Abstract
Mangrove plants comprise a unique group of organisms that grow within the intertidal zones of tropical and subtropical regions and whose distributions are influenced by both biotic and abiotic factors. To understand how these extrinsic and intrinsic processes influence a more fundamental level of the biological hierarchy of mangroves, we studied the genetic diversity of two Neotropical mangrove trees, Avicenniagerminans and A. schaueriana, using microsatellites markers. As reported for other sea-dispersed species, there was a strong differentiation between A. germinans and A. schaueriana populations sampled north and south of the northeastern extremity of South America, likely due to the influence of marine superficial currents. Moreover, we observed fine-scale genetic structures even when no obvious physical barriers were present, indicating pollen and propagule dispersal limitation, which could be explained by isolation-by-distance coupled with mating system differences. We report the first evidence of ongoing hybridization between Avicennia species and that these hybrids are fertile, although this interspecific crossing has not contributed to an increase in the genetic diversity the populations where A. germinans and A. schaueriana hybridize. These findings highlight the complex interplay between intrinsic and extrinsic factors that shape the distribution of the genetic diversity in these sea-dispersed colonizer species.
Introduction
Mangrove plants encompass a polyphyletic and heterogeneous group defined by ecological and physiological traits that are adaptations to life within the intertidal zones of tropical and subtropical regions. These plants establish discrete communities, known as mangrove forests [1], which are globally distributed, covering approximately 137,700 km2 worldwide [2]. Within this larger distribution, mangrove species richness is heterogeneously distributed, such that the Eastern or Indo-West Pacific (IWP) region is one order of magnitude more diverse than the Western or Atlantic, Caribbean and East Pacific (ACEP) region [3]. Moreover, within each of these biogeographic regions, species number decreases as the latitude increases [3,4].
These patterns of species diversity are influenced by abiotic factors, such as oceanography, climate, topography and soil conditions [3]. Additionally, biotic factors play important roles in the maintenance of these patterns. For instance, the limited effective dispersal [3] and establishment of floating, long-lived, salt-tolerant propagules—with variable degrees of “viviparity” and continuous embryonic development without dormancy [5,6]—can affect species composition at many scales [3]. These issues concerning the distribution of mangrove species richness raise further questions regarding other levels of biological hierarchy, such as the organization of genetic diversity.
Since the 1980s, the genetic variation of mangrove plants at the molecular level has been evaluated using a variety of methods in both biogeographic regions [7]. More recently, a wide range of questions have been answered, and new patterns have emerged [8–12]. For instance, it is now clear that large land and ocean barriers to propagule dispersal are important evolutionary factors that differentiate mangrove populations [8]. However, at smaller geographic scales, substantial genetic structure is also observed among different genera [10,13]. Another important recent advance is that ancient and ongoing interspecific hybridization has now been recorded [8,11,14], including between taxa for which no morphologically intermediate individuals have been found [15].
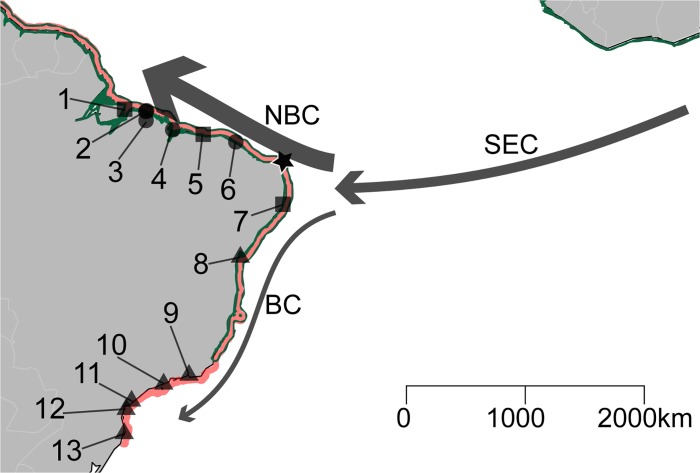
Using two Neotropical species of Avicennia L. (Acanthaceae) as models, we evaluated the extrinsic and intrinsic factors that shape the genetic variation of different species of mangrove plants. Avicennia, the most widely distributed genus of mangrove plants [16], is found in both the IWP (five species) and the ACEP (three species) regions. The ACEP Avicennia species with the largest distribution is A. germinans L., which is distributed across the entire region except for southern Brazil, where A. schaueriana Stapf and Leechman ex Moldenke are dominant (Fig. 1). These species are partially sympatric from the northeastern coast of Brazil to the northern limit of distribution for A. schaueriana in the lower Lesser Antilles [1,16]. Preliminary evidence of chloroplast capture between these species has been found within their sympatry zone [15]. This interaction is biologically feasible as both these species present a generalist pollination system and share pollinators [17,18]. Additionally, the reproductive phenology of these species overlaps in certain localities [19].
Fig 1. Geographic distribution of A. germinans and A. schaueriana samples along the Brazilian coast.
A map of the South Atlantic showing the geographic distribution of A. schaueriana (red) and its sympatry region with A. germinans (green and red). Squares and triangles indicate locations where only A. germinans and or A. schaueriana, respectively, presented reproductive branches during sampling. Circles represent locales where both species presented flowers. The star indicates the northeastern extremity of South America. Sampling locations are displayed according to Table 1. Arrows denote the following near-surface ocean currents that influence the sampling range: the South-Equatorial (SEC), North Brazil (NBC) and Brazil currents (BC). Arrow sizes and line widths illustrate the mean current speed [61].
We hypothesized that multiple geographic scales of genetic structure exist for A. germinans and A. schaueriana, as has been independently observed for many true and associate mangrove plants [8–10,20,21]. To evaluate this hypothesis and the potential intrinsic and extrinsic factors currently influencing these genetic structures, we characterized the population genetics of these species at three geographic scales, the A. schaueriana mating system, and hybridization between these species using microsatellite markers.
Materials and Methods
Plant material and sampling strategy
From June 2008 to December 2010, we sampled 400 individuals of A. schaueriana from 11 localities and 181 individuals of A. germinans from seven locales along the Brazilian coast, covering more than 4900 km of coastline, to evaluate local (thousands of meters, microscale), regional (hundreds of kilometers, mesoscale) and continental geographic scales (thousands of kilometers, macroscale) (Fig. 1, Table 1). Latitude and longitude were recorded using a global positioning system (Garmin 76CSx, WGS-84 standard, Garmin International Inc., Olathe, KS, USA). Licenses (17159 and 17130) to collect the leaves and reproductive branches of these species were obtained from the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA, currently, Instituto Chico Mendes de Conservação da Biodiversidade, ICMBio).
Table 1. Locality descriptions for the samples of Avicennia germinans and A. schaueriana from the Brazilian coast.
| As | Ag | Locality (City, State) | Geographic Coordinate | Location in Fig. 1 |
|---|---|---|---|---|
| AgMRJ (31) | Soure, Pará | 0° 43' 26" S, 48° 29' 24" W | 1 | |
| AsSAL (22) | Salinópolis, Pará | 0° 36' 36" S, 47° 22' 41" W | 2 | |
| AsAJU (46) $ | Bragança, Pará | 0° 49' 12" S, 46° 36' 56" W | 2 $ | |
| AsPRM (47) | Bragança, Pará | 0° 57' 42" S, 46° 37' 5" W | 2 | |
| AgPAa (28)* | Bragança, Pará | 0° 54' 17" S, 46° 41' 15" W | 2 | |
| AgPAb (27) | Bragança, Pará | 0° 56' 21" S, 46° 43' 17" W | 3 | |
| AsALC (30) | AgALC (29) | Alcântara, Maranhão | 2° 24' 37" S, 44° 24' 22" W | 4 |
| AgPNB (29) | Parnaíba, Piauí | 2° 46' 42" S, 41° 49' 20" W | 5 | |
| AsPRC (31) | AgPRC (5) | Paracuru, Ceará | 3° 24' 47" S, 39° 3' 23" W | 6 |
| AgTMD (32) | Tamandaré, Pernambuco | 8° 31' 35" S, 35° 0' 48" W | 7 | |
| AsVER (31) | Vera Cruz, Bahia | 12° 59' 1" S, 38° 41' 5" W | 8 | |
| AsGPM (35) | Guapimirim, Rio de Janeiro | 22° 42' 5" S, 43° 0' 26" W | 9 | |
| AsUBA (32) | Ubatuba, São Paulo | 23° 29' 22" S, 45° 9' 52" W | 10 | |
| AsCNN (32) | Cananéia, São Paulo | 25° 1' 12" S, 47° 55' 5" W | 11 | |
| AsPPR (28) | Pontal do Paraná, Paraná | 25° 34' 30" S, 48° 21' 9" W | 12 | |
| AsFLN (66) | Florianópolis, Santa Catarina | 27° 34' 37" S, 48° 31' 8" W | 13 |
Sampled populations of A. germinans (Ag) and A. schaueriana (As) are indicated by three capital letters. Sample sizes are indicated with parentheses. The city and state in Brazil, geographic coordinate and numbers corresponding to Fig. 1 are denoted for each site.
* indicates a sample under a reduced inundation frequency [19];
$ indicates the locality where the progeny array was sampled.
Each species sample is presented in Table 1, with Ag and As indicating A. germinans and A. schaueriana, respectively; localities from which the samples were obtained are indicated with three letter abbreviations. For A. germinans, we collected individuals from two nearby localities in Bragança, Pará, Brazil that experience different tidal influences: one area in which inundation frequency was reduced due to changes in the hydrographic regime caused by highway construction (AgPAa) [19], and another that experienced a regular tidal pattern (AgPAb) (Fig. 1, Table 1). To minimize misidentification, we distinguished species in the field based on both vegetative and reproductive traits. We identified A. germinans individuals by their ovate leaves that usually present a blunt apex; by their long, exserted stamens; and by their conspicuously hairy petals. We identified A. schaueriana by their obovate leaves; by their inserted stamens; and by their glabrous inner-face corolla [1]. Voucher specimens from every location, except for the AsALC and AgALC samples, were deposited and cataloged in the University of Campinas (UEC) and Embrapa Amazônia Oriental (IAN) herbaria, both of which are located in Brazil. We sampled leaves from flowering trees located at least 20 m from any other tree, and leaves were stored in zip-lock bags containing silica gel. Leaf material was lyophilized and stored at −20°C prior to DNA isolation. Despite our best sampling efforts, we found few individuals (five) of A. germinans presenting flowers at AgPRC (Table 1).
In June 2008, we randomly chose 24 mother trees separated by at least 30 m from AsAJU for the A. schaueriana mating system analyses (Fig. 1, Table 1). In total, 288 healthy propagules attached to the tree were sampled from each of these trees, with a mean of 12 propagules per parental plant (ranging from 8 to 14). Each progeny was stored in an individual zip-lock bag in the field, and when brought to the laboratory, each fruit was stored in distilled water to isolate the pericarp from the embryo, which was stored at −80°C until DNA extraction.
A. schaueriana microsatellite development and molecular biology procedures
For total genomic DNA isolation, we ground leaf samples and whole embryos into fine powder in liquid nitrogen, according to a cetyltrimethyl ammonium bromide protocol. The genetic diversity of each Avicennia species was analyzed using previously published microsatellite markers for A. germinans [22–24] and markers developed for A. schaueriana (described herein), which were isolated to achieve more reliable results using a larger number of loci for both species. The A. schaueriana microsatellites were developed using a method previously used to isolate A. germinans markers [24]. Monomorphic markers for A. germinans [24] were tested using a subsample of eight individuals from three and five locales for A. germinans and A. schaueriana, respectively. Markers showing intra- or interspecific polymorphisms were considered in subsequent examinations (Table 2).
Table 2. Microsatellites markers used in this study.
| Marker | Primer sequence (5’-3’) | Size (bp) | GenBank | Repeat motif | AC | Reference | Ag | As | Both |
|---|---|---|---|---|---|---|---|---|---|
| Agerm1-02 | F: TAACTAGCCGCCCATCCATC | 168 | HM470004 | (ca)11 | 53.4°C | 24 | P | P | P |
| R: ACCAGCCCACATCCAACAAT | |||||||||
| Agerm1-03 | F: CCATGTTTTTGACTTTTTATTTTG | 161 | HM470005 | (ca)9 | 48.2°C | 24 | P | P | P |
| R: TTACGATAGGGTGGATTGAGATTTT | |||||||||
| Agerm1-06 | F: GAATTGGCTGGAATGAGGAA | 175 | HM470008 | (gt)6 | 63.4°C | 24 | P | P | P |
| R: GTGTTTTGGAAGGAGCCTGT | |||||||||
| Agerm1-07 | F: CCTGACACTCTGGGACATCA | 157 | HM470009 | (gt)9 | 50.5°C | 24 | P | P | P |
| R: CCTTTTGACGCATTTGTGG | |||||||||
| Agerm1-08 | F: CTGCCGAGCAAAGGCTTA | 183 | HM470010 | (tg)9 | TD65-55 | 24 | M | P | P |
| R: GCAAGATCCACAGCTTCACA | |||||||||
| Agerm1-12 | F: CAGTTTGGTGAGAAGGATGTT | 127 | HM470014 | (ac)15 | 53.4°C | 24 | P | M | P |
| R: TTTGAGGTCGGCTCGTTAAG | |||||||||
| Agerm1-14 | F: CCAATTGTGTCGTCCTTTTA | 159 | HM470016 | (ca)8(at)6 | 59.6°C | 24 | P | M | P |
| R: AGCCTTACTTTTCCTTTGT | |||||||||
| Agerm1-15 | F: ACTTACACACAAAATGCACA | 248 | HM470017 | (ca)4. . .(ac)13 | 56.7°C | 24 | P | M | P |
| R: CTGAGAGTGCCGACTGAATG | |||||||||
| Agerm1-16 | F: CCTAATACAAATGACACTAAAA | 176 | HM470018 | (tg)9 | 53.4°C | 24 | P | - | - |
| R: TGCATGTCAATTATCAGTCT | |||||||||
| Agerm1-18* | F: CAGCGGGAAAAATCAAACCAA | 243 | HM470020 | (ag)16 | 63°C | 24 | P | P | P |
| R: CCTGTGCACATCCGCCTCTC | |||||||||
| Agerm1-21* | F: GGAGCAATTGTCGAAAGGAC | 150 | HM470023 | (ca)8 | 61.8°C | 24 | P | - | - |
| R: CGTTGCTGAGACAAGGAACA | |||||||||
| Agerm1-22 | F: CACAGGTTCTACTCGGAAGATG | 167 | HM470024 | (tttctt)4 | 63.4°C | 24 | P | M | P |
| R: CGTCCGGGTCTACTCAAAAA | |||||||||
| Agerm1-25 | F: GAGCAAAACTGGATACTCAAATG | 237 | HM470027 | (tg)10. . .(tg)4 | 65.5°C | 24 | P | - | - |
| R: AATAATAAGGCGCCCGTGT | |||||||||
| CTT01 | F: CATCCACATTGCCCTGAT | 102 | DQ240228 | (ctt)8 | TD65-55 | 23 | P | P | P |
| R: GCCTGATAAGTTGAGTTGCTG | |||||||||
| T7* | F: CTAAGTAGGACAGTAATGCGAC | 170 | AY741799 | (cat)2(at)3(gtat)5 | 48.2 | 22 | P | P | P |
| R: AATCATCAGAATCCCTCAAGTGC | |||||||||
| T8 | F: ACACAACGCAGATAAATCC | 112 | AY741802 | (tgta)6 | 59.6 | 22 | P | - | - |
| R: AATGATGCGCTGTCTCCGTC | |||||||||
| Aschau1-01 | F: AACGACAAACCATTAGAAACCAA | 219 | KC783259 | (tg)21. . .(at)6 | 46.7 | This work | P | P | P |
| R: CAATTGAATTTTCTGATTCCCTAA | |||||||||
| Aschau1-02 | F: ACACTACACCCTTCAGCTCAATAA | 150 | KC783260 | (ac)15 | 60 | This work | P | P | P |
| R: ACCCCCAATGGTAGGACAT | |||||||||
| Aschau1-03 | F: GCGGTATCTCCCGTGATTT | 227 | KC783261 | (ca)9gc(ca)14 | 60 | This work | P | P | P |
| R: TAGAGGGGAGATTGGTGTGG | |||||||||
| Aschau1-04 | F: ACGTAAGCTGTGGACGAAGG | 218 | KC783262 | (ct)6(ac)6gc(ac)5 | 60 | This work | P | P | P |
| R: AAGGGATGGGAAGTGGATTC | |||||||||
| Aschau1-05 | F: TCTAATTGGACGATGGCAGA | 179 | KC783263 | (gt)4gc(gt)5tt(gt)4 | 60 | This work | P | P | P |
| R: TGTAGCTGAAATTCCCCTTTTT | |||||||||
| Aschau1-06 | F: AACGTTTTGCCTACACCCTCT | 176 | KC783264 | (ca)14 | 60 | This work | P | P | P |
| R: GCAAGAACTATCGTTCCATCA | |||||||||
| Aschau1-07 | F: TGGCAGATGTGTCTTCCTGA | 209 | KC783265 | (tg)11 | 56.7 | This work | - | P | - |
| R: CCTCAGACTTGAATCAGCAGTG | |||||||||
| Aschau1-08 | F: AATAATTAAGCATCCACTCG | 174 | KC783266 | (gt)14…(gt)6 | TD 65-55 | This work | P | P | P |
| R: TTTAACTTGATGAGGAACTTG | |||||||||
| Aschau1-09 | F: TATCCCTTTGCATTGTTTGAGT | 202 | KC783267 | (ca)21 | 60 | This work | M | P | P |
| R: TTTCAACTCAACTTCATCCT |
Characteristics of the microsatellite markers developed for Avicennia germinans previously developed [22; 23; 24] and for A. schaueriana, described herein. The expected size based on the clone fragment, GenBank accession number, repeat motif, optimal PCR amplification conditions (AC) are shown for each marker. TD65-55 indicates touchdown PCR with temperatures ranging from 65 to 55°C. P indicates polymorphic and M denotes monomorphic marker for A. germinans (Ag) and A. schaueriana (As).
* indicates evidence of null alleles for three or more samples.
Polymerase chain reactions (PCRs) were performed as previously described [24] or with certain modifications [22, 23] (Table 2). Amplification products were visualized via vertical electrophoresis in 1× TBE, 6% polyacrylamide denaturing gels stained with silver nitrate. Product sizes were determined by comparison to a 10 bp DNA ladder (Invitrogen, Carlsbad, CA, USA). Approximately 10% of the total samples were re-genotyped to evaluate scoring genotyping errors. To evaluate the A. schaueriana mating systems, only markers that presented polymorphisms at AsSAL, AsAJU and AsPRM were employed, except for the loci Agerm6 and Agerm8, out of a total of 13 markers.
Intraspecific genetic diversity analyses
After characterizing the A. schaueriana markers, we tested the occurrence of linkage disequilibrium (LD) for all pairs of markers per sample using the FSTAT 2.9.3 software program [25]. As we found consistent evidence of LD for A. germinans and A. schaueriana, we considered 22 and 17 polymorphic markers for further analyses, respectively (Table 2).
Using the MICRO-CHECKER software program [26], we observed evidence of null alleles and stuttering (Table 2). However, despite these indications, we observed no substantial differences between the global and pairwise GST values of samples that were corrected or not corrected for null alleles using the “excluding null alleles” method implemented in the FreeNA software [27] (S1 and S2 Tables); therefore, we used the original dataset for further analyses. We evaluated intraspecific genetic variation based on the average effective number of alleles (Ne), the expected (HE) and observed heterozygosities (HO), and the fixation index (f) for each sample using GenAlEx 6.5 software [28]. The apparent outcrossing rate (t a) was determined as (1 - f)/(1 + f) assuming Wright equilibrium [29] (Table 3). We tested each sample for Hardy-Weinberg equilibrium (HWE) using heterozygote deficiency as the alternative hypothesis in the Genepop 4.0 software program [30].
Table 3. Intraspecific genetic diversity of A. germinans and A. schaueriana samples from the Brazilian coast.
Sample codes are denoted as in Table 1.
| Sample | Ne | HO | HE | f | ta | ||||
|---|---|---|---|---|---|---|---|---|---|
| A. germinans | |||||||||
| AgMRJ | 3.461 | (0.559) | 0.541 | (0.056) | 0.587 | (0.05) | 0.066 | (0.049) | 0.876 |
| AgALC | 2.731 | (0.336) | 0.452 | (0.045) | 0.540 | (0.05) | 0.132 | (0.032) | 0.767 |
| AgPAa | 2.356 | (0.304) | 0.437 | (0.054) | 0.452 | (0.056) | -0.002 | (0.029) | 1.005 |
| AgPAb | 3.259 | (0.506) | 0.522 | (0.056) | 0.569 | (0.053) | 0.083 | (0.048) | 0.846 |
| AgPNB | 1.788 | (0.170) | 0.294 | (0.046) | 0.350 | (0.053) | 0.131 | (0.036) | 0.768 |
| AgPRC | 2.397 | (0.179) | 0.310 | (0.049) | 0.598 | (0.048) | 0.452 | (0.079) | 0.377 |
| AgTMD | 1.174 | (0.064) | 0.025 | (0.012) | 0.110 | (0.037) | 0.662 | (0.083) | 0.203 |
| Average | 2.452 | (0.142) | 0.369 | (0.022) | 0.458 | (0.023) | 0.174 | (0.024) | 0.704 |
| A. schaueriana | |||||||||
| AsSAL | 1.816 | (0.247) | 0.242 | (0.050) | 0.333 | (0.066) | 0.209 | (0.056) | 0.654 |
| AsAJU | 2.002 | (0.349) | 0.245 | (0.051) | 0.346 | (0.066) | 0.308 | (0.075) | 0.529 |
| AsPRM | 2.168 | (0.443) | 0.21 | (0.043) | 0.358 | (0.071) | 0.378 | (0.04) | 0.452 |
| AsALC | 1.879 | (0.273) | 0.255 | (0.051) | 0.336 | (0.068) | 0.207 | (0.061) | 0.656 |
| AsPRC | 1.279 | (0.079) | 0.136 | (0.036) | 0.178 | (0.046) | 0.217 | (0.052) | 0.644 |
| AsVER | 1.455 | (0.170) | 0.144 | (0.036) | 0.213 | (0.058) | 0.187 | (0.073) | 0.685 |
| AsGUA | 1.638 | (0.319) | 0.146 | (0.041) | 0.216 | (0.065) | 0.195 | (0.055) | 0.674 |
| AsUBA | 1.174 | (0.080) | 0.05 | (0.022) | 0.104 | (0.044) | 0.481 | (0.095) | 0.351 |
| AsCNN | 1.529 | (0.202) | 0.193 | (0.060) | 0.215 | (0.067) | 0.112 | (0.077) | 0.799 |
| AsPPR | 1.626 | (0.321) | 0.155 | (0.049) | 0.213 | (0.067) | 0.185 | (0.049) | 0.688 |
| AsFLN | 1.477 | (0.211) | 0.148 | (0.052) | 0.188 | (0.064) | 0.248 | (0.072) | 0.603 |
| Average | 1.64 | (0.081) | 0.175 | (0.014) | 0.246 | (0.019) | 0.242 | (0.02) | 0.610 |
Average effective number of alleles (Ne), expected (HE) and observed (HO) heterozygosities and fixation index (f), with respective standard errors between parentheses, and outcrossing apparent rate (t a) denoted.
Population structure within species
Considering the different sets of microsatellite markers for each species, we used different approaches to evaluate how genetic diversity within each species is organized. We assessed population structure using the summary statistics D [31] and GST [32] using the diveRsity package [33]. The reliability of these statistics was verified using 105 permutations. We also verified the pairwise relations of these statistics between every pair of localities, and we evaluated isolation by distance (IBD) by performing a Mantel test in the ade4 software program [34], considering the approximate linear distance between the sample locales along the coastline and the summary statistics GST [32] and D [31]. The Mantel correlograms were calculated in the vegan package [35], considering 10 classes of geographic distances for A. germinans and 15 for A. schaueriana, with 105 permutations performed to verify the significant correlations.
To further analyze how genetic diversity is structured within each species, we used a multivariate method known as discriminant analysis of principal components (DAPC) [36] implemented in the R package ADEGENET 1.3.7 [37], considering the samples from each locale as different groups. We applied this model-free approach, which provides principal components (PCs) that rely solely on the inter-population variability [36], and we retained seven and six PCs that represented 49.9% and 60.4% of the total genetic information of A. germinans and A. schaueriana samples, respectively, with the number of clusters (k) varying from 1 to 50. The number of PCs was chosen using the optim.a.score function to avoid over-fitting during discrimination. The choice of an optimal k value was based on the Bayesian information criterion provided for each k tested. We also used the model-based clustering method implemented in the Structure 2.3.4 software program, assuming correlated allele frequencies and admixture [38,39], and disregarding any a priori information. We performed 50 independent Markov chain Monte Carlo (MCMC) runs for each k, which ranged from 1 to 10 for A. germinans and from 1 to 15 for A. schaueriana, with 5×105 iterations following a burn-in period of 5×105 iterations. The k value that best explained our data was determined using both maximization of the logarithm likelihood of the data, lnL [38], and the ad hoc statistic ΔK [40]. We used the CLUMPP software [41] to address label switching and multimodality issues using the Greedy algorithm, with 106 random input orders. For A. schaueriana, to determine fine-scale population structure revealed throughout the analyses, we used the same strategy described above for each of the two clusters we obtained (see below), with k ranging from 1 to 10 for each group. We used an extended model-based approach implemented in the InStruct software [42] to consider inbreeding—which is likely to be a violated condition in these species (see below)—without prior information regarding spatial location or sample membership. We performed five independent runs of 106 MCMC repetitions with a 5×105 burn-in period for each. We also performed hierarchical analyses of molecular variance (AMOVA) using permutation procedures (105 iterations) in the Arlequin 3.5 program [43], considering the clusters identified using both multivariate and Bayesian methods. For this purpose, we considered the FST analog estimator under the infinite alleles mutation model (IAM) [44].
Ongoing hybridization between A. germinans and A. schaueriana
To evaluate the ongoing hybridization between these Avicennia species, we used a different set of markers as only microsatellites that yielded PCR products for both species were considered to reduce marker development bias. We evaluated 20 microsatellites in these analyses, including intraspecific monomorphic markers that showed variation between species (Table 2).
The same methods used to study genetic diversity and population structure described above, with the exception of InStruct, were used in this investigation. For DAPC analysis, we used seven PCs, which explained 67.7% of the variance using every species sample as a priori clusters. In addition to using the same model-based clustering approach described above [38,39], with k ranging from 1 to 10, we arbitrarily defined a threshold of 0.15 for membership probability to assess any sign of historical hybridization. We also evaluated the existence and categories (F1, F2 and backcrosses between a “pure species” and a F1) of eventual two-generation hybrids between each “pure species,” employing a different model-based method implemented in the NewHybrids 1.1 beta software [45]. For this purpose, we evaluated the posterior distributions using five independent chains of 106 MCMC following 5×105 burn-in steps without prior allele frequency information, considering Jeffrey-type and uniform distribution priors for θ and π. We also evaluated the groups identified with these approaches using hierarchical AMOVA to evaluate how genetic diversity is organized between and within A. germinans and A. schaueriana.
A. schaueriana mating system
Using 24 open-pollinated progeny arrays composed of 288 individuals, we evaluated the mating system of A. schaueriana under the mixed mating model for unlinked markers [46] using the program MLTR v3.4 [47]. According to this method, considering both Newton-Raphson (NR) and expectation-maximization (EM) methods regarding the absence and presence of null alleles, we estimated the multilocus (t m) and single-locus (t s) outcrossing rates, the outcrossing rate between related individuals (t m-t s), the average single locus inbreeding coefficient of maternal parents (Fm) and the multilocus paternity correlation (r p(m)). Standard error was determined based on 104 bootstraps among families.
Ethics Statement
We confirm that we obtained two licenses (Nos. 17159 and 17130) to collect the leaves and propagules of A. germinans and A. schaueriana from the Brazilian Institute of the Environment and Natural Renewable Resources—IBAMA (currently Chico Mendes Institute for Biodiversity Conservation—ICMBio). We confirm that Avicennia germinans and A. schaueriana are not endangered or protected species.
Results
A. schaueriana microsatellite development and intraspecific genetic diversity
Of the 192 sequenced clones that constituted the constructed library, 52 presented 60 microsatellites. Based on these loci, we designed 43 primer pairs, 16 of which were excluded from further analyses due to amplification failure, unexpected fragment size or nonspecific products. Of the remaining markers, 18 were monomorphic for A. germinans and A. schaueriana, eight were polymorphic for A. germinans, and nine were informative for A. schaueriana (Table 2).
We observed varying degrees of polymorphism among markers within and between samples of each species in terms of Ne, HE and HO (Table 3). We measured significant departures from HWE for every sample of both species. We also observed f values ranging from 0.112 to 0.481 for A. schaueriana and from −0.002 to 0.662 for A. germinans, with average values of 0.242 and 0.174, respectively. Therefore, there was evidence that these species exhibit mixed mating systems as the average ta estimated for the former was 0.610 and 0.704 for the latter (Table 3).
A. schaueriana mating system
Regardless of the method used (NR or EM, with or without considering the presence of null alleles), the results were similar and differed only when thousandths were considered; therefore, only the EM results disregarding null allele outcomes are shown. We observed a predominantly outcrossing mixed-mating system (t m = 0.542 ± 0.062, ts = 0.232 ± 0.065) for this progeny array, which was in accordance with the ta (0.529) estimated for AsAJU, which was composed of only reproductive individuals, with Fm = 0.232 ± 0.065 assuming Wright’s equilibrium. We also observed that biparental inbreeding (t m—ts = 0.151 ± 0.026) contributed to the apparent selfing rate of the progeny, which showed a fraction composed of full-sibs (rp(m) = 0.178 ± 0.057).
Population structure within species
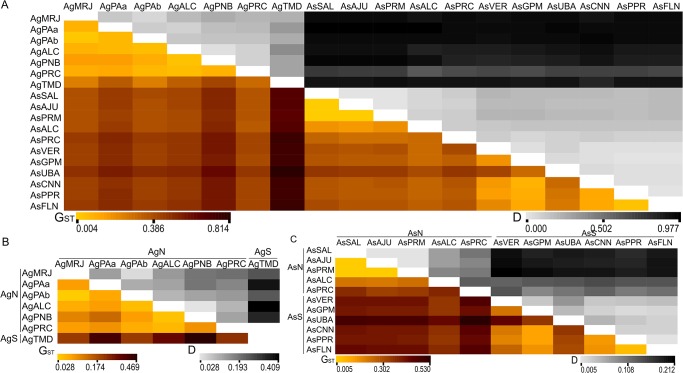
We found substantial evidence for differentiation among the samples with global GST and D values of 0.241 and 0.263 for A. germinans and 0.480 and 0.127 for A. schaueriana, respectively. The pairwise GST and D values for both species were highly correlated, indicating a varying degree of differentiation among different pairs of samples for both species (rho ≥ 9.11 for all cases, Fig. 2 and S3 Table), despite the lower values of D than GST for A. schaueriana. These results indicate that substantial genetic structure exists for both species, primarily when samples north and south of the northeastern extremity of South America (NEESA—Fig. 1) are considered (Fig. 2, S1 and S2 Figs.). For simplicity, we use “Ag” and “As” as acronyms for A. germinans and A. schaueriana, respectively, and “N” or “S” as codes for samples north and south of the NEESA, respectively.
Fig 2. Pairwise comparisons of population genetic differentiation in A. germinans and A. schaueriana.
Pairwise measurements of the genetic variation between samples in terms of Nei’s GST (1973—below diagonal) and D (Jost 2008—above diagonal) values in A) both A. germinans and A. schaueriana, B) A. germinans, and C) A. schaueriana samples. AgN and AgS and AsN and AsS refer to samples of A. germinans and A. schaueriana taken from locales north and south of the NEESA, respectively. All measures were significant (p < 0.05).
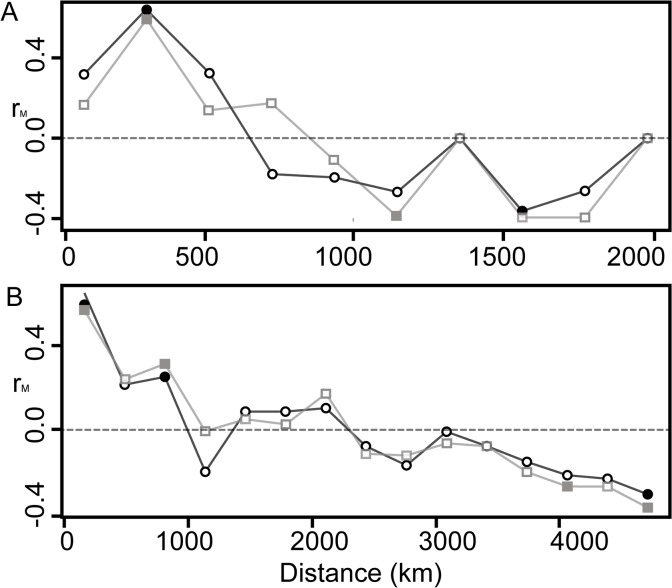
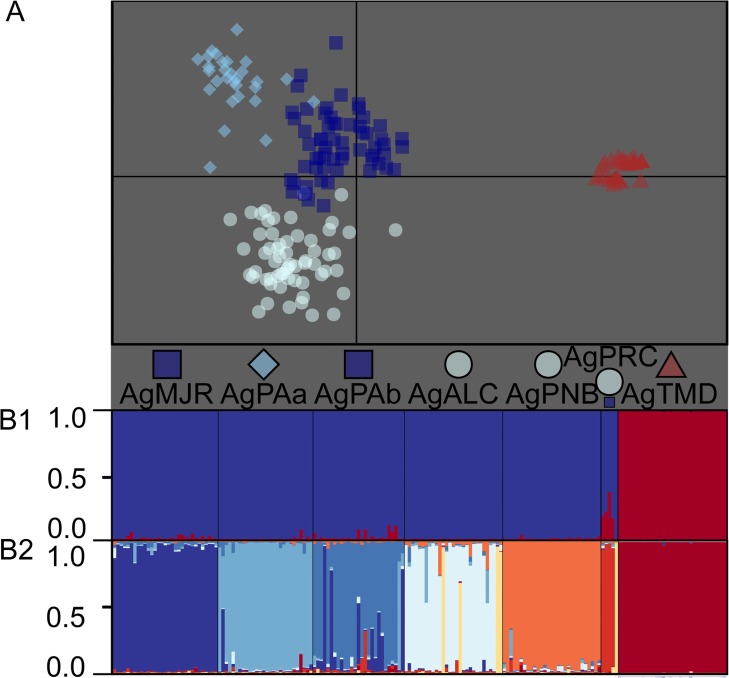
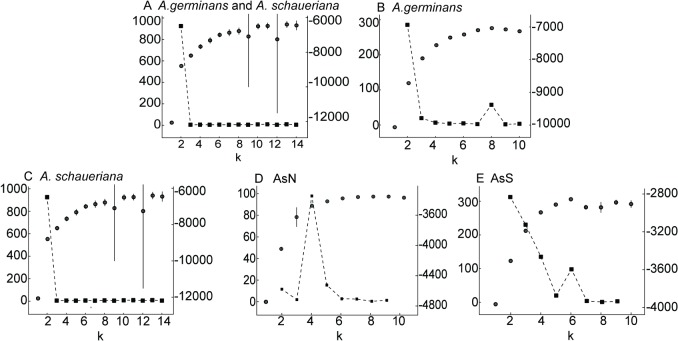
A significant association was found between genetic and geographic distances, as indicated by the D and GST values for A. germinans and A. schaueriana (p < 0.01 for both species, with rM = 0.777 and rM = 0.696 for the former considering GST and D, and rM = 0.755 and rM = 0.830 for the latter). The observed IBD was even more evident when we evaluated the correlograms of rM and classes of geographic distance (Fig. 3), which indicate a significant positive spatial structure. This IBD pattern was particularly evident when the multivariate DAPC analyses were considered. Using this method, we found that k was equal to 4 for A. germinans, with a substantial differentiation between TMD and the remaining samples (Fig. 4A). Also considering the DAPC analyses, for A. schaueriana, the most likely inferred k value was 10, and similarly, a clear pattern of divergence was observed between AsN and AsS (Fig. 5A).
Fig 3. Isolation by distance of A. germinans and A. schaueriana.
Mantel correlograms performed on approximated distances between each pair of sample locales along the coastline for A) A. germinans and B) A. schaueriana. Black interconnected circles refer to D [31], and gray interconnected squares indicate Nei’s genetic distance [69]. Filled squares and circles indicate significant correlations (p < 0.05) after 10,000 permutations.
Fig 4. A. germinans population structure along the Brazilian coast.
A) A scatterplot of the first two principal components of the multivariate analysis of DAPC [36], which resulted in a most likely k value of 4. Symbols indicate the group to which each individual was assigned. The geographic origin of each individual is denoted for each locality sample such that more than 10% of the total number of individuals was composed of the inferred cluster. A larger symbol indicates that a cluster was predominant in the locality sample (pairwise cluster ratio larger than 1:5), whereas equal symbol sizes indicate similar cluster contributions to the total number of individuals in each sample. B1) Model-based clustering analyses [38,39] considering k = 2 and B2) k = 8, where each individual is represented by a vertical line and each color refers to one inferred cluster; the posterior probability of group membership is indicated.
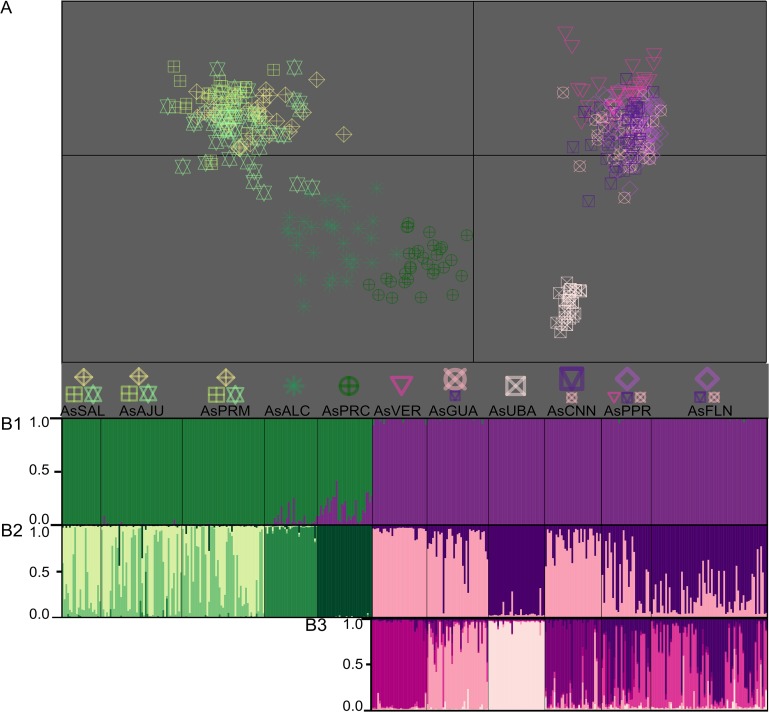
Fig 5. A. schaueriana population structure along the Brazilian coast.
A) A scatterplot of the first two principal components of the multivariate analysis of DAPC [36], which resulted in a most likely k value of 10. Each symbol indicates the group to which each individual was assigned. The geographic origin of each individual is denoted for each locality sample such that more than 10% of the total number of individuals was composed of the inferred cluster. A larger symbol indicates that a cluster was predominant in the locality sample (pairwise cluster ratio larger than 1:5), whereas equal symbol sizes indicate similar cluster contributions to the total number of individuals in each sample. B1) Model-based clustering analyses [38,39] considering k = 2, where each individual is represented by a vertical line and each color refers to one inferred cluster; the posterior probability of group membership is indicated. These Bayesian analyses were extended to the inferred groups observed in B1, which correspond to samples north (green) and south (violet) of the NEESA. The posterior probabilities of group membership in this fine-scale analysis of AsN (k = 4) and AsS (k = 2) are shown in B2; further evaluations of AsS (k = 6) are displayed in B3.
Using the model-based method implemented in the Structure software, we observed two possible scenarios for A. germinans: k = 2 (AgN and AgS), as the most likely number of populations according to the ΔK approach, and k = 8, as an alternative scenario of finer genetic structure based on lnL and a smaller ΔK value peak (Fig. 6B). These results indicate the existence of a multiple-scale genetic structure. The fine-scale scenario suggests the divergence of samples across every evaluated sampling site, including AgPAa and AgPAb, which are separated by thousands of meters. For A. schaueriana, the most likely k value was 2 (AsN and AsS), based only on ΔK (Fig. 6C). Each of these groups was evaluated separately based on the pairwise GST, D (Fig. 2), and DAPC (Fig. 5A) results. Finer-scale genetic structure was evident with AsN presenting k = 4 based on the ΔK values; with AsS presenting k = 2 based on the ΔK value; and with AsS showing k = 6 based on lnL and a smaller ΔK value peak (Fig. 6D and 6E). When inbreeding was considered, similar patterns of genetic structure were observed (S3 and S4 Figs.), despite the different inferred k values for both species (10 for A. germinans and 11 for A. schaueriana). Therefore, both species clearly exhibit variations in genetic structure on different geographic scales, which is in partial agreement with the IBD results as we found two inferred groups (AgPAa and AgPAb) that were clearly distinguished at the genetic level despite their geographic proximity. In all cases, a correspondence was observed between samples from each locality and the inferred clusters, which also allowed for the inference of admixed individuals and migrants (Figs. 4, 5, S1 and S2 Figs.).
Fig 6. Bayesian inference of cluster number (k).
The mean values of log posterior probability of data (lnL—isolated gray circles, left-axis [38]) and the ΔK ad hoc statistic (interconnected squares, right axis [40]) considering A) samples of both A. germinans and A. schaueriana, B) samples of A. germinans, C) individuals of A. schaueriana from all locales sampled, D) individuals of A. schaueriana from the northern cluster (AsN), and E) samples of A. schaueriana from the southern cluster (AsS).
The results of the hierarchical AMOVA indicated that the variation was primarily between AsN and AsS (45.88%). For this species, the among-group fixation indexes were significant, although they were lower in terms of the proportion of variation (Table 4) when fine-scale scenarios were considered (k = 4 for AsN and k = 5 or 8 for AsS). With respect to A. germinans, the variation between AgN and AgS, although substantial, was not supported by the permutation tests. However, when all seven samples were considered, a significant and substantial proportion of the variation could be explained by this level of the hierarchy (15.37%) (Table 4).
Table 4. Analysis of molecular variance (AMOVA) for A. germinans and A. schaueriana samples.
| A) As and Ag | B) AgN, AgS | C) AsN, AsS | D) Ag k = 4, As k = 5 | E) Ag k = 4, As k = 8 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | F-FI | % | F-FI | % | F-FI | % | F-FI | % | F-FI | ||||||
| Among groups | 58.92 | FGT | 0.589 | 27.52 | FGT | 0.275NS | 45.88 | FGT | 0.458 | 49.22 | FGT | 0.492 | 51.23 | FGT | 0.512 |
| Among populations within groups | 17.60 | FSG | 0.428 | 15.37 | FSG | 0.212 | 14.83 | FSG | 0.274 | 5.36 | FSG | 0.105 | 1.33 | FSG | 0.027 |
| Among individuals within populations | 3.54 | FIS | 0.150 | 7.07 | FIS | 0.123 | 9.85 | FIS | 0.250 | 11.38 | FIS | 0.250 | 11.89 | FIS | 0.250 |
| Within individuals | 19.94 | FIT | 0.800 | 50.04 | FIT | 0.499 | 29.44 | FIT | 0.705 | 34.04 | FIT | 0.659 | 35.55 | FIT | 0.644 |
Results of the hierarchical analysis of molecular variance (AMOVA) considering A) both A. germinans (Ag) and A. schaueriana (As) as groups, B) northern (AgN) and southern (AgS) to the northeast extreme of South America (NEESA) of A. germinans as groups, and C) samples of A. schaueriana north (AsN) and south (AsS) to the NEESA; samples of AgN with k = 4 and AsN with k = 2 (D) and k = 5 (E) according to Fig. 3. % indicates total variance; F-FI: fixation indexes considering infinite allele model (IAM). All results are significant (p < 0.005) except for those results presenting NS.
Ongoing hybridization between A. germinans and A. schaueriana
We observed that as expected, the observed differentiation between species was greater than the differentiation among samples within each species, with global GST and D values of 0.583 and 0.549, respectively, which were considerably higher than the values observed for individual species. Pairwise comparisons between samples, both within and between species, also indicated that the interspecific differences were much more pronounced than the intraspecific differences (Fig. 2). These results were also consistent with the hierarchical AMOVA approach, which revealed that the majority of the variation (58.92%) could be explained between species, although considerable and significant genetic variation remained between populations within each species (17.6%) (Table 4).
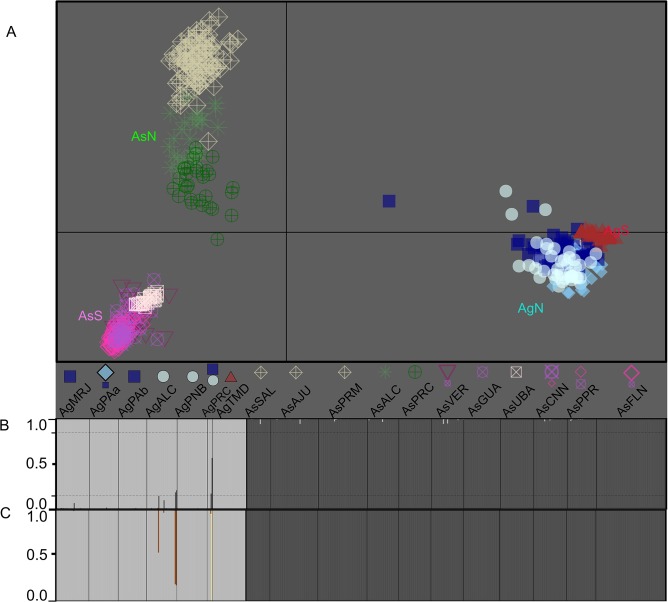
Regarding the DAPC results, we observed an optimum of 11 clusters, revealing the differences between A. germinans and A. schaueriana (Fig. 7A). The patterns of genetic structure within each species were consistent with the multi-scale results described above when each taxon was evaluated separately, including a clear divergence between the northern and southern samples for both A. germinans and A. schaueriana. Using DAPC, one F1 interspecific hybrid from AgPRC (Fig. 7A), which was situated between the two primary clusters, was evident despite the individual assignment. Based on two different model-based approaches, we uncovered more evidence that these species are hybridizing and that these hybrids are fertile. Considering the model implemented in the Structure software [38], the most likely k value was 2 based only ΔK (Fig. 6). The differentiation between A. germinans and A. schaueriana remained obvious; however, compared with the DAPC results, there was more evidence supporting ongoing hybridization between these species. Considering the arbitrary threshold of the assignment probability of 0.15, we identified four individuals that were most likely the result of matings between A. germinans and A. schaueriana. When placed between the two primary clusters in the multivariate analysis (Fig. 7A), the same interspecific hybrid individual presented assignment probabilities of 0.566 and 0.434 for each inferred group, which is a reliable indication of an F1 hybrid (Fig. 7B). Considering each species as a “pure category,” we used another model-based approach to verify the presence of different classes of up to two generation hybrids. As the use of Jeffrey-like and uniform priors yielded slightly different results, with the latter providing more conservative outcomes for the numbers and likelihood of hybrids, we considered only the uniform distribution as prior. The individual inferred as a likely hybrid from PRC considering the Structure and DAPC analyses was unequivocally assigned as an F1 hybrid with a posterior probability of 1.0. We also observed that two plants from AgALC were likely descendants of a cross between an F1 individual and an A. germinans genitor (unidirectional backcross—probability > 0.80) (Fig. 7C). No sign of hybridization was evident when considering individuals identified as A. schaueriana, indicating a unidirectional introgression process (Fig. 7 and S5 Fig.).
Fig 7. Analyses of ongoing hybridization between A. germinans and A. schaueriana.
A) A scatterplot of the first two principal components of the multivariate analysis of DAPC [36]. Each symbol indicates the group to which each individual was assigned. The geographic origin of each individual is denoted for each locality sample such that more than 10% of the total number of individuals is composed of the inferred cluster. A larger symbol indicates that a cluster was predominant in the locality sample (pairwise cluster ratio larger than 1:5), whereas equal symbol sizes indicate similar cluster contributions to the total individuals of each sample. B) Model-based clustering analyses [38,39] considering k = 2, where each individual is represented by a vertical line and each color refers to one cluster; the posterior probability of group membership is indicated. The dashed horizontal lines denote the arbitrary threshold of 0.15, which was used as a sign of possible interspecific hybridization. C) Posterior probability of the model-based approach for identifying species hybrids [45]. Each vertical line also refers to an individual, and different colors indicate distinct classes of individuals: light gray: “pure” A. germinans; dark gray: “pure” A. schaueriana; dark orange: A. germinans second-generation backcrossed individuals; and yellow: F1 interspecific hybrid.
Discussion
A. schaueriana microsatellite markers
We characterized the first set of microsatellites for A. schaueriana to complement the molecular markers developed for A. germinans [22–24]. We observed a variable degree of polymorphism among loci and a high transferability rate between these species (Table 2). The present study demonstrates that these markers are valuable molecular tools that can be used to address a wide range of questions regarding these species.
Mating system and intraspecific genetic diversity and structure in A. germinans and A. schaueriana
Considering the progeny array of A. schaueriana, we found an intermediate outcrossing rate, which significantly departed from both 0 and 1, indicating that this population has a mixed mating system [48]. Comparing the ta values estimated for A. schaueriana from AsAJU (0.529), we observed that this variable closely approximated the t m estimated via the progeny array (0.542 ± 0.062), indicating that inbreeding is a consistent feature of this population across generations. These results are consistent with the reproductive biology of these species as this tree is self-compatible with a pollinator-dependent generalist pollination system [17], similar to the gray mangrove A. marina [49], which is widespread throughout the IWP region. Considering the three A. germinans progeny arrays from the Pacific coast, we found comparable tm values (0.583 ± 0.09, 0.774 ± 0.09 and 0.770 ± 0.12 for each array) and proportions of progeny that shared the same male parent for two of the three progeny, indicating that this species may also exhibit moderate levels of self-fertilization. Considering the evaluations of the A. germinans mating system, substantially lower biparental inbreeding was apparent [50]. Unfortunately, these previous analyses are not easily comparable with ours as we used more loci (13) than Nettel et al. [50] (six) and the difference between tm and ts is sensitive to the number of loci, with more markers resulting in values closer to the true difference between these parameters [47]. However, using a different method, Cerón-Souza and colleagues [10] suggested that biparental inbreeding may indeed be relevant to the A. germinans mating system, indicating that inbreeding in these species could be affected by both selfing and mating among relatives. Taken together, this evidence suggests that A. germinans, whose flowers are visited by a wide range of insects [18], and A. schaueriana generally exhibit similar mixed mating systems.
For both species throughout the sampling range, we also found evidence of a mixed mating system (average ta of 0.704 for A. germinans and 0.610 for A. schaueriana) and varying degrees of inbreeding for most of the evaluated samples (Table 3). Previous studies of A. germinans [10,15], A. marina [51–53] and A. bicolor [15] are consistent with these findings. However, we did not observe evidence for large amounts of inbreeding in the southernmost samples of A. germinans or in the southernmost or northernmost samples of A. schaueriana, similar to the findings reported for A. marina [51], which may be due to ecological, geographical and/or historical differences between these species. This mixed mating system involving a broad range of pollinators may represent an adaptation [54] that may be particularly important to mangrove trees as colonizers [1,55] in terms of reproductive assurance [48]. These findings suggest that self-fertilization—and likely mating between relatives—is a frequent feature throughout the genus, and has had substantial influence on the genetic structure of these species due to pollen dispersal restrictions and, consequently, limited gene flow.
The genetic structure patterns we observed for both species support these reproduction-related findings. We argue that this mixed mating system showing biparental inbreeding influences the substantial genetic variation observed among the samples, despite the different global measures of population differentiation. In particular, D was considerably lower for A. schaueriana, which could be explained by its low diversity within populations, likely due to high levels of inbreeding and/or mating between relatives [31]. These summary values are comparable to those previously reported for A. germinans [10,15], A. bicolor [15] and A. marina [52]. Pairwise comparisons between the different population differentiation measurements were significantly correlated for every pair of samples (Fig. 2). This finding indicates that significant genetic differences exist even for geographically close samples despite low values for GST and D, which is consistent with multivariate and model-based assignment analyses, which showed very similar results.
Considering the fine-scale genetic structures from the DAPC and Bayesian population assignment methods, with k = 8 for A. germinans, k = 4 for AsN, and k = 6 for AsS, we observed well-defined groups at both the local and regional scales. This was true even where there was no clear physical barrier to pollen and propagule dispersal, for instance, among AsSAL, AsAJU, AsPRM and AsALC or among AgPAa, AgPAb or AgALC, which are samples collected within the world’s largest continuous area of mangrove forests [56] (Figs. 4, 5, S1 and S2 Figs.). AMOVA results also supported this fine-scale genetic structure pattern, as both species had significant fixation index values when the variation among populations within groups was considered (Table 4). For instance, the clear differentiation between AgPAa and AgPAb was striking. These populations are geographically close, within a few kilometers of one another, but are subject to distinct hydrographic regimes [19]. In this case, assuming microsatellites as neutral genetic markers, we argue that tide plays an important role in shaping the genetic diversity of Avicennia. Despite the putative neutrality of these microsatellites, this phenomenon may be enhanced by selective pressures acting on these neutral markers via the hitchhiking effect [57], leading to even more differentiated populations. Thus, hydrographic patterns with a low-frequency tide play an important role as barriers to dispersal, as expected based on the water-based dispersal of mangrove propagules. Only one likely admixed individual was identified within the AgPAa samples, indicating that a limited pollen dispersal constraint also exists, which may be enhanced by the mating systems of these species. In this sense, despite the evidence of LDD for A. germinans [20], our results indicate that regardless of their significant evolutionary consequences [58,59], these phenomena are sufficiently rare that A. germinans and A. schaueriana populations present divergent gene pools even at the local and regional geographic scales.
Therefore, both the present and previous analyses indicates that the pollen and propagule dispersal constraints of the mating system of these species influence their genetic structure at small geographic scales. However, despite the genetic divergence at local and regional spatial scales, a positive relation exists between genetic and geographic distances, leading to an IBD pattern (Fig. 3). Similar to that reported for R. mangle [8,9] and Hibiscus pernambucensis [60], substantial divergence exists between samples from locales north and south of the NEESA for both A. germinans and A. schaueriana. According to the majority of the approaches we used, the most pronounced indications of genetic structure emerged when this divergence was considered. Pairwise GST and D values were the highest for pairs of samples between these regions (Fig. 2). The DAPC analyses suggest that the greatest differentiation was found between these northern and southern groups, compared with samples within each cluster (Figs. 4A and 5A). The ΔK values were also the highest for k = 2 for both A. germinans and A. schaueriana (Fig. 6). For A. schaueriana, the amount of variation between these groups was 45.88% according to AMOVA, and lower variation, although significant, was added when we considered more levels of the hierarchy. However, for A. germinans, the between-group fixation indexes were not significant (Table 4), regardless of the mutation model assumed, most likely because AgTMD was the only sample of AgS and because this species exhibits substantial local and regional genetic structure.
As has been previously discussed [8,9,60], the bifurcation of the southern branch of the South Equatorial Current (SEC) into the Brazil Current (BC) and the cross-equatorial North Brazil Current (NBC—northwestward) (Fig. 1–[61]) constrains the movement of propagules between the northern and southern groups. In addition to acting as a barrier, the branching of the SEC, as well as the high velocity of the NBC compared with the BC [61], favor the migration of individuals from the southern to the northern regions, leading to a higher frequency of admixture events in the north. This pattern is readily observed in the Structure software results (Figs. 4, 5, S1 and S2 Figs.). For instance, considering A. germinans, for k = 2, the absence of admixed individuals in the TMD sample and their presence in the AgPRC samples follow the direction of the NBC marine current. A. schaueriana exhibits the same pattern: for k = 2, there is little evidence of admixture in the southern group, whereas substantial evidence of admixed individuals exists in AsALC and AsPRC samples. Moreover, regarding AsN, there are indications that propagule flow follows the direction of the NBC. In contrast, AsS shows a more complex pattern in which no single direction of admixed individuals is apparent for k = 2 or 6 (Fig. 5B). This result may be related to the slower mean velocity of the BC and its seasonal variations in velocity and direction [61] in southern and southeastern Brazil, where AsGPM, AsUBA and AsCNN were sampled. Furthermore, not only the direction but also the speed and seasonal variance of the marine currents play an important role in the genetic diversity of these sea-dispersed plants at multiple geographic scales. However, the role of marine currents in shaping mangrove genetic diversity is not restricted to the Atlantic coast of South America. Along the northwestern coast of Mexico, for instance, the California Current and the El Niño Southern Oscillation could explain the patterns of gene flow observed for R. mangle [62]. Regarding the IWP biogeographic region, links between marine currents and genetic variation have been reported for several different mangrove taxa, including R. mucronata Lam. (Rhizophoraceae) [63], Ceriops tagal (Perr.) (Rhizophoraceae) [64] Kandelia candel (L.) Druce (Rhizophoraceae) [65] and Lumnitzera racemosa Willd (Combretaceae) [66]. Considered together with our findings, this body of evidence suggests that the influence of surface marine currents on genetic variation in mangroves, and likely other sea-dispersed organisms, is a general feature of evolution.
Ongoing hybridization between A. germinans and A. schaueriana
There is evidence of an ancient introgression between A. germinans and A. bicolor, a species with a limited distribution along the Pacific coast of Central America [1], as well as chloroplast capture between A. germinans and A. schaueriana in the Atlantic basin [15]. Although we were unable to evaluate any chloroplast genome markers, we found evidence that ongoing hybridization is indeed occurring between these species within the A. germinans and A. schaueriana sympatry zone. To the best of our knowledge, this is the first report of ongoing hybridization in Avicennia.
These mangrove species present distinct gene pools, which is evident when these species are compared using the pairwise GST and D values between samples or using the DAPC approach, in which the genetic diversity between each species is much greater than the variation within each species (Figs. 2 and 7). Moreover this is consistent with the AMOVA at the interspecific level of the hierarchy (Table 4). In addition to differentiating these two species, these analyses also recapitulate the previously discussed genetic structure within each species. The multivariate assignment approach suggests a likely interspecific hybrid identified as A. germinans from AgPRC, which is graphically located between the clusters of individuals representing each species (Fig. 7A). Further Bayesian evaluations not only corroborated the likely hybrid identified using DAPC as an F1 hybrid but also assigned other A. germinans individuals from AgALC as likely second-generation backcrosses between F1 hybrids and this species (Fig. 7C); these results also indicate that eventual hybrids are fertile, at least when mating with A. germinans. The Bayesian methods also revealed that this introgression is likely unidirectional as only individuals identified as A. germinans showed evidence of ongoing admixture between the species (as illustrated in S5 Fig.). These species share both pollinators [17,18] and flower traits [1,17], and a reproductive phenological overlap has been reported [19], indicating that hybridization is possible between these species. Additionally, during our sampling, A. germinans was more commonly found in Alcântara, Maranhão, Brazil [19], but was much less abundant in Paracuru, Ceará, throughout our sampling. These findings indicate that post-zygotic mechanisms are related to this asymmetric hybridization, as was previously observed for other mangrove genera [12,67]. We consider the mechanisms that generate and maintain this unidirectional introgression worthy of further investigation.
The ancient introgression between A. bicolor and A. germinans observed along the Pacific coast of Central America [15] was related to the higher diversity in that region than the Atlantic coast [10,15]. However, when considering the South American Atlantic coast, this association is not possible as AgAJU and AgPRC do not show higher genetic diversities in terms of the number of alleles or expected heterozygosity than locales with no evidence of hybridization (Table 3). Therefore, we argue that ongoing interspecific hybridization did not increase the genetic diversity of A. germinans in the samples we evaluated. This could indicate that the observed interspecific mating was recent and that too little time has passed for A. schaueriana alleles to spread among the A. germinans populations or, alternatively, that natural selection is acting against the hybrid, reducing the spread of its alleles. Again, although we may hypothesize about the consequences of the observed hybridization, we encourage further investigations into the effects of interspecific hybridization on not only the genetic diversity of populations but also individual phenotype and fitness.
Concluding remarks
Using the first set of microsatellites developed for A. schaueriana as well as markers previously developed for A. germinans, we studied three biological aspects of these species: the existence of hybridization between these species, their mating systems, and the organization of neutral genetic variants at different geographic scales. Our results suggest that an interplay between intrinsic (e.g., mating system, limited pollen and propagule dispersal but not hybridization) and extrinsic factors (e.g., marine currents and tide) shape the genetic diversity of A. germinans and A. schaueriana, leading to genetic diversity structured at the micro-, meso- and macro-scales for both of these Avicennia species.
Similar patterns of neutral genetic variation organization have been observed in different taxa of mangrove species, which is likely related to their shared colonization ability [1,55]. For instance, a large-scale genetic structure was reported for Rhizophora mangle [8,9]; however, its genetic diversity is also locally and regionally organized on smaller scales [10]. This multiple-scale genetic structure is observed in these phylogenetically distant species regardless of the different propagule features of each genus [5,6]. The generality or specificity of these findings and the mechanisms that generate these similar patterns remains to be evaluated in other mangrove species, perhaps using a landscape genetics approach [68]. However, this investigation demonstrates a consistent, multiple-geographic-scale genetic structure pattern for two Neotropical Avicennia species. Moreover, we are aware that these different spatial scales imply distinct temporal scales of evolutionary response. Considering this, future efforts will be focused on elucidating the processes that generate and maintain these patterns.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(XLSX)
Photographs of two geographically close environments under different tide regimes (photos by Gustavo Maruyama Mori).
(PNG)
(PNG)
(PNG)
(PNG)
(PNG)
Acknowledgments
The authors thank I. C. Menezes, I. Sampaio, L. G. Dantas de Oliveira, T. Mori, P. M. Francisco and M. A. Louzada for sampling support and assistance in the field, and P. Zambon for support in the laboratory.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors gratefully acknowledge the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) for financial support (2008/52045-0 and 2010/50178-2) and scholarships to GMM (2007/57021-9 and 2010/50033-4) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for research fellowships to APS and MIZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tomlinson PB (1986) The Botany of Mangroves. Cambridge, UK: Cambridge University Press; 1986. [Google Scholar]
- 2. Giri C, Ochieng E, Tieszen LL, Zhu Z, Singh A, et al. (2011) Status and distribution of mangrove forests of the world using earth observation satellite data. Global Ecology and Biogeography. 2011; 20: 154–159. [Google Scholar]
- 3. Duke NC, Ball MC, Ellison JC (1998) Factors influencing biodiversity and distributional gradients in mangroves. Global Ecology and Biogeography Letters. 1998; 7: 27–47. [Google Scholar]
- 4. Ellison AM (2002) Macroecology of mangroves: large-scale patterns and processes in tropical coastal forests. Trees. 2002; 16: 181–194. [Google Scholar]
- 5. Clarke PJ, Kerrigan RA, Westphal CJ (2001) Dispersal potential and early growth in 14 tropical mangroves: do early life history traits correlate with patterns of adult distribution? J of Ecol. 2001; 89: 648–659. [Google Scholar]
- 6. Rabinowitz D (1978) Dispersal properties of mangrove propagules. Biotropica. 1978; 10: 47–57. [Google Scholar]
- 7. Triest L (2008) Molecular ecology and biogeography of mangrove trees towards conceptual insights on gene flow and barriers: A review. Aquatic Botany. 2008; 89: 138–154. [Google Scholar]
- 8. Takayama K, Tamura M, Tateishi Y, Webb EL, Kajita T (2013) Strong genetic structure over the American continents and transoceanic dispersal in the mangrove genus Rhizophora (Rhizophoraceae) revealed by broad-scale nuclear and chloroplast DNA analysis. Am J Bot. 2013; 100: 1–11. 10.3732/ajb.1200591 [DOI] [PubMed] [Google Scholar]
- 9. Pil MW, Boeger MRT, Muschner VC, Pie MR, Ostrensky A, et al. (2011) Postglacial north-south expansion of populations of Rhizophora mangle (Rhizophoraceae) along the Brazilian coast revealed by microsatellite analysis. Am J Bot; 2011 98: 1031–1039. 10.3732/ajb.1000392 [DOI] [PubMed] [Google Scholar]
- 10. Cerón-Souza I, Bermingham E, McMillan WO, Jones FA (2012) Comparative genetic structure of two mangrove species in Caribbean and Pacific estuaries of Panama. BMC Evol Biol. 2012; 12: 205 Available: 10.1186/1471-2148-12-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun M, Lo EYY (2011) Genomic markers reveal introgressive hybridization in the Indo-West Pacific mangroves: a case study. PloS ONE. 2011; 6: e19671 10.1371/journal.pone.0019671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang R, Liu T, Wu W, Li Y, Chao L, et al. (2013) Molecular evidence for natural hybridization in the mangrove fern genus Acrostichum . BMC Plant Biol. 2013; 13: 74 10.1186/1471-2229-13-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geng Q, Lian C, Goto S, Tao J, Kimura M, et al. (2008) Mating system, pollen and propagule dispersal, and spatial genetic structure in a high-density population of the mangrove tree Kandelia candel . Mol Ecol. 2008; 17: 4724–4739. 10.1111/j.1365-294X.2008.03948.x [DOI] [PubMed] [Google Scholar]
- 14. Cerón-Souza I, Rivera-Ocasio E, Medina E, Jiménez JA, McMillan WO, et al. (2010) Hybridization and introgression in New World red mangroves, Rhizophora (Rhizophoraceae). Am J Bot. 2010; 97: 945–957. 10.3732/ajb.0900172 [DOI] [PubMed] [Google Scholar]
- 15. Nettel A, Dodd RS, Afzal-Rafii Z, Tovilla-Hernández C (2008) Genetic diversity enhanced by ancient introgression and secondary contact in East Pacific black mangroves. Mol Ecol. 2008; 17: 2680–2690. 10.1111/j.1365-294X.2008.03766.x [DOI] [PubMed] [Google Scholar]
- 16. Duke NC (1991) A systematic revision of the mangrove genus Avicennia (Avicenniaceae) in Australasia. Aust Syst Bot. 1991; 4: 299–324. [Google Scholar]
- 17. Nadia TDL, De Menezes NL, Machado IC (2013) Floral traits and reproduction of Avicennia schaueriana Moldenke (Acanthaceae): a generalist pollination system in the Lamiales. Plant Species Biol. 2013; 28: 70–80. [Google Scholar]
- 18. Landry CL (2013) Pollinator-mediated competition between two co-flowering Neotropical mangrove species, Avicennia germinans (Avicenniaceae) and Laguncularia racemosa (Combretaceae). Ann. Bot. 2013; 111: 207–214. 10.1093/aob/mcs265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Menezes MPM, Berger U, Mehlig U (2008) Mangrove vegetation in Amazonia: a review of studies from the coast of Pará and Maranhão States, north Brazil. Acta Amazon. 2008; 38: 403–420. [Google Scholar]
- 20. Nettel A, Dodd RS (2007) Drifting propagules and receding swamps: genetic footprints of mangrove recolonization and dispersal along tropical coasts. Evolution. 2007; 61: 958–971. [DOI] [PubMed] [Google Scholar]
- 21. Takayama K, Kajita T, Murata J, Tateishi Y (2006) Phylogeography and genetic structure of Hibiscus tiliaceus—speciation of a pantropical plant with sea-drifted seeds. Mol Ecol. 2006; 15: 2871–2881. [DOI] [PubMed] [Google Scholar]
- 22. Nettel A, Rafii F, Dodd RS (2005) Characterization of microsatellite markers for the mangrove tree Avicennia germinans L. (Avicenniaceae). Mol Ecol Notes. 2005; 5: 103–105. [Google Scholar]
- 23. Cerón-Souza I, Rivera-Ocasio E, Funk SM, Mcmillan WO (2006) Development of six microsatellite loci for black mangrove (Avicennia germinans). Mol Ecol Notes. 2006; 6: 692–694. [Google Scholar]
- 24. Mori GM, Zucchi MI, Sampaio I, Souza AP (2010) Microsatellites for the mangrove tree Avicennia germinans (Acanthaceae): Tools for hybridization and mating system studies. Am J Bot. 2010; 97: e79–81. 10.3732/ajb.1000219 [DOI] [PubMed] [Google Scholar]
- 25. Goudet J (1995) FSTAT (Version 1.2): A computer program to calculate F-statistics. J Hered. 1995; 86:485–486. [Google Scholar]
- 26. Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004; 4: 535–538. [Google Scholar]
- 27. Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007; 24: 621–631. [DOI] [PubMed] [Google Scholar]
- 28. Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012; 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fyfe J, Bailey NTJ (1951) Plant breeding studies in leguminous forage crops I. Natural cross-breeding in winter beans. J Agric Sci. 1951; 41: 371–378. [Google Scholar]
- 30. Rousset F (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Res. 2008; 8: 103–106. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 31. Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol. 2008; 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- 32. Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 1973; 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity : An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol. 2013; 4: 782–788. [Google Scholar]
- 34. Chessel D, Dufour AB, Thioulouse J (2004) The ade4 package—I: One-table methods. R News. 2004; 4: 5–10. [Google Scholar]
- 35.Oksanen J, Blanchet FG, Kindt R, Simpson GL, Solymos P, et al. (2013) vegan: Community Ecology Package. R package version 2.0-7. 2013.
- 36. Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010; 11: 94 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011; 27: 3070–3071. 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics. 2000; 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003; 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005; 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- 41. Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007; 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- 42. Gao H, Williamson S, Bustamante CD (2007) A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007; 176: 1635–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol EcolRes. 2010; 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 44. Weir B, Cockerham C (1984) Estimating F-statistics for the analysis of population structure. Evolution. 1984; 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- 45. Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002; 160: 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ritland K, Jain S (1981) A model for the estimation of outcrossing rate and gene frequencies using n independent loci. Heredity. 1981; 47: 35–52. [Google Scholar]
- 47. Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Heredity. 2002; 88: 221–228. [DOI] [PubMed] [Google Scholar]
- 48. Goodwillie C, Kalisz S, Eckert CG (1991) The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst. 2005; 36: 47–79. [Google Scholar]
- 49. Clarke PJ, Myerscough PJ (1991) Floral biology and reproductive phenology of Avicennia marina in south-eastern Australia. Aust J Bot. 1991; 39: 283–294. [Google Scholar]
- 50. Nettel A, Dodd RS, Ochoa-Zavala M, Tovilla-Hernández C, Días-Gallegos JR (2013) Mating System Analyses of tropical populations of the black mangrove Avicennia germinans (L.) L. (Avicenniaceae). Bot Sci. 2013; 91: 115–117. [Google Scholar]
- 51. Arnaud-Haond S, Teixeira S, Massa SI, Billot C, Saenger P, et al. (2006) Genetic structure at range edge: low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol Ecol. 2006; 15: 3515–3525. [DOI] [PubMed] [Google Scholar]
- 52. Maguire TL, Saenger P, Baverstock P, Henry R (2000) Microsatellite analysis of genetic structure in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Mol Ecol. 2000; 9: 1853–1862. [DOI] [PubMed] [Google Scholar]
- 53. Duke NC, Benzie JAH, Goodall JA, Ballment ER (1998) Genetic structure and evolution of species in the mangrove genus Avicennia (Avicenniaceae) in the Indo-West Pacific. Evolution. 1998; 52: 1612–1626. [DOI] [PubMed] [Google Scholar]
- 54. Vallejo-Marín M, Uyenoyama MK (2004) On the evolutionary costs of self-incompatibility: incomplete reproductive compensation due to pollen limitation. Evolution. 2004; 58: 1924–1935. [DOI] [PubMed] [Google Scholar]
- 55. Burns BR, Ogden J (1985) The demography of the temperate mangrove [Avicennia marina (Forsk.) Vierh.] at its southern limit in New Zealand. Aust J Ecol. 1985; 10: 125–133. [Google Scholar]
- 56. Nascimento WR, Souza-Filho PWM, Proisy C, Lucas RM, Rosenqvist A (2013) Mapping changes in the largest continuous Amazonian mangrove belt using object-based classification of multisensor satellite imagery. Estuar Coast Shelf Sci. 2013; 117: 83–93. [Google Scholar]
- 57. Kaplan NL, Hudson RR, Langley CH (1989) The “hitchhiking effect” revisited. Genetics. 1989; 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nathan R, Schurr FM, Spiegel O, Steinitz O, Trakhtenbrot A, et al. (2008) Mechanisms of long-distance seed dispersal. Trends Ecol Evol. 2008; 23: 638–647. 10.1016/j.tree.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 59. Duke NC, Lo Y, Sun M (2002) Global distribution and genetic discontinuities of mangroves—emerging patterns in the evolution of Rhizophora . Trees. 2002; 16: 65–79. [Google Scholar]
- 60. Takayama K, Tateishi Y, Murata J, Kajita T (2008) Gene flow and population subdivision in a pantropical plant with sea-drifted seeds Hibiscus tiliaceus and its allied species: evidence from microsatellite analyses. Mol Ecol. 2008; 17: 2730–2742. 10.1111/j.1365-294X.2008.03799.x [DOI] [PubMed] [Google Scholar]
- 61. Lumpkin R, Johnson GC (2013) Global ocean surface velocities from drifters: Mean, variance, El Niño-Southern Oscillation response, and seasonal cycle. J Geophys Res Oceans. 2013; 118: 2992–3006. [Google Scholar]
- 62. Sandoval-Castro E, Muñiz-Salazar R, Enríquez-Paredes LM, Riosmena-Rodríguez R, Dodd RS, et al. (2012) Genetic population structure of red mangrove (Rhizophora mangle L.) along the northwestern coast of Mexico. Aquat Bot. 2012; 99: 20–26. [Google Scholar]
- 63. Wee AKS, Takayama K, Asakawa T, Thompson B, Sungkaew S, et al. (2014) Oceanic currents, not land masses, maintain the genetic structure of the mangrove Rhizophora mucronata Lam. (Rhizophoraceae) in Southeast Asia. J Biogeogr. 2014; 41: 954–964. [Google Scholar]
- 64. Liao P-C, Hwang S-Y, Huang S, Chiang Y-C, Wang J-C (2011) Contrasting demographic patterns of Ceriops tagal (Rhizophoraceae) populations in the South China Sea. Aust J Bot. 2011; 59: 523–532. [Google Scholar]
- 65. Chiang TY, Chiang YC, Chen YJ, Chou CH, Havanond S, et al. (2001) Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Mol Ecol. 2001; 10: 2697–2710. [DOI] [PubMed] [Google Scholar]
- 66. Su G-H, Huang Y-L, Tan F-X, Ni X-W, Tang T, et al. (2006) Genetic variation in Lumnitzera racemosa, a mangrove species from the Indo-West Pacific. Aquat Bot. 2006; 84: 341–346. [Google Scholar]
- 67. Zhou R, Gong X, Boufford D, Wu C-I, Shi S (2008) Testing a hypothesis of unidirectional hybridization in plants: observations on Sonneratia, Bruguiera and Ligularia . BMC Evol Biol. 2008; 8: 149 10.1186/1471-2148-8-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP (2010) Landscape genetics: where are we now? Mol Ecol. 2010; 19: 3496–3514. 10.1111/j.1365-294X.2010.04691.x [DOI] [PubMed] [Google Scholar]
- 69. Nei M (1972) Genetic Distance between Populations. Am Nat. 1972; 106: 283–292. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(XLSX)
Photographs of two geographically close environments under different tide regimes (photos by Gustavo Maruyama Mori).
(PNG)
(PNG)
(PNG)
(PNG)
(PNG)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.