoned that sequential treatment with LTM and PMY would dissociate the elongating ribosomes while leaving the initiating ribosomes insensitive to PMY (Fig. 1a). Indeed, PMY addition in the presence of LTM resulted in polysome disassembly with a corresponding increase of the monosome (Supplementary Fig. 2). To examine whether the increased monosome contains the preserved initiating ribosomes, we purified RPFs followed by deep sequencing. Metagene analysis revealed that the LTM-associated RPF reads were highly enriched at the aTIS codon (Fig. 1b). Additionally, the aTIS ribosome density captured by LTM was highly reproducible (Supplementary Fig. 3). We termed this approach as quantitative translation initiation sequencing (QTI-seq).
Figure 1. QTI-seq captures real-time translation initiation events in a qualitative and quantitative manner.
(a) Schematic of Ribo-seq (left panel) and QTI-seq (right panel) procedures. The red line represents the ribosome density on average based on regular Ribo-seq, whereas the blue line indicates the density of initiating ribosome obtained from QTI-seq.
(b) Meta-gene analysis of CHX-associated ribosome density (top panel) and LTM-associated ribosome density (bottom panel) in HEK293 cells captured by QTI-seq. Normalized RPF reads are averaged across the entire transcriptome and aligned at the annotated start codon. Different reading frames are separated and color coded.
(c) A stacked bar plot showing the relative ratio of different types of TIS identified by GTI-seq or QTI-seq in HEK293 cells.
(d) A scatter plot showing the correlation between LTM-associated aTIS density normalized by upper quartile and CHX-associated CDS ribosome occupancy normalized by RPKM. Genes with single annotated TIS or multiple TISs are shown in blue and red dots respectively.
(e) Examples of single TIS (ABCD3) and multiple TIS (GEMIN6) genes revealed by GTI-seq (top panel) and QTI-seq (bottom panel). The same scale is used for Y-axis. The corresponding gene structure is shown below the X-axis.
Since QTI-seq captures a small population of initiating ribosomes without 5′ end RPF inflation, QTI-seq identified fewer total TISs than GTI-seq in HEK293 cells (5,099 vs. 13,915), especially the upstream TIS codons (uTISs) (Fig. 1c). Consistent with previous reports15,20, codon composition analysis revealed that more than half the TISs used non-AUG as the translation initiator (Supplementary Fig. 4). The associated LTM peak height at the aTISs sites showed a strong correlation with the average ribosome occupancy along the corresponding CDS (r = 0.728, Fig. 1d). In addition to this quantitative feature, QTI-seq retained the high precision in mapping TIS positions at a single nucleotide resolution. For instance, a prominent LTM peak was located exactly at the annotated start codon of ABCD3; and an out-of-frame uTIS was uncovered in GEMIN6 (Fig. 1e). QTI-seq thus offers a promising approach to exploring real-time translation initiation in a qualitative and quantitative manner.
Quantitative TIS profile in response to starvation
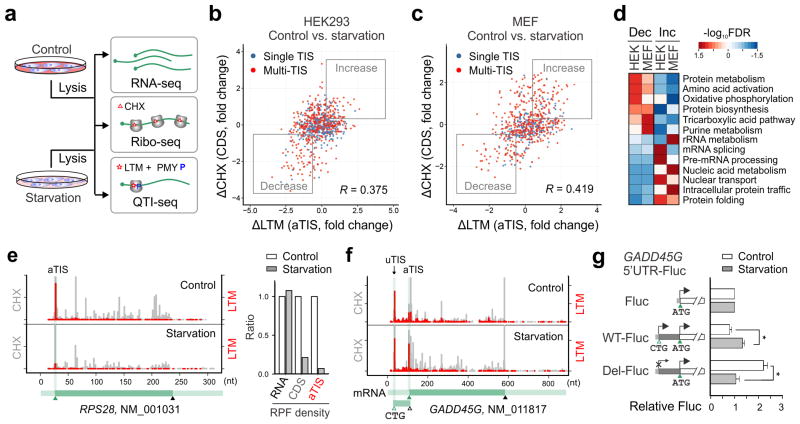
We next applied QTI-seq to HEK293 cells with amino acid deprivation (Supplementary Fig. 5). Total cellular RNA was also collected in parallel for RNA-seq to quantify mRNA abundance. In response to starvation, the changes in the initiation rates showed positive correlation with the differences of ribosome occupancy on the corresponding CDS (r = 0.375, Fig. 2b). The same observation holds true for a mouse embryonic fibroblast (MEF) cell line subjected to starvation (r = 0.419, Fig. 2c). The imperfect correlation is partially due to reduced elongation speed under nutrient starvation that potentially increases the CDS ribosome occupancy (Supplementary Fig. 6). From the comprehensive datasets acquired from QTI-seq and Ribo-seq, we identified a large number of transcripts that undergo 2-fold changes upon starvation (1,073 in HEK293 and 820 in MEF, Supplementary Table 1 and 2). Among the genes showing repressed translation, many of them are involved in protein biosynthesis and metabolism (Fig. 2d). As a typical example, the gene encoding ribosomal protein RPS28 showed a nearly 5-fold decrease in ribosome occupancy on the CDS in response to starvation (Fig. 2e). Remarkably, QTI-seq displayed a greater than 14-fold decrease in the ribosome density at the start codon of RPS28. Without the negative influence imposed by elongation speed variation, QTI-seq sets apart from previous methods by reporting true measures of initiation rates.
Figure 2. QTI-seq reveals pervasive translational regulation in response to starvation.
(a) Schematic of experimental procedures for RNA-seq, Ribo-seq, and QTI-seq in cells with or without starvation.
(b) A scatter plot of fold changes in LTM-associated aTIS density and CHX-associated CDS ribosome occupancy in HEK293 cells before and after amino acid starvation. Genes with single annotated TIS or multiple TISs are shown in blue and red dots respectively. Genes with more than 1.5 fold changes are marked with “Increase” or “Decrease” boxes.
(c) A scatter plot of fold changes in LTM-associated aTIS density and CHX-associated CDS ribosome occupancy in MEF cells before and after amino acid starvation.
(d) A heatmap of false discovery rate (FDR) of enriched GO terms (biological process) for gene groups with translational downregulation (Dec) or upregulation (Inc) in response to amino acid starvation.
(e) An example of genes (RPS28) with translational downregulation after amino acid starvation. The same scale is used for Y-axis. The corresponding gene structure is shown below the X-axis. The right panel is a bar graph depicting the relative RNA abundance, CDS ribosome occupancy, and aTIS ribosome density.
(f) An example of multi-TIS genes (GADD45G) showing an increased aTIS fraction after amino acid starvation.
(g) Experimental validation of translational control of GADD45G by a Fluc reporter bearing the 5′UTR of GADD45G with (WT) or without the uTIS codon CUG (Del). (means ± SEM; n = 3; * p < 0.05 student t-test).
QTI-seq also captured a considerable number of genes whose transcripts experienced increased initiation in response to starvation (638 in HEK293 and 515 in MEF). Interestingly, many of these genes are involved in protein folding and nuclear transport (Fig. 2d). Using a firefly luciferase (Fluc) reporter containing the 5′UTR, we validated the translational up-regulation of a nucleoporin-encoding gene NUP88 (Supplementary Fig. 7). It is noteworthy that many starvation-responsive genes contain multiple TISs (1,286 in HEK293 and 1,343 in MEF), suggesting a regulatory role for alternative TISs in translational control21. To demonstrate the influence of alternative translation on the aTIS initiation, we selected genes with multiple TISs and computed the relative ribosome density at the aTIS codon over the total TISs on the same transcript (Supplementary Fig. 8). This analysis uncovered many genes whose translational regulation is indiscernible by simple comparison of ribosome density changes at either CDS or aTIS. A total of 428 genes in HEK293 and 212 genes in MEF demonstrated an altered aTIS ratio over the total TISs upon amino acid deprivation (FDR < 0.05). This strategy uncovered several stress responsive genes whose transcripts contain previously uncharacterized TISs. For instance, GADD45G bears a CUG start codon in the 5′UTR (Fig. 2f). Fluc reporter assays confirmed the critical role of 5′UTR in the starvation-induced up-regulation of GADD45G (Fig. 2g). In particular, deleting the CUG codon was sufficient to prevent the starvation responsiveness.
Programmatic TIS regulation in response to starvation
Many upstream open reading frames (uORFs) are believed to exert negative effects on the main ORF translation, presumably by capturing the scanning ribosome12,22. It is thus not surprising to find that a large number of multi TIS-containing genes showed increased aTIS initiation when uTIS initiation is repressed under starvation. However, a handful of transcripts exhibited decreased aTIS fraction in spite of the presence of alternative TISs (Supplementary Fig. 8). To identify possible factors governing differential regulation of alternative TISs, we surveyed for consensus sequence motifs in gene groups that respond differently to starvation. Among transcripts with increased aTIS initiation upon starvation, the Kozak consensus motif is prominent (Supplementary Fig. 9a). For transcripts with increased uTIS, an evident purine-rich sequence context emerges above the background (Supplementary Fig. 9b). This finding is reminiscent of the polypurine (A)-rich sequences (PARSs) found in many evolutionarily conserved internal ribosome entry sites23,24. Alternatively, the purine-rich sequence may act in a manner opposite to the 5′ terminal oligopyrimidine (5′-TOP) tract, whose translation is highly sensitive to the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway25,26.
Dissect TIS regulation mechanisms in response to starvation
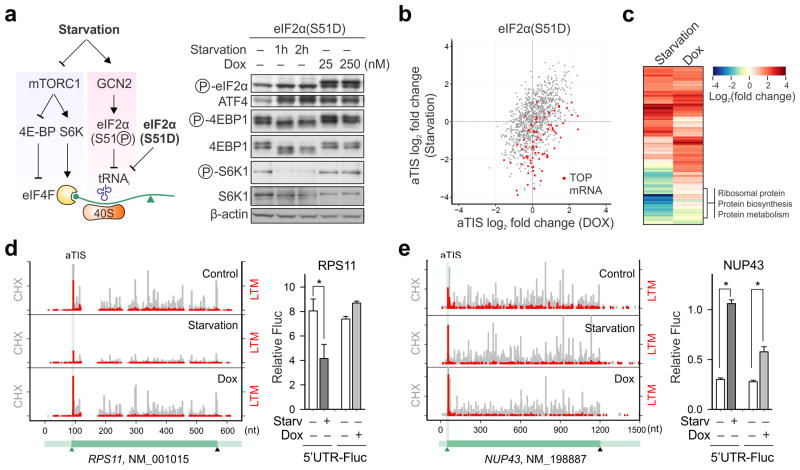
Amino acid deprivation suppresses global protein synthesis by inhibiting the mTORC1 signaling pathway and activating GCN2 kinases27. While the former regulates cap recognition, the latter controls the ternary complex formation (Fig. 3a). Little is known whether these distinct signaling pathways have overlapping or unique contributions to the translational regulation. We took advantage of a HEK293 cell line harboring an inducible phosphor-mimetic allele of eIF2α in which serine 51 was mutated to an aspartic acid (S51D)28. Expression of this mutant upon doxycycline (Dox) addition led to translational attenuation as well as an evident induction of ATF4 (Supplementary Fig. 10a and Fig. 3a). Unlike amino acid deprivation, however, eIF2α(S51D) expression had little effect on mTORC1 signaling (Fig. 3a). Therefore, the direct contribution of eIF2α phosphorylation to translational reprogramming can be dissected without pleiotropic effects caused by nutrient deprivation.
Figure 3. Distinct role of eIF2α phosphorylation in translational response to starvation.
(a) Schematic of mTORC1 signaling and eIF2α phosphorylation in controlling translation initiation. Right panel shows immunoblotting results of eIF2α(S51D) cells with either amino acid starvation or Dox-induced eIF2α(S51D) expression.
(b) A scatter plot of fold changes in LTM-associated aTIS density in eIF2α(S51D) cells between amino acid starvation and Dox-induced eIF2α(S51D) expression. TOP mRNAs are shown in Red dots.
(c) A heatmap of fold change of enriched GO terms (biological process) for gene groups with translational downregulation (green) or upregulation (red) in response to amino acid starvation (left) or Dox-induced eIF2α(S51D) expression (right).
(d) An example of genes (RPS11) showing translational repression in response to amino acid starvation but not Dox-induced eIF2α(S51D) expression. The right panel is validation of RPS11 translational control by a Fluc reporter bearing the 5′UTR of RPS11. (means ± SEM; n = 3; * p < 0.01 student t-test).
(e) An example of genes (NUP43) showing translational upregulation in response to either amino acid starvation or Dox-induced eIF2α(S51D) expression. The right panel is validation of NUP43 translational control by a Fluc reporter bearing the 5′UTR of NUP43. (means ± SEM; n = 3; * p < 0.01 student t-test).
We applied QTI-seq to HEK293/eIF2α(S51D) cells with or without Dox treatment, together with RNA-seq and Ribo-seq. Similar to amino acid starvation, eIF2α(S51D) expression resulted in evident differences for the LTM-associated aTIS ribosome density captured by QTI-seq (Supplementary Fig. 10b and Table 3). A comparative analysis of the aTIS profiles between these two conditions revealed a partially overlapping gene pattern (Fig. 3b). Interestingly, the majority of overlapping genes are translationally up-regulated with increased aTIS ribosome density. For those mRNAs undergoing translational repression under starvation, very few are affected in the presence of eIF2α(S51D) with the most notable gene group as TOP mRNAs (Fig. 3b). Clustering of the transcripts with changed aTIS ribosome density further confirmed the dichotomy of regulation between eIF2α(S51D) expression and starvation (Fig. 3c). In general, the transcripts commonly affected under both conditions are translationally up-regulated and span diverse functions, whereas the starvation-sensitive mRNA subsets are translationally repressed and enriched in functions highly relevant to ribosome biogenesis. These results indicate that, in response to amino acid starvation, reduced mTORC1 signaling is more responsible for the suppression of TOP mRNA translation. In contrast, eIF2α phosphorylation mainly contributes to the selective up-regulation of a subset of transcripts.
To confirm the differential regulation of translation initiation controlled by distinct upstream signaling pathways, we chose two genes for experimental validation. The gene encoding ribosomal protein RPS11 contains a typical TOP sequence on its 5′UTR. Amino acid starvation led to an about 3-fold decrease of the aTIS ribosome density (Fig. 3d). However, the translation of RPS11 exhibited little changes in response to Dox-induced eIF2a(S51D) expression. The Fluc reporter bearing the RPS11 5′UTR behaved exactly like the endogenous RPS11 in transfected cells (Fig. 3d). Only amino acid starvation, but not eIF2a(S51D) expression, caused a significant reduction of Fluc expression. Unlike RPS11, the nucleoporin gene NUP43 exhibited a translational up-regulation under either starvation or eIF2a(S51D) expression as evidenced by a more than 3-fold increase for its aTIS ribosome density (Fig. 3e). In agreement with the QTI-seq data, the Fluc reporter fused with the 5′UTR of NUP43 showed a consistent increase of translation in response to both conditions (Fig. 3e). These results strongly favor the hypothesis that distinct upstream signaling pathways act on different aspects of translation initiation, resulting in a coordinated translational reprogramming to achieve cellular adaptation.
Tissue-specific QTI-seq in liver cells
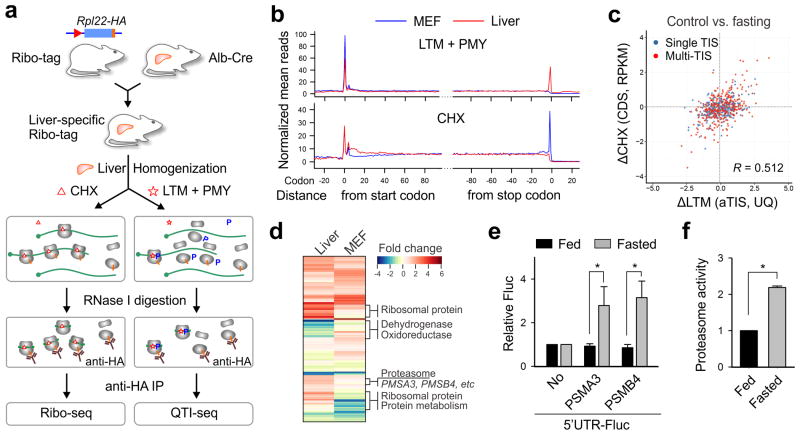
Having elucidated translational regulation from cells in culture, we reasoned that the principle of QTI-seq is applicable to solid tissues because it directly captures initiating ribosomes from lysates. To achieve cell type-specific ribosome profiling, we capitalized on the available RiboTag mouse, which carries a floxed Rpl22HA allele13. As a pilot study, we crossed the RiboTag mouse to a liver cell-specific Cre recombinase-expressing mouse under the control of albumin promoter (Alb-Cre) (Fig. 4a). The tissue specificity of RPL22HA incorporation into the ribosome was confirmed by anti-HA immunoprecipitation (IP) of polysome fractions (Supplementary Fig. 11a and 11b). In particular, the hepatocyte-specific transcripts were highly enriched in the tagged ribosome-associated mRNAs (Supplementary Fig. 11c). Following homogenization of liver tissues in the presence of LTM, PMY treatment led to efficient disassociation of the non-immobilized ribosomes (Supplementary Fig. 12a). The LTM-preserved ribosome complexes bearing RPL22HA were enriched by anti-HA IP after RNase I digestion (Fig. 4a). Metagene analysis revealed an elevated ribosome density at the annotated start codon, suggesting that QTI-seq worked equally well in solid tissue samples (Fig. 4b). Compared to MEF cells, there were fewer TIS sites identified from liver cells (1,770 vs. 5,796) but with similar codon composition and percentage of TIS types (Supplementary Fig. 13). In addition, there is a considerable overlap for the identified TISs between the two samples and the overlap is further increased when only the aTIS is considered (Supplementary Fig. 14). The non-overlapped TISs can be classified into differential aTIS and cell type-specific alternative TISs. For instance, the gene encoding hydroxyacyl-CoA dehydrogenase (HADH) showed a clear LTM peak at the aTIS in both liver and MEF samples (Supplementary Fig. 15). However, an in-frame dTIS was evident in the liver sample, but vaguely present in the MEF. It is likely that liver cells can give rise to different isoforms of HADH enzyme.
Figure 4. Liver-specific QTI-seq reveals translational reprogramming in response to fasting.
(a) Schematic of tissue-specific QTI-seq procedures using liver-specific RiboTag mice.
(b) Meta-gene analysis of LTM-associated ribosome density (top panel) or CHX-associated ribosome density (bottom panel) in MEF cells (blue line) and liver cells (red line). Normalized RPF reads are averaged across the entire transcriptome and aligned at the annotated start codons and stop codons.
(c) A scatter plot of fold changes in LTM-associated aTIS density and CHX-associated CDS ribosome occupancy in mouse liver cells with and without fasting.
(d) A heatmap of fold changes for gene groups with translational downregulation (green) or upregulation (red) in fasted liver (left) or starved MEF cells (right).
(e) Reporter assay using an in vitro translation system reprogrammed from mouse liver lysates with or without fasting. The relative translation efficiency of a synthesized Fluc mRNA containing 5′UTRs of PSMA3 or PSMB4 is shown in bar graph (means ± SEM; n = 3; * p < 0.01 student t-test).
(f) Mice of 8-12 week old were treated with or without overnight fasting. The chymotrypsin activity of liver homogenates was measured by Proteasome-Glo (means ± SEM; n = 3; * p < 0.01 student t-test).
Liver-specific TIS profile in response to fasting
The successful application of QTI-seq in mouse liver offers a unique opportunity to explore the translational response to a fasting condition at the organismal level. To this end, liver-specific RiboTag mice were subject to overnight fasting followed by RNA-seq, Ribo-seq, and QTI-seq. Food deprivation resulted in large disassembly of polysomes in liver tissues (Supplementary Fig. 12a). Consistently, liver-specific QTI-seq uncovered extensive changes of ribosome density at the aTIS sites (Supplementary Fig. 12b and Table S4). Supporting the quantitative feature of liver-specific QTI-seq, the changes of aTIS ribosome density in response to fasting correlated well with the differences of CDS ribosome occupancy (r = 0.512, Fig. 4c). Interestingly, overnight fasting elicited distinct translational responses in liver cells when compared to MEFs under starvation (Fig. 4d). Although MEFs showed prominent repression of ribosome biogenesis in response to starvation, this negative translational response was no longer obvious in liver cells after overnight fasting. In fact, some ribosomal protein genes underwent translational up-regulation in fasted liver. This observation suggests that, contrary to the widely accepted notion, global inhibition of the translation machinery is a transient response upon acute nutrient deprivation. Under prolonged fasting conditions, a continuous supply of ribosomal proteins is likely needed for selective protein synthesis.
For transcripts showing increased aTIS ribosome density, many of them encode components of the protein degradation pathway, in particular the proteasome system (Fig. 4d). This is consistent with the finding that the ubiquitin/proteasome system (UPS) not only helps to remove misfolded or damaged proteins29, but also contributes to the intracellular amino acid recycling during nutrient deprivation30. Our results, for the first time, demonstrate a translational mechanism that promotes the proteasome activity in response to prolonged fasting. To obtain direct evidence for translational control of the proteasome system, we cloned the 5′UTR of several proteasome genes and fused them to the Fluc reporter. We established a reconstituted translation system programmed from the liver homogenates to mimic the in vivo translation environment. Using equal amount of synthesized mRNAs, we found that the presence of 5′UTR of PSMA3 or PSMB4 resulted in a greater than 3-fold increase of Fluc activity in liver cells obtained from fasted mice, but not the one from normally fed mice (Fig. 4e). To ascertain the functional consequence of this translational switch in response to fasting, we directly measured the proteasome activity in liver homogenates using Proteasome-Glo. Overnight fasting resulted in a more than 2 fold increase in the proteasome chymotrypsin activity (Fig. 4f). Our results indicate a coordinated regulation between protein synthesis and degradation, which provides an elegant mechanism for cells and tissues to achieve metabolic homeostasis under nutrient deprivation.
Discussion
Translation initiation is a crucial point of regulation in eukaryotic gene expression, allowing cells to adapt rapidly to changing environmental conditions4-7. The significance of translation initiation is further substantiated by the existence of alternative translation that utilizes one or more potential TIS sites exist in addition to the main start codon12. The ability to monitor quantitatively the engagement of 80S ribosomes at the individual TIS would provide an effective means to evaluate start codon selection and to uncover fundamental principles underlying translational regulation. Compared to GTI-seq we developed previously15, QTI-seq represents a conceptually distinct approach that permits quantitative profiling of initiating ribosomes in samples from cells in culture and solid tissues. Using starvation as a model, the quantitative feature of QTI-seq resolves the complex signaling pathways controlling different aspects of translation initiation. It is clear that eIF2α phosphorylation plays a distinct role in translational regulation by mediating alternative translation. In line with many other studies8,27, QTI-seq reveals that genes involved in ribosome biogenesis are among the gene group strongly suppressed in translation initiation upon starvation. Since inhibiting mTORC1 signaling alone by rapamycin or torin is sufficient to cause translational repression of TOP mRNAs31,32, our finding strongly suggests a coordinated translational regulation between global translational reduction and selective protein synthesis in response to nutrient deprivation.
The applicability of QTI-seq to solid tissues has the potential to greatly advance our understanding of the basic mechanisms of translation initiation from cells to organismal levels. Using RiboTag mice, we for the first time demonstrated liver cell-specific profiling of initiating ribosomes. It serves as a prototype for tissue-specific ribosome profiling using other techniques such as BacTRAP14. Compared to MEFs under acute starvation, QTI-seq revealed distinct pattern of translational response in liver cells after overnight fasting. First, the global reduction of TOP mRNA translation is no longer evident, suggesting that a continuous supply of ribosomal proteins is needed for selective protein synthesis. Second, QTI-seq uncovered a potent translational re-programming for the proteasome system. The ubiquitin-proteasome system targets many proteins for degradation and the lethality of proteasome inhibition has been attributed to amino acid scarcity33. It therefore makes intuitive sense that the up-regulation of proteasome system is essential for cell survival by continuously supplying amino acids. It will be much of interest to demonstrate this phenomenon in other types of tissues, such as skeletal muscle, in response to fasting.
High resolution mapping of start codon positions and quantifying the corresponding initiation efficiency would empower modeling of gene expression, understanding of translational control, and comparison of proteome landscape under different growth conditions at both cellular and organismal levels. QTI-seq represents a powerful platform that can be readily applied to various tissues to uncover how translational regulation contributes to organismal function and disease.
Online Methods
Cells and reagents
HEK293, HEK293Trex, and MEF cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) with 10% fetal bovine serum (FBS). Cycloheximide (CHX), and puromycin were purchased from Sigma. Anti-RPS6 (cat#2217), anti-phospho-eIF2α (cat#3398), anti- eIF2α (cat#5324), anti-phospho-4EBP1 (cat#9459), and anti-4EBP1 (cat#9452) antibodies were purchased from Cell Signaling. Anti-RPL5 antibody (SAB1100578) was from Sigma. Anti-HA antibody (SC-7392) was from Santa Cruz Biotechnology. Plasmids transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instruction.
Animal treatment
The RiboTag mice and the Alb-Cre mice were purchased from Jackson Laboratory. Mice were housed in a temperature- and humidity- controlled facility with a 12-h light/dark cycle. The RiboTag mouse was bred to the Alb-Cre mouse line to obtain Rpl22HA-expressing homozygous mice. Mice at the age of 8-12 weeks were subjected to overnight fasting. All animal procedures were approved by Cornell IACUC (#2008-0167).
Preparation of cell lysates
For cell lysates, two 10-cm dishes of cells were harvested in 400 μL ice-cold polysome buffer (10 mM Hepes, pH 7.4, 100 mM KCl, 5 mM MgCl2) containing cycloheximide (100 μg/mL) or lactimidomycin (5 μM). Cells were then disrupted by vortexing using Lysing Matrix-D (Fisher) six times for 20 s, followed by a 40-s interval on ice. After centrifugation at 4 °C and 12,000 × g for 10 min, the supernatant was saved for puromycin treatment. For liver homogenates, a male RiboTag/Alb-Cre with or without overnight fasting were sacrificed by decapitation and their livers were quickly excised and chilled. Liver was immediately homogenized in 5 volumes of ice-cold polysome buffer containing cycloheximide (100 μg/mL) or lactimidomycin (5 μM) using mortar and pestle. Homogenates were centrifuged at 4 °C and 12,000 × g for 10 min, and the supernatant was subjected to puromycin treatment. To dissociate non-initiating ribosomes, cell lysates or liver homogenates were incubated in a solution containing 16 mM HEPES buffer, pH 7.4, 10 mM creatine phosphate, 0.1 mM spermidine, 40 μg/mL creatine phosphokinase, 0.8 mM ATP, and 25 μM of puromycin for 15 min. Puromycin-treated samples were then subjected to sucrose gradient sedimentation.
Polysome profiling
Sucrose solutions were prepared in polysome gradient buffer [10 mM HEPES, pH 7.4, 100 mM KCl, 5 mM MgCl2, 100 μg/mL cycloheximide, 5 mM DTT, and 20 U/mL SUPERase_In (Ambion)]. Sucrose density gradients [15–45% (wt/vol)] were freshly made in SW41 ultracentrifuge tubes (Fisher) using a BioComp Gradient Master (BioComp) according to the manufacturer's instructions. Puromycin-treated cell lysate was loaded onto sucrose gradients, followed by centrifugation for 100 min at 38,000 rpm, 4 °C, in an SW41 rotor. Separated samples were fractionated at 0.375 mL/min by using a fractionation system (Isco) that continually monitored OD254 values. Fractions were collected into tubes at 1-min intervals.
Immunoprecipitation of ribosomes from liver polysome fractions
The pooled polysome samples were treated with E. coli RNase I (Ambion, 750 U per 100 A260 units) at 4 °C for 1 h to convert the polysome into monosome. Digestion was neutralized with SUPERase inhibitor and the samples were incubated with 30 μL anti-HA beads (Sigma) at 4 °C for overnight. Immuno-precipitates were washed 3 times with high salt wash buffer (10 mM Hepes, pH 7.4, 200 mM KCl, 5 mM MgCl2, 1% Nonidet P-40, 1 mM DTT, 100 μg/mL cycloheximide). The washed beads were subjected to RNA extraction for library construction or immunoblotting analysis.
cDNA library construction of ribosome-protected mRNA fragments
To convert the polysome into monosome, E. coli RNase I (Ambion) was added into the pooled polysome samples (750 U per 100 A260 units) and incubated at 4 °C for 1 h. SUPERase inhibitor (50 U per 100 U RNase I) was then added to stop digestion. Total RNA extraction was performed using TRIzol reagent. Purified RNA samples were dephosphorylated in a 15 μL reaction containing 1× T4 polynucleotide kinase buffer, 10 U SUPERase_In, and 20 U T4 polynucleotide kinase (NEB). Dephosphorylation was carried out for 1 h at 37 °C, and the enzyme was then heat-inactivated for 20 min at 65 °C. Dephosphorylated samples were then mixed with 2× Novex TBE-Urea sample buffer (Invitrogen) and loaded on a Novex denaturing 15% polyacrylamide TBE-urea gel (Invitrogen). The gel was stained with SYBR Gold (Invitrogen) to visualize the RNA fragments. Gel bands containing RNA species corresponding to 28 nt were excised and physically disrupted by using centrifugation through the holes of the tube. RNA fragments were dissolved by soaking overnight in gel elution buffer (300 mM NaOAc, pH 5.5, 1 mM EDTA, 0.1 U/μL SUPERase_In). The gel debris was removed using a Spin-X column (Corning) and RNA was purified by using ethanol precipitation. Purified RNA fragments were resuspended in 10 mM Tris (pH 8) and denatured briefly at 65 °C for 30 s Poly-(A) tailing reaction was performed in a 8 μL with 1 × poly-(A) polymerase buffer, 1 mM ATP, 0.75 U/μL SUPERase_ In, and 3 U E. coli poly-(A) polymerase (NEB). Tailing was carried out for 45 min at 37 °C. For reverse transcription, the following oligos containing barcodes were synthesized:
MCA02, 5′-pCAGATCGTCGGACTGTAGAACTCTØCAAGCAGAAGACGGCATACGATT TTTTTTTTTTTTTTTTTTVN-3′;
LGT03, 5′-pGTGATCGTCGGACTGTAGAACTCTØCAAGCAGAAGACGGCATACGATT TTTTTTTTTTTTTTTTTTVN-3′;
YAG04, 5′-pAGGATCGTCGGACTGTAGAACTCTØCAAGCAGAAGACGGCATACGATT TTTTTTTTTTTTTTTTTTVN-3′;
HTC05, 5′-pTCGATCGTCGGACTGTAGAACTCTØCAAGCAGAAGACGGCATACGATTTTTTTTTTTTTTTTTTTTVN-3′.
In brief, the tailed RNA product was mixed with 0.5 mM dNTP and 2.5 mM synthesized primer and incubated at 65 °C for 5 min, followed by incubation on ice for 5 min. The reaction mix was then added with 20 mM Tris (pH 8.4), 50 mM KCl, 5 mM MgCl2, 10 mM DTT, 40 U RNaseOUT, and 200 U SuperScript III (Invitrogen). RT reaction was performed according to the manufacturer's instructions. Reverse transcription products were separated on a 10% polyacrylamide TBE-urea gel as described earlier. The extended first-strand product band was expected to be approximately 100 nt, and the corresponding region was excised. The cDNA was recovered by using DNA gel elution buffer (300 mM NaCl, 1 mM EDTA). First-strand cDNA was circularized in 20 μL of reaction containing 1× CircLigase buffer, 2.5 mMMnCl2, 1M Betaine, and 100 U CircLigase II (Epicentre). Circularization was performed at 60 °C for 1 h, and the reaction was heat inactivated at 80 °C for 10 min. Circular single-strand DNA was re-linearized with 20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT, and 7.5 U APE 1 (NEB). The reaction was carried out at 37 °C for 1 h. The linearized single-strand DNA was separated on a Novex 10% polyacrylamide TBE-urea gel (Invitrogen) as described earlier. The expected 100-nt product bands were excised and recovered as described earlier.
Deep sequencing
Single-stranded template was amplified by PCR by using the Phusion High-Fidelity enzyme (NEB) according to the manufacturer's instructions. The oligonucleotide primers qNTI200 (5′-CAAGCAGAAGACGGCATA- 3′) and qNTI201 (5′-AATGATACGGCGACCACCG ACAGGTTCAGAGTTCTACAGTCCGACG- 3′) were used to create DNA suitable for sequencing, i.e., DNA with Illumina cluster generation sequences on each end and a sequencing primer binding site. The PCR contains 1× HF buffer, 0.2 mM dNTP, 0.5 μM oligonucleotide primers, and 0.5 U Phusion polymerase. PCR was carried out with an initial 30 s denaturation at 98 °C, followed by 12 cycles of 10 s denaturation at 98 °C, 20 s annealing at 60 °C, and 10 s extension at 72 °C. PCR products were separated on a nondenaturing 8% polyacrylamide TBE gel as described earlier. Expected DNA at 120 bp was excised and recovered as described earlier. After quantification by Agilent BioAnalyzer DNA 1000 assay, equal amount of barcoded samples were pooled into one sample. Approximately 3–5 pM mixed DNA samples were used for cluster generation followed by sequencing by using sequencing primer 5′-CGACAGGTTCAGAGTTC TACAGTCCGACGATC-3′ (Illumina HiSeq).
Preprocessing of ribosome profiling sequencing reads
Raw ribosome profiling sequencing reads were first trimmed by 10nt from the 3′ end and trimmed reads were further processed by removing adenosine (A) stretch from the 3′ end (one mismatch is allowed). The processed reads between 25nt and 35nt were first mapped by Tophat to transcriptome (human : Ensembl release 70; mouse: Ensembl release 66)34. The unmapped reads were then mapped to corresponding genome (human: hg19; mouse: mm10). Non-uniquely mapped reads were disregarded for further analysis due to ambiguity. The 13th position (12nt offset from the 5′ end) of the uniquely mapped read was defined as the ribosome “P-site” position. The RPF density was computed after mapping uniquely mapped reads to each individual mRNA transcript according to the NCBI Refseq gene annotation. For RNA-seq, the raw mRNA sequencing reads (50nt in length) were mapped to human genome and transcriptome using parameters (--bowtie1 –p 10 –no-novel-juncs). Cuffdiff in the cufflinks package were used to calculate FPKM values based on Ensembl gene annotation35.
Prediction of translation initiation sites
Given the fact that many P-sites have a small number of GTI-seq reads and the distribution of P-site read count is apparently Poisson over-dispersed (unequal mean and variance), we applied Zero-Truncated Binomial Negative (ZTNB) model to determine P-sites with statistically significant number of read counts36. A global ZTBN model was first fit over all the non-empty P-sites of the entire transcriptome. Second, for each individual transcript with more than 50 distinct P-site positions, a local ZTNB model was trained on the non-zero P-sites. A putative TIS codon is predicted provided it meets the following criteria: 1) the P-site satisfying p-value cut-offs (global p-value cutoff 0.05 and local p-value cutoff 0.01); 2) it is a local optimum in a 31 nt window (-15, +15) flanking P-site position; 3) it is located in the 5′ UTR or in the first one-third of the CDS region. When predicted codons are within 1 nt offset of ATG or near-cognate codon, their coordinates were manually adjusted accordingly.
Quantification of Ribo-Seq and QTI-Seq
Reads per kilo bases per million reads (RPKM) value was calculated to quantify the ribosome occupancy of mRNA for CHX profiling20. A window centering the predicted TIS codon (-1,+4) was summarized to represent the abundance of translation initiation signal. To facilitate the comparison between different experimental conditions, upper quartile (UQ) normalization was applied to each predicted TIS codons based on the population of total QTI-Seq read count of each individual mRNA37. The fold changes of translational signal between two experimental conditions for both LTM and CHX profiling data were normalized by fold changes of RNA-Seq FPKM values of the corresponding mRNAs.
Ribosome footprint density aggregation plot
A set of longest mRNA transcripts were compiled by comparing different mRNA isoforms of the same gene on CDS length (if CDS lengths are the same, 5′ UTR lengths are compared). For each ribosome “P-site” in an mRNA, a normalized ribosome density value is calculated by dividing the P-site read count by the average P-site read count across the whole mRNA region (including the CDS and UTRs). Next, all the transcripts were aggregate by averaging each P-site position (relative to start codon or stop codon for the mouse liver data) across all the available transcripts embracing the P-site position.
Volcano plot of aTIS fraction differences
Genes bearing the aTIS and at least another alternative TIS site were retained for drawing volcano plot. The fraction of aTIS among all TIS sites of a gene was derived by dividing aTIS read count by the sum of read counts of all predicted TIS sites in the same gene. The difference of aTIS fraction between starvation and control samples was then calculated. To measure the significance of aTIS fraction differences, permutation test was conducted by randomly redistributing all the TIS reads over predicted TIS sites. This procedure was repeated for 1 million times and during each time permutated aTIS fraction difference is compared to aTIS fraction difference in real data at both sides (greater if aTIS fraction difference > 0 or smaller if aTIS fraction difference < 0). A p-value was inferred by dividing number of occurrences where permutated aTIS fraction difference is higher or smaller than the real aTIS fraction differences by the total number of permutations (1 million). The “Benjamini-hochberg” algorithm was used to adjust p-value for estimating false discovery rate (FDR).
Gene ontology analysis
DAVID is used for gene ontology (GO) enrichment analysis based on PANTHER annotation. The expressed genes (FPKM >1) were used as the background gene set.
Motif analysis
Annotated TIS and upstream TIS above the change cutoff (2-fold change for increased TIS and 0.5 fold change for decreased TIS) after starvation in MEF cell lines were used for motif analysis. A 100nt sequence flanking the aTIS or uTIS (±50nt) were retrieved within the context of mRNA. MEME was used to conduct discriminative motif analysis on increased TIS using decreased TIS as the background signal 38. At each position over the 100nt flanking sequence of TIS, a motif occurrence frequency is calculated by dividing the number of genes containing enriched motif in the 11nt window centering current position (±5nt) by the total number of genes. A scatter plot (x-axis: relative position to TIS; y-axis: motif occurrence frequency) was then drawn to reflect the positional distribution of the enriched motif.
Plasmid construction
The 5′UTRs of interest were amplified by RT-PCR reaction and cloned into pGL3-control vector (Promega). RPS11-promoter and RPS11-promoter-UTR were amplified from human genome and cloned into pGL3-basic vector (Promega). The sequences of all primers are listed below:
| Gene name | Sense primer | Anti-sense primer |
|---|---|---|
| NUP88-UTR | GGCCAAGCTTaatcctgagctgttagttcaagatggCCATGGATGC | GCATCCATGGccatcttgaactaacagctcaggattAAGCTTGGCC |
| NUPR1-UTR | GGCCAAGCTTcacaggcaagactttg | GCATCCATGGtggccattatgcctag |
| NUPR1-mut | cctagtctgctctctcagtcttgcctgtgaag | cttcacaggcaagactgagagagcagactagg |
| GADD45G-WT | GGCCAAGCTTggcgcactcgctggtg | GCATCCATGGgagtcattgtgcgatc |
| GADD45G-Mut | gggcgcgcatcggactggaatctttacctg | caggtaaagattccagtccgatgcgcgccc |
| RPS11-promoter | GGCCAAGCTTtgcaatggcgcgatctcg | GGAATTCaacgcagcacggccttg |
| RPS11-promoter-UTR | GGCCAAGCTTtgcaatggcgcgatctcg | GGAATTCcatcttcccggccgcctg |
| NUP43-UTR | GGCCAAGCTTgttccgcgggaagag | GCATCCATGGcctccatgccgaaagc |
| PSMA3-UTR | GGCCAAGCTTaagaggaaaggctattg | GCATCCATGGagctcatggtgctgac |
| PSMB4-UTR | GGCCAAGCTTcgtttccggtgacgct | GCATCCATGGcttccatcttagtcac |
Luciferase assay
Real time measurements of Fluc activity were recorded at 37°C with 5% CO2 using KronosDio Luminometer (Atto) as previously described 39. In brief, cells were plated on 35-mm dishes and transfected with plasmids encoding Fluc fused with 5′UTR of interest. Cells were incubated in HBSS medium for starvation or treated with DOX to induce eIF2α(S51D) expression. 1 mM luciferase substrate D-luciferin (Regis Tech) was added into the culture medium before Luminometer measurement.
Immunoblotting
Proteins were resolved on SDS-PAGE and transferred to Immobilon-P membranes (Millipore). Membranes were blocked for 1-h in TBS containing 5% BSA, followed by incubation with primary antibodies at 1:1000 dilutions. After incubation with horseradish peroxidase-coupled secondary antibodies, immunoblots were developed using enhanced chemiluminescence (ECLPlus, GE Healthcare).
Liver proteasome activity assay
The proteasome chymotrypsin-like activity of liver was detected using Proteasome-Glo Chymotrypsin-Like Assay kit (Promega) following the manufacturer's instruction. In brief, protein concentration of liver lysates from control mice and fasted mice was measured by Bradford assay. Equal amount of liver extract was diluted with polysome buffer to a final volume of 100 μL and loaded to a white 96-well plate. 100 μL of Proteasome-Glu reagent was added to each sample and incubated at room temperature for 10 mins. Luminescence was measured by a plate-reading luminometer (BioTek).
In vitro translation assay
Fluc, PSMA3-Fluc and PSMB4-Fluc mRNA was synthesized through in vitro transcription using mMESSAGE mMACHINE T7 ULTRA Kit (Ambion). Programmed in vitro translation was performed as described previously 40. A typical reaction contains equal amount of liver extract, 1× translation buffer (16 mM HEPES, pH 7.6, 10 mM creatine phosphate, 40 μg/mL creatine kinase, 0.1 mM spermidine, 10 μM each amino acid), 80mM potassium acetate, 1 mM magnesium acetate, 20U RNase inhibitor (Ambion), 0.8 mM ATP, 0.1 mM GTP and 50 μg/mL mRNA template. In vitro translation was incubated at 37°C for 2 hour and luciferase activity was measured with luciferase substrate (Promega).
Supplementary Material
Acknowledgments
We'd like to thank Qian lab members for helpful discussion and Drs. John Parker and Robert Weiss for critical reading of the manuscript. We also thank Dr. Peter Walter (University of California at San Francisco) sending us eIF2α(S51D) cell line. We thank Cornell University Life Sciences Core Laboratory Center for performing deep sequencing. This work was supported by grants to B.S from US National Institutes of Health (CA106150) and to S.-B.Q. from US National Institutes of Health (DP2 OD006449, R01AG042400), Ellison Medical Foundation (AG-NS-0605-09), and US Department of Defense (W81XWH-14-1-0068).
Footnotes
Accession codes. NCBI Sequence Read Archive: raw sequence data have been deposited under accession SRA160745.
Author Contributions: X.G. and S.-B.Q. conceived the original idea. X.G. designed experimental approaches and performed the experiments. J.W. analyzed the data. B.L. assisted data interpretation. M.M. and B.S. synthesized LTM. S.-B.Q. wrote the paper.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 2.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 3.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 6.Liu B, Qian SB. Translational reprogramming in cellular stress response. Wiley Interdiscip Rev RNA. 2014;5:301–315. doi: 10.1002/wrna.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 10.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S, Liu B, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2012;109:E2424–2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritsch C, et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. doi: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci U S A. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David A, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorokhov YL, et al. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc Natl Acad Sci U S A. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 25.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPs and their regulation. Biochem Soc Trans. 2006;34:12–16. doi: 10.1042/BST20060012. [DOI] [PubMed] [Google Scholar]
- 27.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 28.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 30.Vabulas RM, Hartl FU. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- 31.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suraweera A, Munch C, Hanssum A, Bertolotti A. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell. 2012;48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilbe JM. Negative binomial regression 1 online resource. Cambridge University Press; Cambridge, UK ; New York: 2011. pp. xviii–553. [Google Scholar]
- 37.Dillies MA, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Briefings in bioinformatics. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 38.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J Biol Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu B, Han Y, Qian SB. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.