Highlights
-
•
SLC25A12 gene expression is regulated in SH-SY5Y cells through CREB.
-
•
SLC25A12 gene is up-regulated in SH-SY5Y cells under conditions of differentiation.
-
•
Ca2+ interaction with CREB further enhances SLC25A12 gene expression.
-
•
SLC25A12 gene is down-regulated in neuroinflammation.
-
•
SLC25A12 gene is up-regulated in SH-SY5Y cells by neuregulin-1.
Abbreviations: Aβ, β-amyloid; AGC1, aspartate/glutamate carrier isoform 1; APP, amyloid precursor protein; BAPTA-AM, 1,2-bis(2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester); ChIP, chromatin immunoprecipitation; P-CREB, phospho-cAMP-response-element-binding protein; T-CREB, total-cAMP-response-element-binding protein db-cAMP, dibutyryl-cAMP; EMSA, electrophoreticmobility-shift assay; IBMX, isobutylmethylxanthine; IFNγ, interferon gamma; LUC, luciferase; NAA, N-acetylaspartate; NRG-1, neuregulin-1; PKA, cAMP-dependent protein kinase; SAP97, synapse-associated protein-97; siCREB, siRNA targeting CREB; siRNA, small interfering RNA; SYP, synaptophysin; TNFα, tumor necrosis factor alpha; WB, Western-blotting
Keywords: Aspartate/glutamate carrier, CREB, Ca2+
Abstract
The aspartate/glutamate carrier isoform 1 is an essential mitochondrial transporter that exchanges intramitochondrial aspartate and cytosolic glutamate across the inner mitochondrial membrane. It is expressed in brain, heart and muscle and is involved in important biological processes, including myelination. However, the signals that regulate the expression of this transporter are still largely unknown. In this study we first identify a CREB binding site within the aspartate/glutamate carrier gene promoter that acts as a strong enhancer element in neuronal SH-SY5Y cells. This element is regulated by active, phosphorylated CREB protein and by signal pathways that modify the activity of CREB itself and, most noticeably, by intracellular Ca2+ levels. Specifically, aspartate/glutamate carrier gene expression is induced via CREB by forskolin while it is inhibited by the PKA inhibitor, H89. Furthermore, the CREB-induced activation of gene expression is increased by thapsigargin, which enhances cytosolic Ca2+, while it is inhibited by BAPTA-AM that reduces cytosolic Ca2+ or by STO-609, which inhibits CaMK-IV phosphorylation. We further show that CREB-dependent regulation of aspartate/glutamate carrier gene expression occurs in neuronal cells in response to pathological (inflammation) and physiological (differentiation) conditions. Since this carrier is necessary for neuronal functions and is involved in myelinogenesis, our results highlight that targeting of CREB activity and Ca2+ might be therapeutically exploited to increase aspartate/glutamate carrier gene expression in neurodegenerative diseases.
1. Introduction
The mitochondrial aspartate/glutamate carrier isoform 1 (AGC1), encoded by the SLC25A12 gene, is a member of the solute carrier family 25 (Palmieri, 2004, 2013). This transporter catalyzes an exchange between intramitochondrial aspartate and cytosolic glutamate plus a proton across the mitochondrial membrane (Palmieri et al., 2001). It plays an important role in the malate/aspartate shuttle, in urea synthesis and in gluconeogenesis from lactate. As a component of the malate/aspartate shuttle, AGC1 transfers the reducing equivalents of NADH + H+ from the cytosol into mitochondria (Indiveri et al., 1987; Palmieri, 2004). Two AGC isoforms, AGC1 and AGC2, are present in man; AGC1 is expressed in heart, skeletal muscle and brain, while AGC2 is expressed in many tissues, particularly in the liver (Iijima et al., 2001). AGC1 is the main AGC isoform in brain, in particular in neurons (del Arco et al., 2002; Contreras et al., 2010). The N-terminal domain of its 678-amino acid sequence contains four EF-hand Ca2+-binding sites, which were conclusively shown to bind Ca2+ in vitro and in vivo (del Arco and Satrustegui, 1998; Lasorsa et al., 2003). Through this interaction, cytosolic Ca2+ stimulates AGC1 and mitochondrial metabolism activity (Palmieri et al., 2001; Lasorsa et al., 2003; Contreras et al., 2007). Instead, the C-terminal domain of AGC1 contains six transmembrane domains and a characteristic mitochondrial carrier family (MCF) signature motif, like all the other members of the SLC25 or MC family (Palmieri, 2004).
Studies in animal models have highlighted the relevance of AGC1 in the physiology of neurons. AGC1 knockout mice showed a dramatic drop in brain aspartate levels, with a concomitant reduction in N-acetylaspartate (NAA) synthesis and hypomyelination (Jalil et al., 2005). The connection between lack of AGC1 and drop in NAA synthesis may due to the lack of mitochondrial aspartate output, which in turn would limit availability of NAA-derived acetate needed for lipid biosynthesis resulting in hypomyelination. Numerous studies have indeed demonstrated that acetate moieties of NAA are incorporated into brain lipids during the development of the central nervous system, hence strongly suggesting that AGC1 may be crucially involved in the myelination process (D’Adamo and Yatsu, 1966; D’Adamo et al., 1968; Patel and Clark, 1979; Burri et al., 1991; Mehta and Namboodiri, 1995; Chakraborty et al., 2001; Ledeen et al., 2006; Namboodiri et al., 2006; Moffett et al., 2013). In support of this conclusion, children harboring mutations of the SLC25A12 gene display severe developmental delay, epilepsy, hypotonia hallmarked by hypomyelination and decreased NAA in the brain (Wibom et al., 2009; Falk et al., 2014). The chromosomal region containing the gene encoding AGC1 has also been identified as a putative autism susceptibility locus (Ramoz et al., 2004; Turunen et al., 2008; Palmieri et al., 2010). In addition, interest in the involvement of mitochondria in neurodegenerative and neuroinflammamtory disorders, such as Parkinson's and Alzheimer's disease, and multiple sclerosis is emerging (Lin and Beal, 2006)
Despite the well-established role of NAA in myelin biosynthesis, it is still unknown in which subcellular compartment the biosynthesis occurs. Different studies have provided evidence that the aspartate-N-acetyltransferase (Asp-NAT), the enzyme that catalyzes the biosynthesis of NAA, is localized in the mitochondria (Patel and Clark, 1979; Madhavarao et al., 2003; Arun et al., 2009). However, other studies performed in primary neuronal cultures established that Asp-NAT is located in the endoplasmic reticulum as well (Wiame et al., 2009; Tahay et al., 2012). A colocalization was reported by other authors (Lu et al., 2004; Ariyannur et al., 2010).
The cAMP response element-binding protein (CREB) has been widely investigated as a key metabolic sensor and regulator of energetic homeostasis (Iacobazzi et al., 2005; Altarejos and Montminy, 2011). Importantly, CREB protein is also one of the major transcriptional factors that regulates the expression of genes necessary for the development and function of the nervous system and such activities require CREB binding to – and transcription regulation of genes containing the cAMP response elements (CRE) (Lonze and Ginty, 2002).
The transcriptional activity of CREB is induced through serine phosphorylation in its conserved kinase inducible domain by the cAMP-dependent protein kinase (PKA) (Sands and Palmer, 2008), Ca2+/calmodulin protein kinase (Enslen et al., 1994), ribosomal S6 kinase (RSK) and mitogen/stress-activated kinase (MSK) families (Deak et al., 1998). Furthermore, the phosphorylation-dependent activation of CREB involves its interaction with basal transcription factors, adaptor(s), constitutive and inducible coactivators, which contribute to form a transcriptional complex (Sheng and Greenberg, 1990). Although the transport activity and the functional role of some mutations of AGC1 have been investigated, nothing is known about the molecular mechanisms of its gene expression in any cell (Palmieri et al., 2001; Lasorsa et al., 2003; Ramoz et al., 2004; Jalil et al., 2005; Contreras et al., 2007; Wibom et al., 2009).
In this work we demonstrate that in neuronal cells CREB is a main activating transcription factor of AGC1 gene expression and cytosolic Ca2+ stimulates AGC1 gene expression through CREB. Furthermore, we also shown that AGC1 is down-regulated in neuroinflammation and is up-regulated in neuronal differentiation. Therefore our data provide an important link for the activation of AGC1 in physiological and pathological conditions.
2. Experimental
2.1. Construction of plasmids
To analyze the promoter of the human AGC1 gene, reporter plasmid constructs were prepared. Progressive deletion fragments of the region from −1995 to −15 bp of the AGC1 gene promoter were amplified by PCR and cloned into the pGL3 basic-luciferase (LUC) vector (Promega) upstream of the LUC gene-coding sequence as previously described (Iacobazzi et al., 2009a). The CREB expression vector (pcDNA3-CREB) was obtained by cloning the human CREB (GenBank Accession No. NM_004379.3) cDNA into the pcDNA3.1 vector (Invitrogen). The sequences of all constructs were verified by DNA sequencing.
2.2. Cell culture, RNA interference and transient transfection
Human neuroblastoma SH-SY5Y cells (ATCCID: CRL-2266) were grown as described (Infantino et al., 2013a). Transient transfection experiments were performed using FuGENE® HD Transfection Reagent (Promega) and 0.5 μg of each construct reported above, as previously described (Iacobazzi et al., 2009b). LUC activity in cell extracts was measured in a 96-well plate format by using a VICTOR3 multi-label plate reader (PerkinElmer, Waltham, MA, USA). The extent of transfection was normalized by β-galactosidase activity (Infantino et al., 2011). In RNA interference experiments, the specific pre-designed small interfering RNA (siRNA) targeting human CREB (s3489, Ambion) was transfected in SH-SY5Y cells (Iacobazzi et al., 2009c).
2.3. SH-SY5Y differentiation
For SH-SY5Y cell differentiation, growth medium was replaced with serum-free medium for 16 h. The cells were then stimulated with the differentiation medium consisting of serum-free medium supplemented with 0.5 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma–Aldrich, St. Louis, MO, USA) and 1 mM dibutyryl-cAMP (db-cAMP; Sigma) (Deng et al., 2001). SH-SY5Y cell differentiation was monitored by evaluating cell morphology under phase-contrast microscopy and by measuring the expression levels of neuron-specific synaptophysin (SYP) and synapse-associated protein-97 (SAP97), two neuronal differentiation markers. Cells were harvested at the indicated time points and total RNA and proteins were isolated and analyzed.
2.4. Treatments
Where indicated, SH-SY5Y cells were incubated with 10 μM forskolin (Sigma) (Monaghan et al., 2008), 20 ng/ml recombinant human neuregulin-1 (NRG-1, Immuno-Tools GmbH, Friesoythe, Germany) (Echave et al., 2009), 100 nM thapsigargin (Sigma) (Pulver et al., 2004) or 10 μM 1,2-bis(2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester) (BAPTA-AM) (Sigma) (Petroni et al., 2013) 16 h after having been depleted of serum; 10 μM H89 (Sigma) (Monaghan et al., 2008) or 10 μg/ml STO-609 (Sigma) (Tokumitsu et al., 2002) were added 1 h prior. Cells were harvested and total RNA and proteins were isolated and analyzed.
2.5. Activating stimuli
SH-SY5Y cells were treated up to 96 h with a combination of 500 U/ml IFNγ (Immuno-Tools GmbH, Friesoythe, Germany), 1000 U/ml TNFα (Sigma–Aldrich) and 10 ng/ml IL-1β (Immuno-Tools GmbH, Friesoythe, Germany). Cells were harvested at the indicated time points and total RNA and proteins were isolated and analyzed.
2.6. Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as previously reported (Iacobazzi et al., 2008). Briefly, 2 × 107 SH-SY5Y cells (exposed up to 72 h to 0.5 mM IBMX and 1 mM db-cAMP) were fixed by 1% formaldehyde at 37 °C for 10 min; afterwards, the cells were lysed and fragments of 400–500 bp. The chromatin was immunoprecipitated for 14–16 h at 4 °C using a specific antibody to CREB (Thermo Fisher Scientific, Catalog No. MA5-15154). After reverse cross-linking, chromatin immunoprecipitates were purified, then 2 μl of each sample was analyzed by PCR (35 cycles) using a forward primer (5′-CGTCCCATGCCAATTTAGGAGCAT-3′) and a reverse primer (5′-AGGTTCCGACGGATCAAAGAGCAC-3′) suitable to amplify the −565/−409 bp region of the AGC1 gene promoter.
2.7. Other methods
Electrophoretic mobility shift assays (EMSA) were performed according to Menga et al. (Menga et al., 2013). Briefly, a biotin 3′-end labeled DNA probe from the −531 to −511 bp region of the human AGC1 gene promoter was incubated with 10 μg of SH-SY5Y nuclear extracts (exposed up to 24 h to 0.5 mM IBMX and 1 mM db-cAMP) for 20 min at room temperature. Total RNA was extracted from 3 × 106 SH-SY5Y cells, and reverse transcription was performed as described previously (Infantino et al., 2007). Real-time PCR was conducted as described (Convertini et al., 2011). Assay-on-demand for human AGC1(Hs00186535_m1) and human β-actin (4326315E) was purchased from Applied Biosystems. Western blotting analysis was performed according to Infantino et al. (2013b). Anti-AGC1 (sc-271056, Santa Cruz Biotechnology), anti-β-actin (Santa Cruz Biotechnology), anti-T-CREB, anti-P-CREB (Ser 133) (sc-101663, Santa Cruz Biotechnology), anti-SAP97 (sc-9961, Santa Cruz Biotechnology), anti-SYP (sc-9116, Santa Cruz Biotechnology) or anti-β-amyloid (sc-9129, Santa Cruz Biotechnology) antibodies were used for immunoreaction. Band intensities were analyzed densitometrically using a Fluor-S MultiImager and Quantity One software (Bio-Rad). Cell viability was evaluated by the CellTiter 96® Non-Radioactive Cell Proliferation Assay (Promega). In brief, cells were seeded onto 96-well microtiter plates and treated with siRNA targeting CREB (siCREB) for up to 72 h. After incubation, 15 μl of the Dye Solution was added to each well and the cells were incubated for 4 h at 37 °C. Subsequently, solubilization solution/stop mix (100 μl) was added to each well and the cells were incubated for 1 h at 37 °C to promote the solubilization of formazan crystals. The level of MTT formazan was determined by measuring its absorbance at 570 nm using a 96-well plate reader (VICTOR3, PerkinElmer).
2.8. Statistical analysis
All data are presented as means ± S.D. for the number of experiments indicated in each case. Statistical analysis was performed using one-way ANOVA. p-Value <0.05 was considered statistically significant.
3. Results
3.1. AGC1 expression is downregulated by inflammatory cytokines
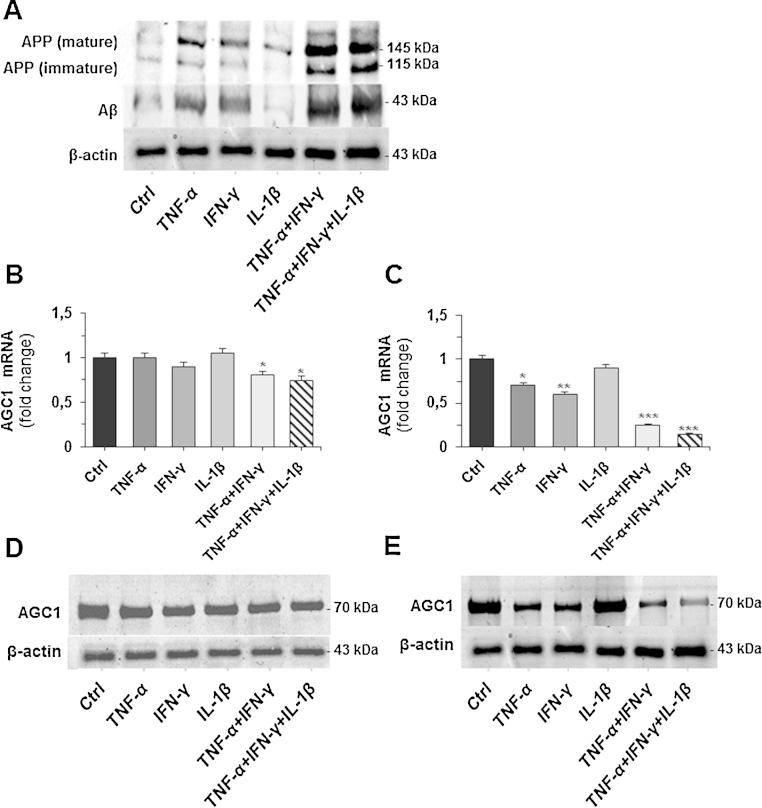
A typical feature of neuroinflammation is the presence in neuronal cells of pro-inflammatory cytokines, such as TNFα, IFNγ and IL-1β (McGeer and McGeer, 1995; Akiyama et al., 2000; Heneka et al., 2010; Zhao et al., 2011). Therefore, we asked whether the expression of the AGC1 gene is influenced by TNFα, IFNγ and IL-1β (individually or in combination). First, we measured the expression of the Aβ peptide, a typical marker of neurodegeneration (Blasko et al., 1999; Zhao et al., 2011) after 24 and 96 h of treatment with TNFα 1000 U/ml, IFNγ 500 U/ml and IL-1β 10 ng/ml alone or in combination. No significant effect was observed after 24 h of treatment (data not shown). After 96 h, the Aβ peptide was evident in TNFα and IFNγ individual treatments and more significantly in the cytokines combination treatment (Fig. 1A). SH-SY5Y cells were subsequently treated with the cytokines individually or in combination for 24 or 96 h. Real-time PCR performed on mRNA extracted from TNFα-, IFNγ- and IL-1β treated and untreated cells did not show any change in AGC1 mRNA after 24 h; whereas a decrease of about 20% was observed in cells treated with combination of TNFα/IFNγ and TNFα/IFNγ/IL-1β as compared with untreated cells (Fig. 1B). A major reduction of AGC1 gene expression was found in cells treated with individual TNFα (about 30%) and IFNγ (about 40%) after 96 h. A more significant reduction (about 80%) was seen in the cytokines combination treatment as compared with untreated cells (Fig. 1C). Accordingly, Western blot analysis demonstrated a major reduction of AGC1 protein level after 96 h of TNFα/IFNγ and TNFα/IFNγ/IL-1β treatment as compared with 24-h cytokines-treated cells (Figs. 1D and E).
Fig. 1.
Effect of pro-inflammatory cytokines on AGC1 gene expression. (A) SH-SY5Y cells were treated with IFNγ (500 U/ml), TNFα (1000 U/ml) and IL-1β (10 ng/ml) alone or in combination for 24 and 96 h. Cells lysates were subjected to immunoblots with mature and immature APP, amyloid beta protein (Aβ) and β-actin antibodies. (B) AGC1 mRNA in SH-SY5Y cells treated for 24 h with the same cytokines as in (A) was quantified by real-time PCR. Data are expressed as means ± S.D. of five duplicate independent experiments. *P < 0.05 versus control, (one-way ANOVA). (C) AGC1 mRNA of SH-SY5Y cells treated for 96 h as above was quantified by real-time PCR. Data are expressed as means ± S.D. of five duplicate independent experiments. *P < 0.05, ***P < 0.001 versus control. (D) AGC1 and β-actin proteins of SH-SY5Y cells treated or untreated with cytokines for 24 h as above, were immunodetected with specific antibodies. Similar results were obtained in three independent experiments. (E) AGC1 and β-actin proteins of SH-SY5Y cells treated or untreated with cytokines for 96 h as above, were immunodetected with specific antibodies. Similar results were obtained in three independent experiments.
These data show that AGC1 protein levels are significantly down-regulated following prolonged exposure to stimuli that mimic neuroinflammation.
3.2. CREB is a main regulator of AGC1 gene expression in neuronal cells
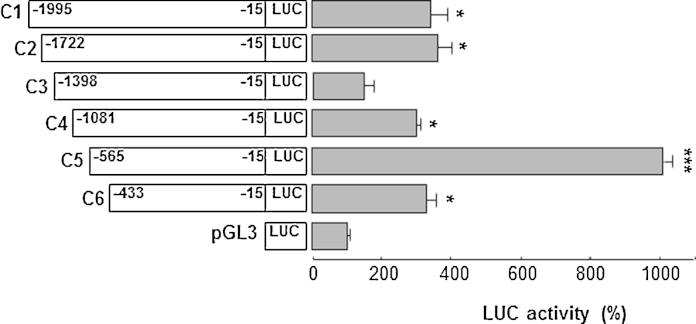
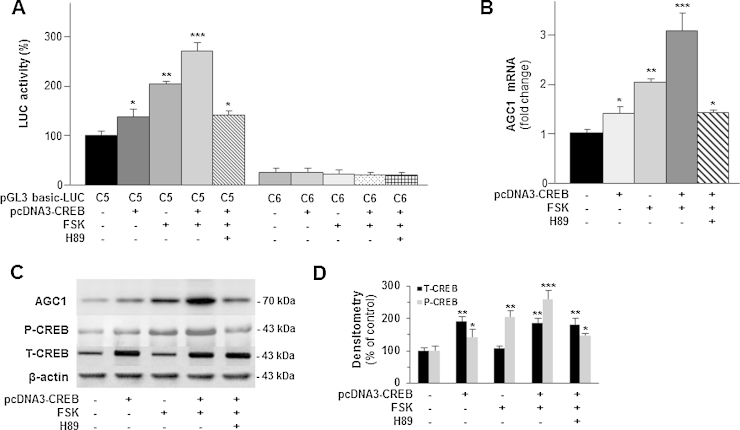
To investigate the regulation of AGC1 gene expression in neuronal cells, different deletion fragments of the AGC1 gene promoter were amplified, cloned into the pGL3 basic-LUC vector and transiently transfected into SH-SY5Y cells. The resulting pattern of reporter LUC activity is shown in Fig. 2. Deletion of the 5′-flanking region from position −1722 bp to position −1398 bp (pGL3-C3-LUC vector) caused a decrease in gene reporter activity of about 50% as compared to the activities of the constructs pGL3-C1-LUC from −1995 bp and pGL3-C2-LUC from −1722 bp. In contrast, deletion from −1081 bp to −565 bp (pGL3-C5-LUC construct) resulted in a drastic increase (>200%) in reporter gene activity, suggesting that a strong enhancer is located within this region. Basal LUC activity was restored with deletion from −565 bp to −433 bp (pGL3-C6-LUC vector). Preliminary, the transcription start site was found at position −82 bp upstream the ATG codon. (http://dbtss.hgc.jp/index.html). To identify the transcription factors present in construct pGL3-C5-LUC that strongly activate AGC1 gene expression, an in silico analysis was performed using different programs: the TFSEARCH Program (http://www.cbrc.jp/research/db/TFSEARCH.html), the Alibaba 2.1 (http://www.gene-regulation.com/pub/programs/alibaba2/index.html), and the LASAGNA-Search (http://biogrid-head.engr.uconn.edu/lasagna_search/). Among several potential factor-binding motifs, a CREB binding site was present at position −525/− 517 bp upstream the ATG translation codon. Because CREB is a key factor that regulates many neural functions (Lonze and Ginty, 2002), we investigated CREB transcriptional activity on the AGC1 gene by overexpressing CREB in SH-SY5Y cells. SH-SY5Y cells transfected with the pcDNA3-CREB and pGL3 basic-LUC vectors harboring the C5 fragment of the AGC1 gene promoter were incubated in the presence or absence of forskolin, an activator of adenylate cyclase (Monaghan et al., 2008). LUC activity was enhanced by about 35% in cells transfected with the pcDNA3-CREB and pGL3-C5-LUC vectors as compared to cells transfected with the pGL3-C5-LUC vector alone used as control (Fig. 3A). Forskolin added to SH-SY5Y cells transfected with the pGL3-C5-LUC vector alone increased reporter activity by about 100% and to cells transfected with both the pGL3-C5-LUC and pcDNA3-CREB vectors by about 200%, as compared to control (Fig. 3A). Furthermore, forskolin-mediated increase of the reporter activity was strongly reduced by H89, a PKA inhibitor (Monaghan et al., 2008) (Fig. 3A). In contrast, LUC activity was not affected by expression of CREB or by forskolin when the control construct pGL3-C6-LUC (without the CREB binding site of the AGC1 gene promoter) was used instead of construct pGL3-C5-LUC. The results of these luciferase assays were further corroborated by monitoring the levels of the AGC1 mRNA with real-time PCR (Fig. 3B). Furthermore, to demonstrate the involvement of CREB, in its active form, in the regulation AGC1 gene expression, we compared the levels of phosphorylated CREB (P-CREB) protein relative to total CREB (T-CREB) and AGC1 protein expression. This analysis showed that AGC1 and P-CREB progressively and simultaneously increased as a consequence of both CREB over-expression and treatment with forskolin alone or in combination (Fig. 3C and quantified in Fig. 3D). Significantly, the addition of H89 abolished any of these increasing effects (Fig. 3C). The analysis of the relative ratio of P-CREB versus T-CREB with densitometry further indicated that forskolin specifically induced the accumulation of the active phosphorylated form of CREB. Specifically, P-CREB increased progressively of 50, 100 and 150%, in presence of pcDNA3-CREB, forskolin, and foskolin/pcDNA3-CREB, respectively, relative to untreated cells (Fig. 3D). All of the above-reported results provide evidence for a direct involvement of active, phosphorylated CREB in the regulation of AGC1 expression in SH-SY5Y cells.
Fig. 2.
Luciferase activity from AGC1 gene promoter constructs. Deletion analysis of the 5′-flanking region of the human AGC1 gene. The deletion fragments named C1–C6 were cloned into the pGL3 basic-LUC vector and tested for LUC expression activity in transfected SH-SY5Y cells. pGL3 indicates the pGL3 basic-LUC vector alone. Numbering indicates the extent of fragments, while gray bars indicate LUC activity. The values were set relative to the pGL3. Means ± S.D. of five duplicate independent experiments are shown. The differences compared to the pGL3 were analyzed by one-way ANOVA. *P < 0.05, ***P < 0.001.
Fig. 3.
Functional characterization of CREB.
(A) SH-SY5Y transfected with pGL3-basic-Luc vector containing (C5) or not (C6) the CREB binding site. Where indicated a cotransfection with pcDNA3-CREB and/or a treatment with 10 μM forskolin for 3 h, and addition of 10 μM H89 1 h before forskolin were performed before luciferase assay. Data are expressed as means ± S.D. of five duplicate independent experiments.*P < 0.05, **P < 0.01, ***P < 0.001 versus control (one-way ANOVA). (B) Total RNA extracted from SH-SY5Y cells, which had been (where indicated) transfected with pcDNA3-CREB and/or treated with 10 μM forskolin for 1 h was used to quantify AGC1 mRNA by real-time PCR. Where indicated 10 μM H89 was added 1 h before forskolin. Data are expressed as means ± S.D. of five duplicate independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus untreated control (one-way ANOVA). (C) AGC1, P-CREB, T-CREB and β-actin proteins of SH-SY5Y cells treated as above (B) were immunodetected with specific antibodies. P-CREB blot was reprobed for T-CREB quantification. A representative of four blots is shown. (D) Bands intensities of T-CREB and P-CREB were analyzed densitometrically using a Fluor-S MultiImager and Quantity One software (Bio-Rad) and corrected for β-actin. Data are expressed as means ± S.D. of four individual experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus untreated control.
3.3. The decrease in AGC1 expression in neuroinflammation is most likely caused by the downregulation of CREB
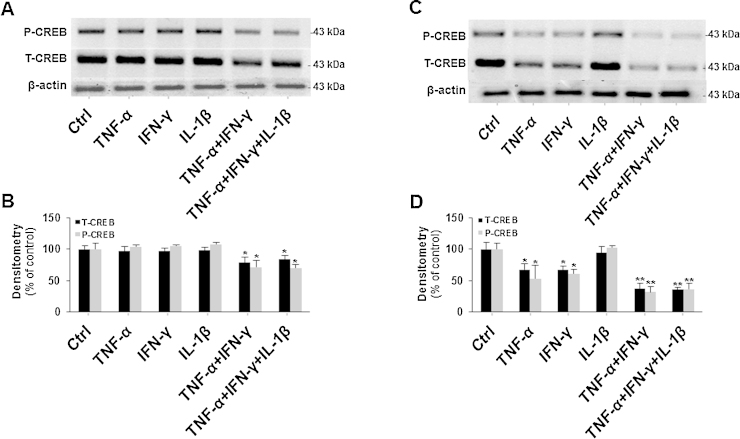
Having established that CREB is a main regulator of AGC1 expression, we asked whether the decrease in AGC1 expression induced by the pro-inflammatory cytokines might be due to a decrease in CREB expression. To this end, SH-SY5Y cells were treated with TNFα, IFNγ and IL-1β individually or in combination for 24 and 96 h; then cell lysates were prepared for immunoblot analysis of the cellular P-CREB and T-CREB content. When SH-SY5Y were exposed to individual cytokines for 24 h no effect was found on P-CREB and T-CREB (Fig. 4A), as confirmed also by densitometric analysis (Fig. 4B). A reduction of P-CREB and T-CREB protein (20%) was evident only in the treatment with combination of cytokines after 24 h (Fig. 4A and B). Both P-CREB and T-CREB decreased significantly (40 and 30%, respectively), after 96 h of treatment with TNFα and/or IFNγ alone. A dramatic decrease (70%) was observed upon treatment with combination of cytokines, TNFα + IFNγ and TNFα + IFNγ + IL-1β (Fig. 4C and D). No effect was evident upon IL-1β treatment. This reduction of CREB expression is fully consistent with previous reports showing that chronic exposure to TNFα, IFNγ, IL-1β, decreases CREB expression (Jambal et al., 2003). Furthermore, Aβ accumulation was shown to be inversely correlated with CREB levels in the brain of patients with Alzheimer Disease (Pugazhenthi et al., 2011). Other studies also reported that CREB-mediated expression of brain-derived neurotrophic factor (BDNF), B-cell lymphoma (Bcl-2) and baculovirus inhibitor of apoptosis proteins repeat-containing proteins (BIRC) 3 and 4 is impaired in neuroinflammation (Pugazhenthi et al., 2011). On this bases we argue that the decrease in CREB content induced by TNFα/IFNγ and TNFα/IFNγ/IL-1β could be likely linked to the reduction of AGC1 expression in neuroinflammation. However, at present other factors cannot be excluded to be involved in gene downregulation in neuroinflammation.
Fig. 4.
Downregulation of CREB in cytokine-treated SH-SY5Y cells. P-CREB, T-CREB and β-actin proteins of SH-SY5Y cells treated or untreated with 500 U/m IFNγ, 1000 U/ml TNFα and 10 ng/ml IL-1β individually or in cambination for 24 h were immunodetected with specific antibodies. P-CREB blot was reprobed for T-CREB quantification. Similar results were obtained in four independent experiments. (B) The intensities of P-CREB and T-CREB in (A) were analyzed densitometrically using a Fluor-S MultiImager and Quantity One software (Bio-Rad) and corrected for β-actin. Data are expressed as means ± S.D. of four individual experiments. *P < 0.05, **P < 0.01 versus untreated control (one-way ANOVA). (C) P-CREB, T-CREB and β-actin proteins of SH-SY5Y cells treated or untreated as above for 96 h were immunodetected with specific antibodies. P-CREB blot was reprobed for T-CREB quantification. Similar results were obtained in four independent experiments. (D) The intensities of P-CREB and T-CREB in (C) were analyzed densitometrically using a Fluor-S MultiImager and Quantity One software (Bio-Rad) and corrected for β-actin. Data are expressed as means ± S.D. of four individual experiments. *P < 0.05, **P < 0.01 versus untreated control.
3.4. Cytosolic Ca2+ level affects AGC1 gene expression via CREB
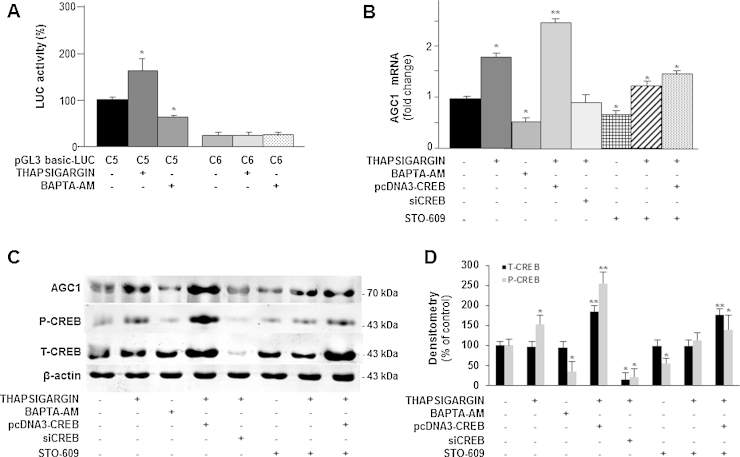
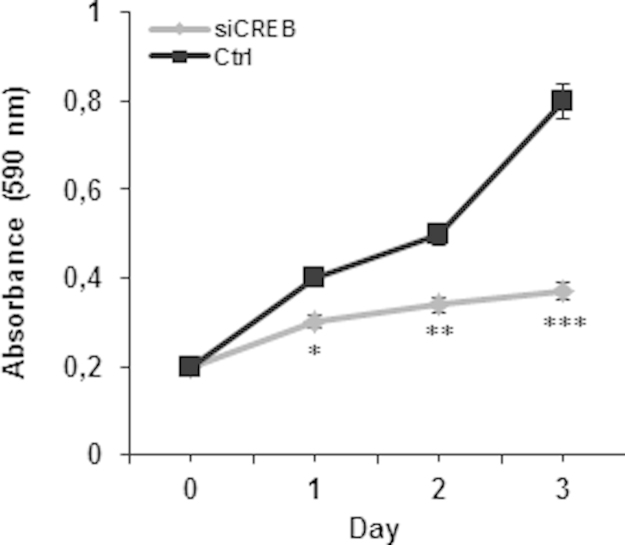
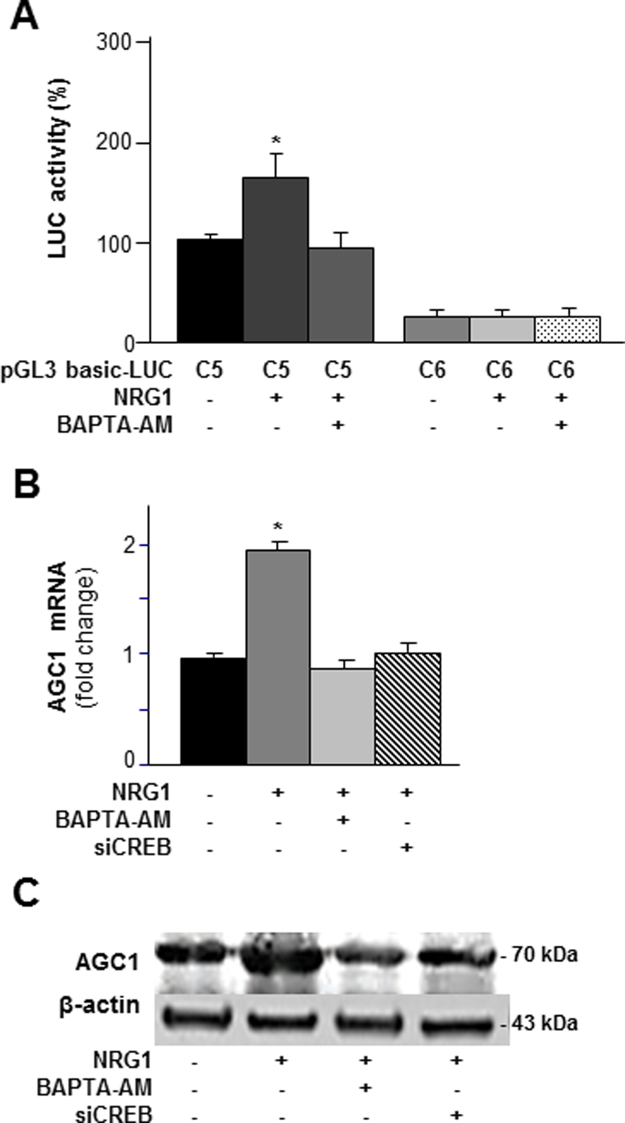
Because Ca2+ is a well-known CREB inducer (Sheng and Greenberg, 1990) and is also known to stimulate AGC1 activity (Palmieri et al., 2001; Lasorsa et al., 2003; Contreras et al., 2007), we investigated whether alterations in the pool of intracellular Ca2 affect AGC1 gene expression. The influence of intracellular Ca2+ on AGC1 gene expression was studied by using two Ca2+ modulators: BABTA-AM, a calcium chelator that penetrates live cells and decreases intracellular Ca2+ concentration (Petroni et al., 2013), and thapsigargin, which causes an increase in cellular Ca2+ level by inhibiting the (sarco) endoplasmatic reticulum Ca2+-ATPase (SERCA) (Pulver et al., 2004). SH-SY5Y cells were transfected with the pGL3-C5-LUC construct and incubated with 10 μM BABTA-AM for 3 h or 100 nM thapsigargin for 6 h. Because of its toxicity, it was not possible to prolong treatment with BAPTA-AM for more than 3 h (data not shown). BAPTA-AM decreased LUC activity by about 40%, whereas thapsigargin increased it by about 80% (Fig. 5A). Notably, when the cells were transfected with the reporter luciferase construct lacking the CREB binding site, the luciferase activity was negligible (Fig. 5A), thus strongly arguing that Ca2+ acts via CREB. The involvement of Ca2+ in the induction of AGC1 through CREB was further demonstrated by using STO-609, a selective inhibitor of Ca2+/calmodulin-dependent protein kinase kinase (CaM-KK) (Tokumitsu et al., 2002). RT-PCR and Western blot showed that both AGC1 mRNA and protein levels were reduced by BAPTA-AM, by CREB silencing and by STO-609, but they were increased by thapsigargin and by combination of thapsigargin with pcDNA3-CREB (Fig. 5B and C). The pre-incubation with STO-609 significantly inhibited AGC1 mRNA and protein levels induced by thapsigargin and thapsigargin/pcDNA3-CREB (Fig. 5B and C). Consistently, P-CREB levels mirrored AGC1 expression. In fact, phosphorylated CREB increased significantly when intracellular Ca2+ was enhanced (thapsigargin) and CREB was overexpressed in presence of thapsigargin while it was reduced when Ca2+ was blocked by BAPTA-AM, when Ca2+/calmodulin-dependent protein kinase kinase (CaM-KK) was inhibited by STO-609 and when CREB was silenced (Fig. 5C and 5D). T-CREB increased only in presence of CREB overexpressing plasmid. Of note, in presence of BAPTA, T-CREB content was unaffected whereas P-CREB decreased suggesting the Ca2+ signaling pathway in the activation of CREB. We next tested the effect of siCREB on cell viability and growth was tested (Fig. S1). In another set of experiments AGC1 gene expression was investigated in conditions in which the levels of cellular N-acetylaspartate is increased by treatment with neuregulin-1, a key regulator of the myelination process (Birchmeier and Nave, 2008), which triggers an increase of intracellular Ca2+mediated by phospholipase C-γ. When SH-SY5Y cells were transfected with pGL3-C5-LUC construct in the presence of neuregulin (20 ng/ml) a significant increase of LUC activity was observed, whereas in the presence of BAPTA the activation was abolished. Accordingly, neuregulin-1 also increased the amount of AGC1 transcript and protein levels. As expected, BAPTA-AM abolished this up-regulation, further suggesting the involvement of Ca2+ in the induction of AGC1 expression (Fig. S2). Viewed together these results provide the first evidence that the levels of intracellular Ca2+ play a key role in regulation of AGC1 gene promoter activity, transcript and protein expression. Furthermore, the finding that AGC1 is also induced by neuregulin is fully consistent with the proposed role of this transporter in myelination and further suggest that such activity is also dependent upon Ca2+.
Fig. 5.
Effect of Ca2+ dysregulation on AGC1 expression. (A) SH-SY5Y cells transfected with pGL3-LUC constructs containing (C5) or not (C6) the CREB binding site were treated with 100 nM thapsigargin for 6 h or 10 μM BAPTA-AM for 3 h and then assayed for LUC expression activity. Data are expressed as means ± S.D. of five duplicate independent experiments. *P < 0.05, versus control (one-way ANOVA). (B) AGC1 mRNA from SH-SY5Y cells exposed to the following treatment: 3 h with 100 nM thapsigargin or 10 μM BAPTA-AM, transfection with pcDNA3-CREB and or with siCREB for 48 h and treated with 100 nM thapsigargin, preincubation with 10 μg/ml STO-609 for 1 h followed by addition of 100 nM thapsigargin and, where indicated, by pcDNA3-CREB were quantified by real-time PCR. Data are expressed as means ± S.D. of five duplicate independent experiments. *P < 0.05, **P < 0.01 (one-way ANOVA). (C) AGC1, P-CREB, T-CREB and β-actin proteins of SH-SY5Y cells treated as above. P-CREB blot was reprobed for T-CREB quantification, (B) were quantified by Western blotting. A representative of four blots is shown. (D) The intensities of P-CREB and T-CREB in (C) were analyzed densitometrically using a Fluor-S MultiImager and Quantity One software (Bio-Rad) and corrected for β-actin. Data are expressed as means ± S.D. of four individual experiments. *P < 0.05, **P < 0.01, versus untreated control.
Supplementary Figs. S1 and S2 related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.biocel.2015.01.004.
3.5. CREB induces AGC1 gene expression during SH-SY5Y cell differentiation
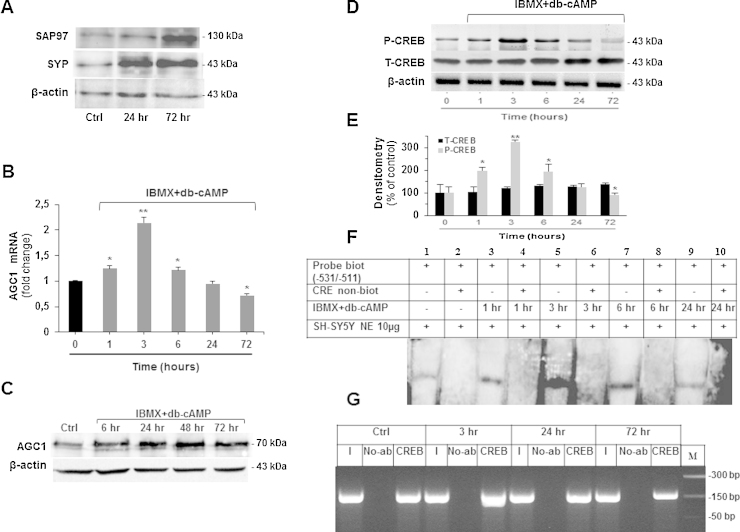
Since neurite outgrowth and differentiation require an increase in energy production (Cheng et al., 2010) and AGC1 is one of the most enriched proteins during neuronal cell differentiation (Watkins et al., 2008), we next asked whether AGC1 expression changes during differentiation of SH-SY5Y cells induced by the IBMX + db-cAMP combination (Deng et al., 2001) as inducer of the CREB pathway. SH-SY5Y cell differentiation was induced with 500 μM IBMX + 1 mM db-cAMP. Successful differentiation was confirmed by the increase of neuronal markers SAP97 and SYP assayed by immunoblot analysis (Fig. 6A). Real-time PCR performed on mRNA extracted from IBMX + db-cAMP-treated cells showed a significant increase of AGC1 mRNA level up to 3 h of incubation (>100%) followed by a progressive decline until 72 h (Fig. 6B). Conversely, immunoblot analysis performed on extracts derived from SH-SY5Y cells during differentiation showed an increase in AGC1 expression level after 6 h, which remained elevated for 72 h (Fig. 6C), suggesting a different mechanism of regulation for AGC1 transcription and translation. P-CREB and T-CREB immunoblots and densitometric quantification revealed that P-CREB increased and reached its maximum (>200%) for the first 3 h subsequently followed by progressive decrease; whereas T-CREB content was unchanged over the time (Fig. 6D and E). Of note, the levels of P-CREB mirrored the AGC1 transcript profile (Fig. 6B). To determine whether these changes were reflected in a time-dependent differential recruitment of CREB on its DNA-binding consensus sites during differentiation, we performed Electromobility Shift Assays (EMSA) and Chromatin immunoprectipitation experiments (ChIP). For the formers, cellular lysates derived from IBMX and db-cAMP-treated SH-SY5Y cells were prepared at 1, 3, 6 and 24 h and incubated with a biotynilated probe containing the CREB binding site of the AGC1 promoter. In agreement with the results of real time PCR (Fig. 6B), the activation of CREB binding activity was maximally stimulated within 3 h after IBMX + db-cAMP treatment, (Fig. 6F). In full agreement with the results obtained with EMSA, the ChIP assays similarly demonstrated optimal CREB DNA binding activity on the endogenous promoter following 3 h of induction of differentiation. After this time, a progressive reduction of CREB binding was evident for up to 72 h, likely as a consequence of a reduction of P-CREB level (Fig. 6G). In summary, our results show that CREB is actively involved in AGC1 expression during SH-SY5Y cell differentiation.
Fig. 6.
Regulatory role of CREB during SH-SY5Y cell differentiation. (A) The two neuronal differentiation markers synaptophysin (SYP) and synapse-associated protein-97 (SAP97) and β-actin of differentiated up to 72 h or undifferentiated SH-SY5Y cells were immunodetected with specific antibodies. Similar results were obtained in three independent experiments. (B) SH-SY5Y cells were seeded onto 6-well plates and grown to 50% confluence. The growth medium was replaced with serum-free medium for 16 h and the cells were then stimulated with differentiation medium consisting of serum-free medium supplemented with 0.5 mM 3-isobutyl-1-methylxanthine (IBMX) and 1 mM dibutyryl-cAMP (db-cAMP) for 1, 3, 6, 24 and 72 h. Once the time of exposure was completed the cells were lysed and used to quantify AGC1 mRNA by real-time PCR. Data are expressed as means ± S.D. of five duplicate independent experiments. *P < 0.05, **P < 0.01 versus control (one-way ANOVA). (C) AGC1 and β-actin proteins of differentiated (up to 72 h) or undifferentiated SH-SY5Y cells were immunodetected with specific antibodies. Similar results were obtained in three independent experiments. (D) P-CREB, T-CREB and β-actin proteins of differentiated (up to 72 h) or undifferentiated SH-SY5Y cells were immunodetected with specific antibodies. P-CREB blot was reprobed for T-CREB quantification. Similar results were obtained in three independent experiments. (E) The intensities of P-CREB and T-CREB in (D) were analyzed densitometrically using a Fluor-S MultiImager and Quantity One software (Bio-Rad) and corrected for β-actin. Data are expressed as means ± S.D. of four individual experiments. *P < 0.05, **P < 0.01, versus untreated control. (F) The biotinylated probe from −531 to −511 bp of AGC1 gene promoter was incubated for 30 min with 10 μg of SH-SY5Y nuclear extracts (SH-SY5Y NE) in the presence (lanes 3, 5, 7 and 9) or absence (lane 1) of 0.5 mM IBMX and 1 mM db-cAMP for the indicated time periods. Where indicated 10-fold excess of non-biotinylated probe was added (lanes 2, 4, 6, 8 and 10). (G) Chromatin of differentiated (up to 72 h) or undifferentiated SH-SY5Y cells was immunoprecipitated by anti-CREB (CREB lanes) antibody. PCR was performed using forward and reverse primers encompassing the AGC1 gene promoter from −565 to −409 bp. Lanes No-Ab, PCR of the precipitates without antibody; lanes I, PCR of input DNA dilutions (1/10).
4. Discussion
In the present study, we have identified for the first time a regulatory element within the human AGC1 gene promoter (comprised within 565–433 bp) that contains a CRE site and whose activity is exquisitely regulated by CREB during physiological and pathological signals. Among the reported results the following supporting evidence can be mentioned. In neuronal SH-SY5Y cells the overexpression of CREB enhances gene reporter expression activity. Vice versa silencing of CREB reduces gene reporter activity. Consistently, the levels of both AGC1 transcript and protein were increased by overexpression of CREB and decreased by CREB knock-down.
Our data also provide evidence that the CREB-induced activation of AGC1 gene transcription in SH-SY5Y cells is accomplished by two known mechanisms leading to phosphorylation of CREB: the cAMP-dependent protein kinase pathway and the Ca2+ signaling to CREB pathway. The relevance of the cAMP-protein kinase pathway in the regulation of AGC1 gene transcription is demonstrated by the observation that forskolin, an activator of adenylate cyclase (Monaghan et al., 2008), increases, and H89, an inhibitor of the cAMP-dependent protein kinase pathway (Monaghan et al., 2008), decreases the luciferase reporter activity, AGC1 mRNA and AGC1 protein levels. The role of Ca2+ signaling via the CREB pathway is further demonstrated by the inhibition of AGC1 gene expression and protein levels by BAPTA-AM, which reduces intracellular Ca2+ (Petroni et al., 2013), as well as by its activation by thapsigargin, which raises cytosolic Ca2+ (Pulver et al., 2004), and by inhibition of Ca2+/calmodulin-dependent protein kinase-kinase [which inhibits the phosphorylation of CaMKIV pathway (Tokumitsu et al., 2002)]. Furthermore, when CREB was silenced, changing intracellular Ca2+ had no effect on AGC1 mRNA and protein. Therefore, cytosolic Ca2+ regulates not only the activity of AGC1 by reacting with the EF-hand Ca2+ binding sites present in the AGC1 N-terminal domain as previously shown (Palmieri et al., 2001; Lasorsa et al., 2003; Contreras et al., 2007), but also activates AGC1 gene expression by phosphorylating CREB. Interestingly, the higher levels of AGC1 transcript and protein, together with higher Ca2+ levels, in post-mortem brains from patients affected by autism as compared to control subjects (Palmieri et al., 2010; Napolioni et al., 2011) can be hypothesized to be due to a CREB-mediated increased AGC1 gene expression. Furthermore, it is likely that excessive increase in AGC1 protein and AGC1 activity as a consequence of sustained elevation of cellular Ca2+ may affect global metabolism leading to oxidative stress and cellular dysfunction (Napolioni et al., 2011).
AGC1 plays an important role in the central nervous system particularly in the process of myelination through the production of NAA which is the precursor of myelin (Jalil et al., 2005; Madhavarao et al., 2005; Wibom et al., 2009). This involvement is further showed in this study since SH-SY5Y cells treated with Neuregulin, a key regulator of myelination process, which triggers an increase of intracellular Ca2+ mediated by phospholipase C, up-regulated AGC1 expression. In addition, CREB, which was shown here to be a key positive transcriptional regulator of AGC1 gene expression, is known to activate transcription of target genes involved in the growth, survival and synaptic plasticity of neurons, memory, learning and mitochondrial biogenesis (Lonze and Ginty, 2002). Most of CREB-dependent genes that encode enzymes involved in energy production are down-regulated in neuroinflammation and neurodegenerative disorders (Tong et al., 2001, 2004; Saura and Valero, 2011). For these reasons, in the present study the regulation of AGC1 gene expression has been explored in neuronal cells under pathological (inflammation) and physiological (differentiation) conditions.
Oxidative stress induced by cytokines is known to play an important role in neurodegenerative process, such as in Alzheimer's (Hensley et al., 1994; Nicolle et al., 2001). Since AGC1 is involved in energy production, in this work we evaluated the effect of cytokines, TNFα, IFNγ and IL-1β, on AGC1 expression. Among the different treatments, a pronounced decrease of AGC1 mRNA and protein in neuronal cells was observed upon treated with the combination of pro-inflammatory cytokines, TNFα and IFNγ, and TNFα, IFNγ and IL-1β. Of note, the cytokines-mediated down-regulation of AGC1 via CREB is exacerbated by prolonged pro-inflammatory stimuli. Although the mechanism is not known, it is very likely that oxidative stress, due to free radical generation and inflammatory responses, induces change in transcriptional events including down regulation of CREB and CREB-regulated genes. It is noteworthy that in the oxidative stress CREB decreases especially in the regions where astrocytes are abundant. Since astrocytes play an important role in neurodegeneration through release of cytokines (Wyss-Coray, 2006; Glass et al., 2010), it will be interesting to evaluate the specific AGC1 expression in this region.
Neuronal differentiation is associated with increased mitochondrial biogenesis and increased demand of metabolic energy (Cheng et al., 2010). During neuronal cell differentiation we have found a CREB-dependent activation of AGC1 gene expression. In SH-SY5Y cells the level of AGC1 protein was increased after 6 h of the CREB pathway induction with the IBMX + db-cAMP combination and remained elevated until 72 h. The maximum increase in AGC1 transcript was reached after 3 h of induction, as shown by real-time PCR, and parallels the increases of P-CREB, the binding of the DNA CRE site to nuclear extracts and the anti-CREB antibody immunoprecipitated DNA. This finding suggests that the half-life time of AGC protein is markedly high.
In conclusion, we have demonstrated that CREB regulates AGC1 gene expression in neuronal cells via PKA/Ca2+-mediated activation. Since AGC1 is involved in myelinogenesis, CREB might be used as drug target to increase AGC1 gene expression in neurodegenerative diseases.
Acknowledgements
This work was supported by grants from the Ministero dell’Università e della Ricerca (MIUR), the Center of Excellence on Comparative Genomics (GEGBA), the Comitato Telethon Fondazione Onlus N. GGP11139, and the Universities of Bari “Aldo Moro” and Basilicata.
References
- Akiyama H., Barger S., Barnum S., Bradt B., Bauer J., Cole G.M. Inflammation and Alzheimer's disease. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos J.Y., Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyannur P.S., Moffett J.R., Manickam P., Pattabiraman N., Arun P., Nitta A. Methamphetamine-induced neuronal protein NAT8L is the NAA biosynthetic enzyme: implications for specialized acetyl coenzyme A metabolism in the CNS. Brain Res. 2010;1335:1–13. doi: 10.1016/j.brainres.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Arun P., Moffett J.R., Namboodiri A.M. Evidence for mitochondrial and cytoplasmic N-acetylaspartate synthesis in SH-SY5Y neuroblastoma cells. Neurochem Int. 2009;55(4):219–225. doi: 10.1016/j.neuint.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Nave K.A. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia. 2008;56:1491–1497. doi: 10.1002/glia.20753. [DOI] [PubMed] [Google Scholar]
- Blasko I., Marx F., Steiner E., Hartmann T., Grubeck-Loebenstein B. TNFalpha plus IFNgamma induce the production of Alzheimer beta-amyloid peptides and decrease the secretion of APPs. FASEB J. 1999;13:63–68. doi: 10.1096/fasebj.13.1.63. [DOI] [PubMed] [Google Scholar]
- Burri R., Steffen C., Herschkowitz N. N-acetyl-l-aspartate is a major source of acetyl groups for lipid synthesis during rat brain development. Dev Neurosci. 1991;13(6):403–411. doi: 10.1159/000112191. [DOI] [PubMed] [Google Scholar]
- Chakraborty G., Mekala P., Yahya D., Wu G., Ledeen R.W. Intraneuronal N-acetylaspartate supplies acetyl groups for myelin lipid synthesis: evidence for myelin-associated aspartoacylase. J Neurochem. 2001;78:736–745. doi: 10.1046/j.1471-4159.2001.00456.x. [DOI] [PubMed] [Google Scholar]
- Cheng A., Hou Y., Mattson M.P. Mitochondria and neuroplasticity. ASN Neuro. 2010;2:e00045. doi: 10.1042/AN20100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras L., Gomez-Puertas P., Iijima M., Kobayashi K., Saheki T., Satrustegui J. Ca2+ Activation kinetics of the two aspartate–glutamate mitochondrial carriers, aralar and citrin: role in the heart malate–aspartate NADH shuttle. J Biol Chem. 2007;282:7098–7106. doi: 10.1074/jbc.M610491200. [DOI] [PubMed] [Google Scholar]
- Contreras L., Urbieta A., Kobayashi K., Saheki T., Satrustegui J. Low levels of citrin (SLC25A13) expression in adult mouse brain restricted to neuronal clusters. J Neurosci Res. 2010;88:1009–1016. doi: 10.1002/jnr.22283. [DOI] [PubMed] [Google Scholar]
- Convertini P., Infantino V., Bisaccia F., Palmieri F., Iacobazzi V. Role of FOXA and Sp1 in mitochondrial acylcarnitine carrier gene expression in different cell lines. Biochem Biophys Res Commun. 2011;404:376–381. doi: 10.1016/j.bbrc.2010.11.126. [DOI] [PubMed] [Google Scholar]
- D’Adamo A.F., Jr., Yatsu F.M. Acetate metabolism in the nervous system. N-acetyl-l-aspartic acid and the biosynthesis of brain lipids. J Neurochem. 1966;13(10):961–965. doi: 10.1111/j.1471-4159.1966.tb10292.x. [DOI] [PubMed] [Google Scholar]
- D’Adamo A.F., Jr., Gidez L.I., Yatsu F.M. Acetyl transport mechanisms. Involvement of N-acetyl aspartic acid in de novo fatty acid biosynthesis in the developing rat brain. Exp Brain Res. 1968;5(4):267–273. doi: 10.1007/BF00235902. [DOI] [PubMed] [Google Scholar]
- Deak M., Clifton A.D., Lucocq L.M., Alessi D.R. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Arco A., Satrustegui J. Molecular cloning of Aralar, a new member of the mitochondrial carrier superfamily that binds calcium and is present in human muscle and brain. J Biol Chem. 1998;273:23327–23334. doi: 10.1074/jbc.273.36.23327. [DOI] [PubMed] [Google Scholar]
- del Arco A., Morcillo J., Martinez-Morales J.R., Galian C., Martos V., Bovolenta P. Expression of the aspartate/glutamate mitochondrial carriers aralar1 and citrin during development and in adult rat tissues. Eur J Biochem. 2002;269:3313–3320. doi: 10.1046/j.1432-1033.2002.03018.x. [DOI] [PubMed] [Google Scholar]
- Deng W., Obrocka M., Fischer I., Prockop D.J. In vitro differentiation of human marrow stromal cells into early progenitors of neural cells by conditions that increase intracellular cyclic AMP. Biochem Biophys Res Commun. 2001;282:148–152. doi: 10.1006/bbrc.2001.4570. [DOI] [PubMed] [Google Scholar]
- Echave P., Machado-da-Silva G., Arkell R.S., Duchen M.R., Jacobson J., Mitter R. Extracellular growth factors and mitogens cooperate to drive mitochondrial biogenesis. J Cell Sci. 2009;122:4516–4525. doi: 10.1242/jcs.049734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enslen H., Sun P., Brickey D., Soderling S.H., Klamo E., Soderling T.R. Characterization of Ca2+/calmodulin-dependent protein kinase IV. Role in transcriptional regulation. J Biol Chem. 1994;269:15520–15527. [PubMed] [Google Scholar]
- Falk M.J., Li D., Gai X., McCormick E., Place E., Lasorsa F.M. AGC1 deficiency causes infantile epilepsy, abnormal myelination, and reduced N-acetylaspartate. JIMD Rep. 2014;14:77–85. doi: 10.1007/8904_2013_287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K., Saijo K., Winner B., Marchetto M.C., Gage F.H. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140(6):918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M.T., O’Banion M.K., Terwel D., Kummer M.P. Neuroinflammatory processes in Alzheimer's disease. J Neural Transm. 2010;117:919–947. doi: 10.1007/s00702-010-0438-z. [DOI] [PubMed] [Google Scholar]
- Hensley K., Carney J.M., Mattson M.P., Aksenova M., Harris M., Wu J.F. A model for beta-amyloid aggregation and neurotoxicity based on free radical generation by the peptide: relevance to Alzheimer disease. Proc Natl Acad Sci U S A. 1994;91(8):3270–3274. doi: 10.1073/pnas.91.8.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobazzi V., Infantino V., Costanzo P., Izzo P., Palmieri F. Functional analysis of the promoter of the mitochondrial phosphate carrier human gene: identification of activator and repressor elements and their transcription factors. Biochem J. 2005;391:613–621. doi: 10.1042/BJ20050776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobazzi V., Infantino V., Palmieri F. Epigenetic mechanisms and Sp1 regulate mitochondrial citrate carrier gene expression. Biochem Biophys Res Commun. 2008;376:15–20. doi: 10.1016/j.bbrc.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Iacobazzi V., Infantino V., Bisaccia F., Castegna A., Palmieri F. Role of FOXA in mitochondrial citrate carrier gene expression and insulin secretion. Biochem Biophys Res Commun. 2009;385:220–224. doi: 10.1016/j.bbrc.2009.05.030. [DOI] [PubMed] [Google Scholar]
- Iacobazzi V., Convertini P., Infantino V., Scarcia P., Todisco S., Palmieri F. Statins, fibrates and retinoic acid upregulate mitochondrial acylcarnitine carrier gene expression. Biochem Biophys Res Commun. 2009;388:643–647. doi: 10.1016/j.bbrc.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Iacobazzi V., Infantino V., Convertini P., Vozza A., Agrimi G., Palmieri F. Transcription of the mitochondrial citrate carrier gene: identification of a silencer and its binding protein ZNF224. Biochem Biophys Res Commun. 2009;386:186–191. doi: 10.1016/j.bbrc.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Iijima M., Jalil A., Begum L., Yasuda T., Yamaguchi N., Xian Li M. Pathogenesis of adult-onset type II citrullinemia caused by deficiency of citrin, a mitochondrial solute carrier protein: tissue and subcellular localization of citrin. Adv Enzyme Regul. 2001;41:325–342. doi: 10.1016/s0065-2571(00)00022-4. [DOI] [PubMed] [Google Scholar]
- Indiveri C., Kramer R., Palmieri F. Reconstitution of the malate/aspartate shuttle from mitochondria. J Biol Chem. 1987;262:15979–15983. [PubMed] [Google Scholar]
- Infantino V., Iacobazzi V., De Santis F., Mastrapasqua M., Palmieri F. Transcription of the mitochondrial citrate carrier gene: role of SREBP-1, upregulation by insulin and downregulation by PUFA. Biochem Biophys Res Commun. 2007;356:249–254. doi: 10.1016/j.bbrc.2007.02.114. [DOI] [PubMed] [Google Scholar]
- Infantino V., Convertini P., Iacobazzi F., Pisano I., Scarcia P., Iacobazzi V. Identification of a novel Sp1 splice variant as a strong transcriptional activator. Biochem Biophys Res Commun. 2011;412:86–91. doi: 10.1016/j.bbrc.2011.07.047. [DOI] [PubMed] [Google Scholar]
- Infantino V., Convertini P., Menga A., Iacobazzi V. MEF2C exon alpha: role in gene activation and differentiation. Gene. 2013;531:355–362. doi: 10.1016/j.gene.2013.08.044. [DOI] [PubMed] [Google Scholar]
- Infantino V., Iacobazzi V., Palmieri F., Menga A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem Biophys Res Commun. 2013;440:105–111. doi: 10.1016/j.bbrc.2013.09.037. [DOI] [PubMed] [Google Scholar]
- Jalil M.A., Begum L., Contreras L., Pardo B., Iijima M., Li M.X. Reduced N-acetylaspartate levels in mice lacking aralar, a brain- and muscle-type mitochondrial aspartate–glutamate carrier. J Biol Chem. 2005;280:31333–31339. doi: 10.1074/jbc.M505286200. [DOI] [PubMed] [Google Scholar]
- Jambal P., Masterson S., Nesterova A., Bouchard R., Bergman B., Hutton J.C. Cytokine-mediated down-regulation of the transcription factor cAMP-response element-binding protein in pancreatic beta-cells. J Biol Chem. 2003;278(25):23055–23065. doi: 10.1074/jbc.M212450200. [DOI] [PubMed] [Google Scholar]
- Lasorsa F.M., Pinton P., Palmieri L., Fiermonte G., Rizzuto R., Palmieri F. Recombinant expression of the Ca(2+)-sensitive aspartate/glutamate carrier increases mitochondrial ATP production in agonist-stimulated Chinese hamster ovary cells. J Biol Chem. 2003;278:38686–38692. doi: 10.1074/jbc.M304988200. [DOI] [PubMed] [Google Scholar]
- Ledeen R.W., Wang J., Wu G., Lu Z.H., Chakraborty G., Meyenhofer M. Physiological role of N-acetylaspartate: contribution to myelinogenesis. Adv Exp Med Biol. 2006;576:131–143. doi: 10.1007/0-387-30172-0_9. [DOI] [PubMed] [Google Scholar]
- Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lonze B.E., Ginty D.D. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lu Z.H., Chakraborty G., Ledeen R.W., Yahya D., Wu G. N-Acetylaspartate synthase is bimodally expressed in microsomes and mitochondria of brain. Brain Res Mol Brain Res. 2004;122(1):71–78. doi: 10.1016/j.molbrainres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Madhavarao C.N., Chinopoulos C., Chandrasekaran K., Namboodiri M.A. Characterization of the N-acetylaspartate biosynthetic enzyme from rat brain. J Neurochem. 2003;86(4):824–835. doi: 10.1046/j.1471-4159.2003.01905.x. [DOI] [PubMed] [Google Scholar]
- Madhavarao C.N., Arun P., Moffett J.R., Szucs S., Surendran S., Matalon R. Defective N-acetylaspartate catabolism reduces brain acetate levels and myelin lipid synthesis in Canavan's disease. Proc Natl Acad Sci U S A. 2005;102:5221–5226. doi: 10.1073/pnas.0409184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeer P.L., McGeer E.G. The inflammatory response system of brain: implications for therapy of Alzheimer and other neurodegenerative diseases. Brain Res Brain Res Rev. 1995;21:195–218. doi: 10.1016/0165-0173(95)00011-9. [DOI] [PubMed] [Google Scholar]
- Mehta V., Namboodiri M.A. N-Acetylaspartate as an acetyl source in the nervous system. Brain Res Mol Brain Res. 1995;31(1–2):151–157. doi: 10.1016/0169-328x(95)00044-s. [DOI] [PubMed] [Google Scholar]
- Menga A., Infantino V., Iacobazzi F., Convertini P., Palmieri F., Iacobazzi V. Insight into mechanism of in vitro insulin secretion increase induced by antipsychotic clozapine: role of FOXA1 and mitochondrial citrate carrier. Eur Neuropsychopharmacol. 2013;23:978–987. doi: 10.1016/j.euroneuro.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Moffett J.R., Arun P., Ariyannur P.S., Namboodiri A.M. N-Acetylaspartate reductions in brain injury: impact on post-injury neuroenergetics, lipid synthesis, and protein acetylation. Front Neuroenergetics. 2013;5:11. doi: 10.3389/fnene.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan T.K., Mackenzie C.J., Plevin R., Lutz E.M. PACAP-38 induces neuronal differentiation of human SH-SY5Y neuroblastoma cells via cAMP-mediated activation of ERK and p38 MAP kinases. J Neurochem. 2008;104:74–88. doi: 10.1111/j.1471-4159.2007.05018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namboodiri A.M., Moffett J.R., Arun P., Mathew R., Namboodiri S., Potti A. Defective myelin lipid synthesis as a pathogenic mechanism of Canavan disease. Adv Exp Med Biol. 2006;576:145–146. doi: 10.1007/0-387-30172-0_10. [DOI] [PubMed] [Google Scholar]
- Napolioni V., Persico A.M., Porcelli V., Palmieri L. The mitochondrial aspartate/glutamate carrier AGC1 and calcium homeostasis: physiological links and abnormalities in autism. Mol Neurobiol. 2011;44:83–92. doi: 10.1007/s12035-011-8192-2. [DOI] [PubMed] [Google Scholar]
- Nicolle M.M., Gonzalez J., Sugaya K., Baskerville K.A., Bryan D., Lund K. Signatures of hippocampal oxidative stress in aged spatial learning-impaired rodents. Neuroscience. 2001;107(3):415–431. doi: 10.1016/s0306-4522(01)00374-8. [DOI] [PubMed] [Google Scholar]
- Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflug Arch. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Asp Med. 2013;34:465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Pardo B., Lasorsa F.M., del Arco A., Kobayashi K., Iijima M. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001;20:5060–5069. doi: 10.1093/emboj/20.18.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L., Papaleo V., Porcelli V., Scarcia P., Gaita L., Sacco R. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry. 2010;15:38–52. doi: 10.1038/mp.2008.63. [DOI] [PubMed] [Google Scholar]
- Patel T.B., Clark J.B. Synthesis of N-acetyl-l-aspartate by rat brain mitochondria and its involvement in mitochondrial/cytosolic carbon transport. Biochem J. 1979;184(3):539–546. doi: 10.1042/bj1840539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni D., Tsai J., Mondal D., George W. Attenuation of low dose methylmercury and glutamate induced-cytotoxicity and tau phosphorylation by an N-methyl-d-aspartate antagonist in human neuroblastoma (SHSY5Y) cells. Environ Toxicol. 2013;28:700–706. doi: 10.1002/tox.20765. [DOI] [PubMed] [Google Scholar]
- Pugazhenthi S., Wang M., Pham S., Sze C.I., Eckman C.B. Downregulation of CREB expression in Alzheimer's brain and in Abeta-treated rat hippocampal neurons. Mol Neurodegener. 2011;6:60. doi: 10.1186/1750-1326-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver R.A., Rose-Curtis P., Roe M.W., Wellman G.C., Lounsbury K.M. Store-operated Ca2+ entry activates the CREB transcription factor in vascular smooth muscle. Circ Res. 2004;94:1351–1358. doi: 10.1161/01.RES.0000127618.34500.FD. [DOI] [PubMed] [Google Scholar]
- Ramoz N., Reichert J.G., Smith C.J., Silverman J.M., Bespalova I.N., Davis K.L. Linkage and association of the mitochondrial aspartate/glutamate carrier SLC25A12 gene with autism. Am J Psychiatry. 2004;161:662–669. doi: 10.1176/appi.ajp.161.4.662. [DOI] [PubMed] [Google Scholar]
- Sands W.A., Palmer T.M. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008;20:460–466. doi: 10.1016/j.cellsig.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Saura C.A., Valero J. The role of CREB signaling in Alzheimer's disease and other cognitive disorders. Rev Neurosci. 2011;22:153–169. doi: 10.1515/RNS.2011.018. [DOI] [PubMed] [Google Scholar]
- Sheng M., Greenberg M.E. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- Tahay G., Wiame E., Tyteca D., Courtoy P.J., Van Schaftingen E. Determinants of the enzymatic activity and the subcellular localization of aspartate N-acetyltransferase. Biochem J. 2012;441(1):105–112. doi: 10.1042/BJ20111179. [DOI] [PubMed] [Google Scholar]
- Tokumitsu H., Inuzuka H., Ishikawa Y., Ikeda M., Saji I., Kobayashi R. STO-609, a specific inhibitor of the Ca(2+)/calmodulin-dependent protein kinase kinase. J Biol Chem. 2002;277(18):15813–15818. doi: 10.1074/jbc.M201075200. [DOI] [PubMed] [Google Scholar]
- Tong L., Thornton P.L., Balazs R., Cotman C.W. Beta-amyloid-(1-42) impairs activity-dependent cAMP-response element-binding protein signaling in neurons at concentrations in which cell survival is not compromised. J Biol Chem. 2001;276:17301–17306. doi: 10.1074/jbc.M010450200. [DOI] [PubMed] [Google Scholar]
- Tong L., Balazs R., Thornton P.L., Cotman C.W. Beta-amyloid peptide at sublethal concentrations downregulates brain-derived neurotrophic factor functions in cultured cortical neurons. J Neurosci. 2004;24:6799–6809. doi: 10.1523/JNEUROSCI.5463-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen J.A., Rehnstrom K., Kilpinen H., Kuokkanen M., Kempas E., Ylisaukko-Oja T. Mitochondrial aspartate/glutamate carrier SLC25A12 gene is associated with autism. Autism Res. 2008;1:189–192. doi: 10.1002/aur.25. [DOI] [PubMed] [Google Scholar]
- Watkins J., Basu S., Bogenhagen D.F. A quantitative proteomic analysis of mitochondrial participation in p19 cell neuronal differentiation. J Proteome Res. 2008;7:328–338. doi: 10.1021/pr070300g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiame E., Tyteca D., Pierrot N., Collard F., Amyere M., Noel G. Molecular identification of aspartate N-acetyltransferase and its mutation in hypoacetylaspartia. Biochem J. 2009;425:127–136. doi: 10.1042/BJ20091024. [DOI] [PubMed] [Google Scholar]
- Wibom R., Lasorsa F.M., Tohonen V., Barbaro M., Sterky F.H., Kucinski T. AGC1 deficiency associated with global cerebral hypomyelination. N Engl J Med. 2009;361:489–495. doi: 10.1056/NEJMoa0900591. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- Zhao J., O’Connor T., Vassar R. The contribution of activated astrocytes to Aβ production: implications for Alzheimer's disease pathogenesis. J Neuroinflammation. 2011;8:150. doi: 10.1186/1742-2094-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]