Abstract
Heat shock factor 2 (HSF2) is a member of the heat shock transcription factor family, which appears to be activated during differentiation and development rather than on cellular stress. Here we report the isolation and characterization of the human hsf2 gene and its 5′-flanking region. The transcription unit of the human hsf2 gene consists of 13 exons dispersed over 33 kbp of genomic DNA on chromosome 6. The hsf2 mRNA is transcribed from multiple start sites, and initiation from the major site results in a transcript of 2.45 kb. A functional promoter, as determined by the ability to direct expression of a transiently transfected luciferase reporter gene, resides in a 950-bp upstream region of the human hsf2 gene. Examination of the core promoter sequence revealed a high GC content and lack of a canonical TATA box. This feature seems to be common among various species, as comparison of the hsf2 proximal promoter sequences from human, mouse, and rat showed distinct conserved regions. Moreover, the overall architecture of the human hsf2 gene is similar to its mouse counterpart. A comparison between human hsf2 gene and other hsf genes showed striking similarities in exon size. However, the exons are assembled in an hsf-specific manner.
INTRODUCTION
The transcriptional regulation of an evolutionary conserved family of proteins known as the heat shock proteins (Hsps) is mediated by the heat shock factors (HSFs). Since the simultaneous cloning of human and murine HSF1 and HSF2 in 1991 (Rabindran et al 1991; Sarge et al 1991; Schuetz et al 1991), much effort has been put on resolving the transcriptional regulation of the stress response. Unexpectedly, only 1 of the factors, HSF1, was shown to be activated on heat shock. HSF2, on the other hand, was not activated by the classical stress stimuli but rather during erythroid differentiation of K562 cells induced by hemin (Sistonen et al 1992). Despite this early finding, the function and regulation of HSF2 activity have remained elusive. Two additional members of the HSF family have subsequently been identified, of which HSF3 is an avian-specific stress-responsive factor and HSF4 is still largely uncharacterized (Nakai and Morimoto 1993; Nakai et al 1997).
HSF1 being the prototype of the HSF family has been most extensively investigated. HSF1 is activated within minutes of an increase in temperature, exposure to oxidants, heavy metals, and bacterial or viral infections. Activation of HSFs is a multistep process, including trimerization of the inactive monomer, inducible phosphorylation, localization to the nucleus, and binding to DNA at highly conserved heat shock response elements (HSE), consisting of multiple inverted repeats of the sequence nGAAn (for review, see Morimoto 1998). The overall activation pattern of HSF1 and HSF2 seem to be distinct (for review, see Pirkkala et al 2001). Although HSF2 binds to the HSE in response to hemin treatment, the activation kinetics is slow, ranging from hours to days (Sistonen et al 1992). HSF2 exists as inactive homo- or heterodimers that trimerize on activation and translocate to the nucleus (Sistonen et al 1994). The activity of HSF2 is further influenced by the existence of 2 isoforms, the longer HSF2-α isoform, which is transcriptionally more active than the shorter HSF2-β (Fiorenza et al 1995; Goodson and Sarge 1995; Leppä et al 1997). In contrast to HSF1, regulation of HSF2 by phosphorylation has not been reported, but the increase in protein levels seems to be central for HSF2 activity (Sistonen et al 1994). Specifically, on hemin activation, the HSF2 expression is up-regulated both at the transcriptional level and by mRNA stabilization (Pirkkala et al 1999). To better understand the complexity of its expression, we performed genomic cloning and characterization of the human hsf2 gene and promoter.
MATERIALS AND METHODS
Isolation of genomic clones
A human genomic P1 library was screened by hybridization with a 931-bp fragment (HindIII/PstI) coding for a part of human hsf2 cDNA (a kind gift from Robert E. Kingston, Harvard Medical School, Boston, MA, USA; Schuetz et al 1991). The hybridization was performed at Genome Systems Inc. (Incyte Genomics Inc., Palo Alto, CA, USA). We obtained 3 P1 clones (21178, 21179, and 21180) that were verified by partial sequencing.
Fluorescence in situ hybridization
Human peripheral blood lymphocytes were cultured according to standard protocols, and the cells were treated with 5-bromodeoxyuridine (BrdU) at early replicating phase to induce the banding pattern as described earlier (Lemieux et al 1992). Three P1 probes (21178, 21179, and 21180) were labeled with biotin 11-dUTP (Sigma Chemicals) according to standard protocols and hybridized on metaphase chromosomes derived from a normal lymphocyte cell culture. The identification of the chromosomes was based on DAPI banding pattern, which resembles G-bands after BrdU incorporation at the early replicating phase. The hybridization was carried out in 50% formamide and 10% dextrane sulfate in 2xSSC, and the signals were detected by a conventional detection method as described earlier (Pinkel et al 1986; Lichter et al 1988). A multicolor image analysis was used for acquisition, display, and quantification of hybridization signals of metaphase chromosomes as earlier described (Heiskanen et al 1996).
Primer extension
Primer extension analysis was performed as previously described (Carey and Smale 2000). Briefly, total RNA from K562 and HeLa cells was extracted using RNAzol™B (Tel-Test Inc.). A primer (5′-GCA GGG ATT CCA AAT TCT ACA CC-3′) −13 to −35 relative to the translation start site of hsf2 was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Thirty micrograms of total RNA were hybridized with the labeled oligonucleotide, at both 45°C and 60°C for 90 minutes. The same amount of yeast tRNA was used as a negative control. Reverse transcription was carried out at 37°C for 60 minutes using M-MLV RT (H-) Point mutant (Promega). Primer extension products were separated on a 6% denaturing polyacrylamide gel, and the gels were fixed for 10 minutes in 10% methanol and 10% acetic acid, dried, and exposed to Fuji-RX film with an intensifying screen at −70°C.
5′rapid amplification of cDNA ends
Total RNA from K562 cells was extracted using RNAzol™B, and the RACE reaction was performed using the 5′/3′ RACE kit from Boehringer-Mannheim. The reverse transcription step was carried out with 2 μg of total RNA with a reverse primer (5′-ACA CCT GCG AAC ACC TCC T-3′) +40 to +58 relative to the translation initiation site of hsf2. Following poly(dA) tailing, double-stranded DNA was obtained with a nested primer (5′-TAC TTC GTC TCA AGC TTG CAC G-3′) annealing at the translation initiation site. A second round of amplification was performed with the adapter primer and a second nested primer (5′-GCA GGG ATT CCA AAT TCT ACA CC-3′) (−13 to −35). The PCR products were TA-cloned into pGEM-T (Promega) and sequenced.
Plasmid construction
The promoter region was isolated from the P1 clone 21179 by PCR, using an oligonucleotide specific for the hsf2 ATG region (5′-TCC TCC ACA AGC GTC CAC A-3′) and an SP6 promoter primer for the P1 plasmid (5′-ATT TAG GTG ACA CTA TAG AAT AC-3′). Briefly, a 50-μL PCR reaction containing primers (300 nM); 0.2–0.3-μg P1 genomic plasmid; 1X EXT-PCR buffer (Finnzymes); MgCl2 (2.3 mM, Finnzymes); dimethyl sulfoxid (2%); dATP, dGTP, dCTP, and dTTP (0.5 mM each, Promega); and Dynazyme EXT DNA polymerase (1.5 U, Finnzymes) was used with a reaction cycle of 1 initial denaturation at 94°C for 4 minutes, followed by 10 cycles of 94°C for 1 minute, 46.7°C for 1 minute, and 70°C for 20 minutes, after which 20 cycles of 94°C for 1 minute, 46.7°C for 1 minute, and 70°C for 20 minutes + 20 s/cycle, with final extension of 10 minutes at 70°C. The PCR product was purified with PCR Kleen Spin Columns (Bio-Rad), TA-cloned into pGEM-T, and sequenced. The sequence can be found in GenBank under accession number AF331667.
The reporter plasmids for the luciferase assay were constructed by subcloning putative promoter fragments from the genomic P1 clone 21179 into the promoterless pGL3 basic vector upstream of the firefly luciferase gene (Promega). PCR was performed using sequence-specific primers containing splice sites for XhoI and MluI. The primer sequences are as follows: Pr.1R(XhoI) 5′-CAT TGT TAA CTC GAG GCA GGG AT-3′, Pr.2F(MluI) 5′-AGC TGT TTC CAC GCG TAA CAT C-3′, Pr.3F(MluI) 5′-GAG ATC TAC TGA CGC GTT TTC CAT-3′, Pr.4R(MluI) 5′-CAT TGT TAA CGC GTG CGC-3′, Pr.5F(XhoI) 5′-CAC AGC CTC GAG ACC ATA AGC-3′, and Pr.6F(XhoI) 5′-TGT CCT CGA GGC TTT ATC TAA ACT G-3′. The PCR reaction was carried out under the following conditions: primers (300 nM); 0.2–0.3 μg genomic P1 plasmid; 1X Pfu-PCR buffer containing MgCl2 (1.5 mM, Promega); dATP, dGTP, dCTP, and dTTP (0.2 mM each); and Pfu DNA polymerase (1.25 U, Promega) in a volume of 50 μL. A reaction cycle of 1 initial denaturation at 95°C for 2 minutes, followed by 35 cycles of 95°C for 1 minute, 55°C for 1 minute, and 70°C for 2 minutes, with a final extension of 5 minutes at 70°C, was performed. The PCR product was purified with QIAquick PCR Purification Kit (Qiagen), digested with the appropriate restriction enzymes, and repurified with the QIAquick kit. After that, the PCR products were ligated into pGL3 plasmid, which was cleaved with the same restriction enzymes, and transformed into competent bacteria. The inserted PCR products were verified by DNA sequencing.
Cell culture and transient transfections
K562 cells were grown in RPMI-1640 (Sigma) with supplements (10% FCS, glutamine, and antibiotics). HeLa cells were cultured in DMEM (Sigma) with supplements (5% FCS, glutamine, and antibiotics). Cells were maintained at 37°C in a humidified incubator in an atmosphere of 5% CO2. K562 cells were seeded at 2 × 105 cells/mL and allowed to recover for 1 day before transfection by electroporation (975 μF, 250 V) using a Bio-Rad Gene Pulser electroporator. The indicated DNA constructs (5 μg of reporter plasmid and 0.5 μg of Renilla luciferase plasmid, a kind gift from Michael J. Courtney, AIV-Institute, University of Kuopio, Finland) were mixed with cells (4.8 × 106) suspended in 400 μL of optiMEM (Gibco-BRL) and placed in a 0.4-cm-gap electroporation cuvette (BTX) and subjected to a single electric pulse. Cells were diluted in 12 mL RPMI-1640 medium with supplements and plated in 6-well tissue culture dishes. Confluent HeLa cells in 6-well plates were transfected with the same amounts of plasmids using the Lipofectin procedure (Life Technologies Inc).
Luciferase assay
Transiently transfected cells were cultured for 24 hours, washed with phosphate-buffered saline, and lysed by 3 freeze-thaw cycles in Passive lysis buffer (Promega). Firefly luciferase and Renilla (sea pansy) luciferase activities were measured sequentially using a Dual-Luciferase Reporter assay system (Promega) and a Labsystems Luminoskan. All the obtained counts were normalized using an internal control containing the SV40 promoter in front of the Renilla luciferase gene. As a positive control for the assay, the Rous sarcoma virus (RSV) promoter in front of the luciferase gene was used (a kind gift from Päivi J. Koskinen, Turku Centre for Biotechnology, Finland).
RESULTS AND DISCUSSION
The human hsf2 gene consists of 13 exons located on chromosome 6
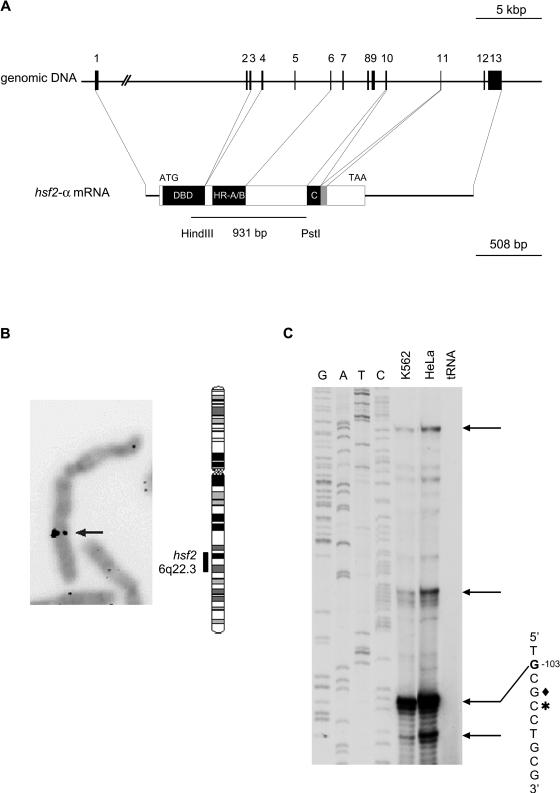
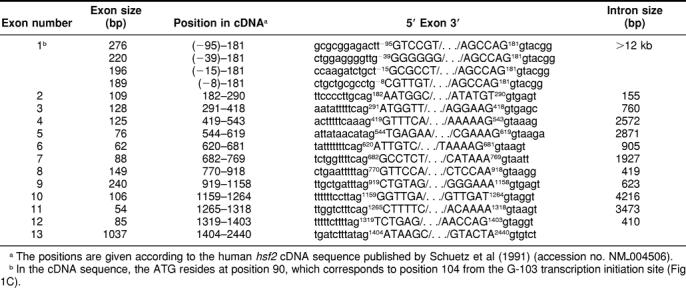
The structure of the human hsf2 gene was deduced by comparing the cDNA and the genomic sequence available from GenBank (accession nos. NM_004506 and Z99129, respectively). The gene was found to be composed of 13 exons and to span approximately 33 kbp of the genome (Fig 1A). The functional domains of the HSF2 protein originated from defined groups of exons; the DNA-binding domain consists of exons 1–3 (DBD), the 3 first hydrophobic heptad repeats (HR-A/B) are encoded by exons 4–6, and exon 10 codes for the fourth heptad repeat (HR-C). Exon 11, which is the shortest exon, is differentially spliced, as it is present in the 54-bp longer hsf2-α isoform, but is absent in the shorter hsf2-β isoform. As summarized in Table 1, splice sites in accordance to the GT-AG rule can be found on both sides of the internal exons.
Fig 1.
The human hsf2 gene. (A) The hsf2 gene spans approximately 33 kbp in the human genome, and the first intron is over 12 kbp. The exons of hsf2 are represented by numbered black boxes and the introns by lines. The functional domains of the HSF2 protein originate from defined groups of exons (DBD: DNA-binding domain; HR-A/B: hydrophobic repeat A/B; HR-C: hydrophobic repeat C). Exon 11 (gray box) is alternatively spliced. The 5′- and 3′-UTRs are 100–200 b and 740 b, respectively. The 931-bp cDNA fragment (Schuetz et al 1991) used as a probe for the P1 screening is shown in the lower panel. The scale of the genomic DNA and mRNA are indicated. (B) To determine the chromosomal localization of the human hsf2 gene, P1 genomic clone was hybridized using fluorescence in situ hybridization to metaphase chromosomes derived from a lymphocyte cell culture. The arrow shows the specific signal of the labeled probe hybridized to chromosome 6, which was identified based on DAPI banding pattern. A schematic representation of human chromosome 6 with the localization of hsf2 is shown. (C) A radiolabeled oligonucleotide, corresponding to positions 13–35 bp upstream of the translation initiation codon, was annealed to total RNA from K562 or HeLa cells or to yeast tRNA. Reverse transcription was carried out, and the extension products were resolved by electrophoresis on a 6% denaturing polyacrylamide gel. A sequencing ladder of human hsf2 5′-flanking region was prepared using the same primer. Arrows indicate the extension products of the most intensive bands obtained in 3 independent experiments. The sequence of the most intense extension product is shown with the start nucleotide in bold (G −103). The asterisk indicates the longest 5′RACE reaction product sequenced, and the diamond indicates the guanine corresponding to the start site deduced for mouse hsf2 (Manuel et al 1999)
Table 1.
Organization of the human hsf2 gene
The chromosomal localization of the human hsf2 gene was analyzed by using 3 hsf2-specific genomic P1 clones as probes in fluorescence in situ hybridization (FISH). In 30 metaphases out of 45 (68%), a probe derived from the P1 clone 21178 showed specific signals on chromosome 6q22.3 (Fig 1B). In 8 metaphases out of 10 (80%), a probe derived from the P1 clone 21179 also showed specific signals on chromosome 6q22.3. Surprisingly, a probe derived from the P1 clone 21180 showed specific signals, in 25 metaphases out of 30 (83%), on chromosome 12q13 (data not shown), suggesting the existence of a potential pseudogene.
The chromosomal region of 6q22.3, where the human hsf2 gene is localized, is homologous to the region of mouse chromosome 10, harboring the hsf2 gene (Manuel et al 1999). Several diseases, including hereditary persistence of fetal hemoglobin (HPFH, OMIM 142470 at http://www.ncbi.nlm.nih.gov), have been linked to the chromosomal locus of 6q22.3. Localization of HPFH and the human hsf2 gene in proximal regions on chromosome 6 addresses a question of whether HSF2 plays a role in this disease state, considering the up-regulation of HSF2 and fetal globins on hemin treatment of K562 cells (Pirkkala et al 1999). However, no mutation in the human hsf2 gene has so far been connected to any disease.
The 5′-flanking region of hsf2 contains multiple transcription initiation sites and a functional promoter
Primer extension analyses of the hsf2 gene revealed multiple transcription initiation sites, as shown in Figure 1C. The most intense extension product ends at a guanine residue 14 bp upstream of the previously reported cDNA (Schuetz et al 1991) or 103 bp upstream of the initial ATG codon (G −103). This start site is also in accordance with the results from the 5′RACE analysis, in which the longest sequence obtained from 5′RACE analysis corresponded to a cytosine 100 bp upstream of the ATG codon (Fig 1C; data not shown). The transcription initiation site at G −103 also corresponds to the transcription start site determined for the mouse hsf2 gene (Fig 1C; Manuel et al 1999). Depending on the transcription start site, hsf2 mRNA can be up to 2534 b, consisting of 1610 b of coding sequence and 5′- and 3′-UTRs, which are 100–200 b and 740 b, respectively.
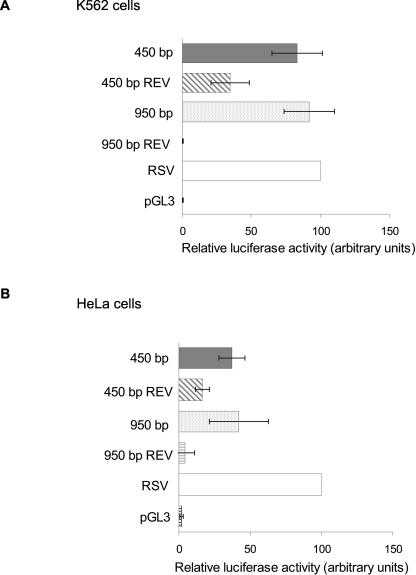
The 5′-flanking region of hsf2 was subcloned from the genomic P1 clone 21179 and sequenced. To determine whether the cloned 5′-flanking region of the hsf2 gene contained a functional promoter, we performed luciferase reporter assays. We generated reporter constructs containing 450 bp and 950 bp of the 5′-flanking region inserted into pGL3 basic vector, upstream of the firefly luciferase reporter gene. The plasmids were transiently transfected into K562 and HeLa cells to test whether they would be capable of directing gene expression. In K562 cells, both 5′-flanking fragments exhibited strong activation of the reporter gene when compared to the negative control consisting of a promoterless pGL3 vector (Fig 2A). In addition, reversion of the upstream region resulted in a marked reduction (450 bp REV) or a complete loss (950 bp REV) of the promoter activities, showing that these fragments contain functional, orientation-dependent promoters (Fig 2A). The same applies to HeLa cells, but the levels of reporter gene expression were approximately 2-fold lower compared to the K562 cells (Fig 2B). The functionality of the assay was assured by using the RSV promoter in front of the luciferase gene as a positive control.
Fig 2.
Luciferase reporter gene analysis of the 5′-flanking region human hsf2. (A) K562 and (B) HeLa cells were transiently transfected with plasmids, in which either a 950-bp or a 450-bp fragment of the 5′-region of the human hsf2 gene was inserted upstream of the luciferase reporter gene in pGL3. The SV40 promoter driving Renilla luciferase gene (SV40-pRL) was transfected into both cell lines as an internal control. Negative controls were provided by vectors containing the promoter fragments in reverse orientation (450 bp REV and 950 bp REV) and by an empty vector (pGL3). The RSV promoter driving the luciferase gene was used as a positive control. The result obtained with RSV was arbitrarily set to 100. The data represent the mean values (±standard deviation) of at least 3 independent experiments in duplicate. All the results are relative to the internal SV40-pRL control plasmid
Computer-aided analysis of the 5′-flanking region of human and murine hsf2 genes
Computer-aided analysis of 1.4 kbp of the human hsf2 promoter revealed multiple putative recognition sites for sequence-specific transcription factors (Fig 3A). For example, putative binding sites for the transcription factor TCF-11 were found on the human hsf2 promoter. The DNA sequence recognized by TCF-11 is also present in the binding site for erythroid-specific activator NF-E2, the antioxidant response element, and the heme-responsive element (Johansen et al 1998). The presence of such a DNA element is of interest, as hsf2 mRNA levels have been shown to be up-regulated after hemin-induced erythroid differentiation of K562 cells (Sistonen et al 1992; Pirkkala et al 1999). Since hemin treatment of K562 cells transiently transfected with the 950-bp hsf2 promoter containing luciferase constructs did not show any marked increase in the reporter gene levels (data not shown), the levels of HSF2 are likely to be regulated posttranscriptionally. However, a putative element further upstream could still contribute to hemin responsiveness of the hsf2 promoter.
Fig 3.
Computer-aided analysis of the 5′-flanking region of the human hsf2 gene. (A) The nucleotide sequence of 1.4 kbp of the human hsf2 5′-flanking region. The ATG codon and the transcription initiation sites are boxed. The first nucleotide upstream of the major transcription initiation site is designated as −1 and indicated with an arrow. Putative transcription factor binding sites, as determined using the MatInspector software V2.2 (Quandt et al 1995) connected to the TRANSFAC database (Heinemeyer et al 1998), are marked by gray boxes, pointing either to the right for binding to the sense strand or to the left for binding to the antisense strand. The quality rating used for choosing the putative transcription factor binding sites is a core similarity of 1.000 and a matrix similarity of ≥0.950. Pr. 1–6 indicate forward (F) and reverse (R) oligonucleotides used for luciferase constructs. (B) Alignment of the proximal promoters of mouse, rat, and human hsf2 promoters by Clustal W 1.8 (−529 bp, −564 bp, and −571 bp relative to ATG, respectively; Thompson et al 1994). The initial methionine ATG codon is shown in the white box, and the identical nucleotides between the different promoters are shaded gray. The 4 transcription initiation nucleotides in human (Fig 1C) and the one determined for mouse (Manuel et al 1999) are highlighted in black background. Arrow marks the major transcription initiation site for human hsf2 (G −103). Data for the transcription initiation sites for rat hsf2 are not available. Putative transcription factor binding sites in the conserved areas are framed
HSF2 has also been implicated in mouse heart development (Eriksson et al 2000), and putative binding sites for 2 transcription factors, Nkx-2.5 and S8, which are believed to be important for heart development, were found in the promoter region (Fig 3A; Leussink et al 1995; Shiojima et al 1996). In addition, the LIM-only protein, Lmo2, and GATA factor binding sites are of interest in the context of hsf2 regulation. Both Lmo2 and GATA-3 are present in the intraembryonic regions known to give rise to hematopoietic precursors in vitro and in vivo, suggesting that they act together at key points of hematopoietic development (Manaia et al 2000). No consensus binding sites for HSFs (Amin et al 1988) could be found in the 5′-flanking region of hsf2 by computer-aided analysis, but several units of the nGAAn sequence were detected in the hsf2 promoter by manual examination. Therefore, the possibility that some nGAAn units could form a functional cluster (ie, an HSE) cannot be excluded.
Since the consensus binding sites of a transcription factor often consist of very short stretches of DNA readily found in multiple sequences, results from computer-aided analysis might contain many false positives. Comparison of promoter sequences among different species offers one way of narrowing down the possibilities of important regulatory sequences. Therefore, a comparison between the proximal promoter regions of human, mouse, and rat hsf2 promoters (accession nos. AF331667, AF045614, and AF172641, respectively) was performed with Clustal W 1.8 (Thompson et al 1994; http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html). As shown in Figure 3B, the proximal regions of the human, mouse, and rat hsf2 promoters are highly conserved. The sequences are rich in Gs and Cs, and they lack consensus TATA boxes. Other core promoter elements found in the 3 species include an initiator-like sequence (Smale and Baltimore 1989), located around the start site of the longest human transcript and a TFIIB recognition element (BRE; Lagrange et al 1998). Other common sites include a classical GC box located next to the major transcription initiation sites of human and mouse hsf2 (Fig 3B; Manuel et al 1999), an E box, and 2 CAAT boxes. The conserved binding sites on the human, mouse, and rat promoters indicate a housekeeping type of regulation on the hsf2 gene. This notion is further strengthened by the fact that human hsf2 mRNA is expressed in all cell lines and tissues tested so far (data not shown).
Conservation of the hsf gene structure in evolution
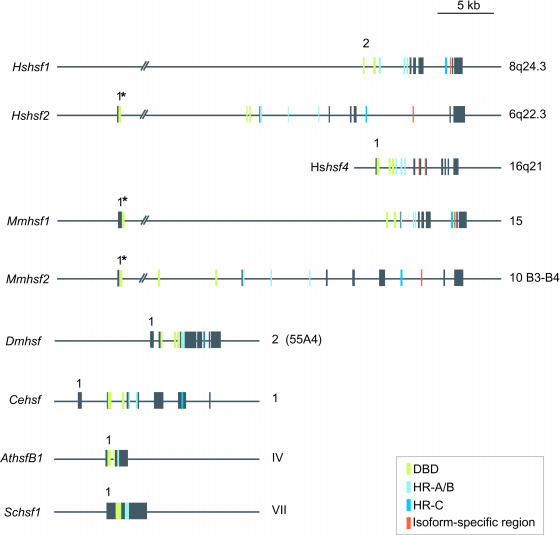
Because of the enormous influx of genomic sequences in publicly available databases, estimations of different hsf gene structures are now feasible. In Figure 4, we have compared previously published gene structures of the Homo sapiens hsf4 (Tanabe et al 1999), Mus musculus hsf1 and hsf2 (Zhang et al 1998; Manuel et al 1999), and Arabidopsis thaliana hsfB1 (Hsf4) (Prändl et al 1998; Nover et al 2001) genes, as well as novel gene structures constructed with computer-aided analysis, that is, the Homo sapiens hsf1, Drosophila melanogaster hsf, Caenorhabditis elegans hsf, and Saccharomyces cerevisiae hsf-1 genes. The high interspecies conservation seen at the sequence level among different HSFs is also evident in their respective gene structures (for sequence comparison, see Pirkkala et al 2001). In human and mouse, hsf1 genes exhibit a distinct pattern of more tightly assembled exons compared to the more widely dispersed hsf2 genes. All the known human and mouse hsf genes consist of 13 exons strikingly similar in size. Another common feature for these genes is the organization of functional domains into distinct exons, that is, the DNA-binding domain (DBD) being encoded by exons 1–3, the hydrophobic oligomerization domain HR-A/B by exons 4–6, and the HR-C by exon 10 (Fig 4). This feature seems to be specific for human and mouse since the functional domains of the Drosophila HSF are not delineated by exon boundaries. Although Drosophila and C. elegans hsf genes contain only 8 exons, as found by computer-aided analysis, the larger overall exon size results in transcripts of comparable sizes to the human and mouse hsf transcripts (Mmhsf1 2062 b vs Cehsf 2019 b). Sequence comparison of the Arabidopsis genome revealed the existence of 21 ORFs corresponding to hsfs (Nover et al 2001). Represented here by the AthsfB1 gene, all ORFs contain a single intron that separates the DNA-binding domain into 2 parts.
Fig 4.
Schematic representation of hsf genes. Structures of different hsf genes, as deduced either by comparison between previously published cDNA sequences and genomic sequences available from the GenBank (Hshsf1, Hshsf2, Dmhsf, and Schsf1, http://www.ncbi.nlm.nih.gov) or from previously published genomic structures (Hshsf4, Mmhsf1, and Mmhsf2). The exons are represented by boxes and the introns by lines. Exon 1 is indicated in all genes except in human hsf1. Experimentally determined 5′UTRs are indicated with an asterisk. The chromosomal localization indicated to the right is either verified by experimental data or obtained from various genome projects. The genomic structure of the C. elegans is derived from a computer-generated cDNA sequence compared to the corresponding unspliced genomic sequence found at http://wormbase.sanger.ac.uk. The exons corresponding to the functional domains in the putative C. elegans hsf gene are concluded according to homology to the functional domains of Drosophila. Accession numbers: Hshsf1, M64673 Rabindran et al 1991 (cDNA), AF205589 (genomic); Hshsf2, NM_004506 Schuetz et al 1991 (cDNA), Z99129 (genomic); Hshsf4, NM_001538 Nakai et al 1997 (cDNA), Tanabe et al 1999, AC074143 (genomic); Mmhsf1, X61753 Sarge et al 1991 (cDNA), AF059275/AF061503 Zhang et al 1998 (genomic); Mmhsf2, NM_008297 Sarge et al 1991 (cDNA), AF045615–27 Manuel et al 1999 (exons); Dmhsf, M60070 Clos et al 1990 (cDNA), AE003800 Adams et al 2000 (genomic); Cehsf, AL033536/Y53C10A.12, http://wormbase.sanger.ac.uk (cDNA, genomic); AthsfB1 (Hsf4), Y14069 Prändl et al 1998 (cDNA), Nover et al 2001, Z99707 (genomic) CAB16764 (protein); Schsf-1, M22040 Wiederrecht et al 1988 (cDNA), NC_001139 Tettelin et al 1997 (genomic)
The difference in intron numbers among the species raises a question of whether the primordial hsf gene was intronless or rich in introns. Currently, 2 theories concerning the structure of ancestral genes exist. According to the “introns early” model, genes originated as interrupted sequences and genes without introns have lost them during evolution. In contrast, the “introns late” model supposes that the ancestral gene consisted of uninterrupted sequence and that introns were subsequently inserted (for review, see Lewin 2000). Based on these 2 theories, the primordial hsf gene would be more similar either to the dispersed mammalian gene or to the condensed Saccharomyces and Arabidopsis hsf genes.
In conclusion, the 5′-flanking region of the human hsf2 gene contains a functional promoter that can drive constitutive expression, and the transcription initiates at multiple start sites. The overall architecture of human and mouse hsf genes exhibits distinct features, including a remarkable conservation of exon sizes and an hsf-specific placement of exons.
Acknowledgments
We are grateful to Robert E. Kingston (Harvard Medical School, Boston, MA, USA), Michael J. Courtney (AIV-Institute, University of Kuopio, Finland), and Päivi J. Koskinen (Turku Centre for Biotechnology, Finland) for providing the human hsf2 cDNA, the SV40 Renilla plasmid, and the RSV firefly luciferase plasmid, respectively. Lutz Nover and coworkers (Biocenter of the Geothe University, Germany) are thanked for generously providing the Arabidopsis HSF review prior to publication. We thank Carina I. Holmberg (Turku Centre for Biotechnology, Finland) for critical reading of the manuscript. This work was supported by the Academy of Finland, the Sigrid Jusélius Foundation, the Finnish Cancer Organizations, the Wihuri Foundation, and the Maud Kuistila Foundation. P.N. and T.-P.A. are supported by the Turku Graduate School of Biomedical Sciences (TuBS).
REFERENCES
- Adams MD, Celniker SE, and Holt RA. et al. 2000 The genome sequence of. Drosophila melanogaster. Science. 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of the heat shock gene regulatory elements. Mol Cell Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Smale ST 2000 Transcription initiation site mapping. In: Transcriptional Regulation in Eukaryotes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 116–123. [Google Scholar]
- Clos J, Westwood T, Becker PB, Wilson S, Lambert K, Wu C. Molecular cloning and expression of hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63:1085–1097. doi: 10.1016/0092-8674(90)90511-c. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Jokinen E, Sistonen L, Leppä S. Heat shock factor 2 is activated during mouse heart development. Int J Dev Biol. 2000;44:471–477. [PubMed] [Google Scholar]
- Fiorenza MT, Farkas T, Dissing M, Kolding D, Zimarino V. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 1995;23:467–474. doi: 10.1093/nar/23.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson ML, Sarge KD. Regulated expression of heat shock factor 1 isoforms with distinct leucine zipper arrays via tissues-dependent alternative splicing. Biochem Biophys Res Commun. 1995;211:943–949. doi: 10.1006/bbrc.1995.1903. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, and Reuter I. et al. 1998 Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 26:264–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiskanen M, Kallioniemi O, Palotie A. Fiber-FISH: experiences and a refined protocol. Genet Anal. 1996;5–6:179–184. [PubMed] [Google Scholar]
- Johansen Ø, Murphy P, Prydz H, Kolstø AB. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T, Kapanidis AN, Tang H, Reinber D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux N, Dutrillaux B, Viegas-Pequignot E. A simple method for simultaneous R- or G-banding and fluorescence in situ hybridization of small single-copy genes. Cytogenet Cell Genet. 1992;4:311–312. doi: 10.1159/000133277. [DOI] [PubMed] [Google Scholar]
- Leppä S, Pirkkala L, Saarento H, Sarge KD, Sistonen L. Overexpression of HSF2-β inhibits hemin-induced heat shock gene expression and erythroid differentiation of K562 cells. J Biol Chem. 1997;272:15293–15298. doi: 10.1074/jbc.272.24.15293. [DOI] [PubMed] [Google Scholar]
- Leussink B, Brouwer A, el Khattabi M, Poelmann RE, Gittenberger-de Groot AC, Meijlink F. Expression patterns of the paired-related homeobox genes MHox/Prx1 and S8/Prx2 suggest roles in development of the heart and the forebrain. Mech Dev. 1995;52:51–64. doi: 10.1016/0925-4773(95)00389-i. [DOI] [PubMed] [Google Scholar]
- Lewin B 2000 From genes to genomes. In: Genes VII, Oxford University Press, New York, NY, 58–62. [Google Scholar]
- Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988;3:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Manaia A, Lemarchandel V, Klaine M, Max-Audit I, Romeo P, Dieterlen-Lievre F, Godin I. Lmo2 and GATA-3 associated expression in intraembryonic hemogenic sites. Development. 2000;127:643–653. doi: 10.1242/dev.127.3.643. [DOI] [PubMed] [Google Scholar]
- Manuel M, Sage J, Mattéi MG, Morange M, Mezger V. Genomic structure and chromosomal localization of the mouse Hsf2 gene and promoter sequences. Gene. 1999;232:115–124. doi: 10.1016/s0378-1119(99)00092-x. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Nakai A, Morimoto RI. Characterization of a novel chicken heat shock transcription factor, Heat Shock Factor 3, suggests a new regulatory pathway. Mol Cell Biol. 1993;13:1983–1997. doi: 10.1128/mcb.13.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Ganguli A, and Scharf K-D 2001 Arabidopsis and the Hsf world: how many heat stress transcription factors do we need? Cell Stress Chap, 6: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Gray JW, Trask B, van den Engh G, Fuscoe J, van Dekken H. Cytogenetic analysis by in situ hybridization with fluorescently labeled nucleic acid probes. Cold Spring Harb Symp Quant Biol. 1986;1:151–157. doi: 10.1101/sqb.1986.051.01.018. [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Alastalo T-P, Nykänen P, Seppä L, Sistonen L. Differentiation lineage-specific expression of human heat shock factor 2. FASEB J. 1999;13:1089–1098. doi: 10.1096/fasebj.13.9.1089. [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykänen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Prändl R, Hiderhofer K, Eggers-Schumacher G, Schöffl F. Hsf3,a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen Genet. 1998;258:269–278. doi: 10.1007/s004380050731. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector—new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran SK, Giorgi G, Clos J, Wu C. Molecular cloning and expression of human heat shock factor, HSF1. Proc Natl Acad Sci U S A. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- Schuetz TJ, Gallo GJ, Sheldon L, Tempst P, Kingston RE. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci U S A. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I, Komuro I, Mizuno T, Aikawa R, Akazawa H, Oka T, Yamazaki T, Yazaki Y. Molecular cloning and characterization of human cardiac homeobox gene CSX1. Circ Res. 1996;79:920–929. doi: 10.1161/01.res.79.5.920. [DOI] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Morimoto RI. Human heat shock factor 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;1:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Tanabe M, Sasai N, Nagata K, Liu X-D, Liu PCC, Thiele DJ, Nakai A. The mammalian hsf4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J Biol Chem. 1999;274:27845–27856. doi: 10.1074/jbc.274.39.27845. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Agostoni Carbone ML, and Albermann K. et al. 1997 The nucleotide sequence of Saccharomyces cerevisiae chromosome VII. Nature. 387:81–84. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988;54:841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Koushik S, Dai R, Mivechi N. Structural organization and promoter analysis of murine heat shock transcription factor-1 gene. J Biol Chem. 1998;49:32514–32521. doi: 10.1074/jbc.273.49.32514. [DOI] [PubMed] [Google Scholar]