Abstract
Neural restrictive silencer factor, NRSF (also known as REST) binds a neuronal cell type selective silencer element to mediate transcriptional repression of neuron-specific genes in non-neuronal cells and neuronal progenitors. Two repression domains (RD-1 and RD-2) occur in its N-terminal and C-terminal regions, respectively. RD-1 recruits mSin3 and HDAC, thereby inhibiting transcription by inducing reorganization of the chromatin structure. However, little is known about how such global repression becomes promoter-specific repression or whether the NRSF–HDAC complex can interact with transcriptional core factors at each specific promoter. Here we show evidence that NRSF interacts with core promoter factors, including TATA-binding protein (TBP). The NRSF–TBP interaction occurred between the linear segments of the N- and C-terminal-most portions of NRSF and the C-terminal half of TBP. A RD-2 mutant of NRSF lost the TBP-binding activity and was unable to repress transcription at an exogenously introduced TGTA promoter. These results indicate that the direct interaction between the NRSF C-terminal domain and TBP is essential for the C-terminal repression mechanism of NRSF. Thus, the RD-1 and RD-2 repression domains of NRSF utilize both chromatin-dependent and chromatin-independent mechanisms, which may be segregated at various stages of neural development and modulation.
INTRODUCTION
The regulation of chromatin structure is crucial for controlling gene expression by altering the accessibility of promoter elements to DNA-binding factors including enhancer and silencer factors and the general transcriptional machinery. Two classes of complexes work together to modulate the chromatin structure: ATP-dependent chromatin remodeling complexes and histone-modifying complexes. Histone acetyltransferase (HAT) or histone deacetylase (HDAC) complexes can change the chromatin folding through covalent modification of the histone tail. A simple model of transcription would be that sequence-specific regulators first bind to the promoter, in conjunction with chromatin remodeling or modifying complexes, and then the core promoter factors are recruited to form an active initiation complex. However, the assembly of these protein complexes varies among different promoters and how chromatin-remodeling events regulate the promoter specificity or cell type specificity of transcription is obscure (1–3).
Neural restrictive silencer factor (NRSF) (4), also known as RE-1 silencing factor (REST) (5), functions as a transcriptional repressor of multiple neuron-specific genes in non-neuronal cells and tissues during neural development and in adulthood (6–8). Many target genes of NRSF relate directly to neuronal function, including ion channels, neurotransmitter synthetases, receptors, synaptosomal proteins, neuronal cell adhesion molecules, neuronal cytoskeleton, neurotrophic factors and neuronal growth-associated proteins (9). NRSF is composed of an N-terminal repression domain (RD-1), a DNA-binding domain with eight consecutive zinc fingers followed by a highly basic region and a C-terminal repression domain (RD-2) containing a single zinc finger motif (10–12). There are several lines of evidence indicating that transcriptional regulation is concerned with the reorganization of chromatin structure through histone acetylation (13,14). Recent studies revealed that NRSF repressed transcription by binding to co-repressor mSin3, thereby recruiting HDAC, through its N-terminal RD-1 (12,15–18), whereas the C-terminal RD-2 was shown to bind to another co-repressor, CoREST (15). More recent studies indicated that CoREST is a component of a novel HDAC complex (19,20) and recruits HDAC2 to the NRS/RE1 of the Nav1.2 sodium channel gene (21). Thus, both RD-1 and RD-2 are involved in HDAC-mediated chromatin remodeling, which could be a primary cause of the initiation of transcriptional repression of specific target genes. In spite of this notion, however, trichostatin A (TSA), an HDAC inhibitor, failed to derepress NRSF C-terminal domain-mediated repression of the transcription of the GluR2 glutamate receptor gene (17) and SCG10 gene (12), thus suggesting that some unknown HDAC-independent repression mechanism(s) may exist for the C-terminus of NRSF in addition to a HDAC-dependent mechanism. Since HDAC generally affects only one or two histone octamers in chromatin (22), the HDAC complex would be required to be brought to chromatin located near the target gene transcription initiation site(s). As to the repression activity of NRSF, it is thus anticipated that the NRSF–mSin3–HDAC complex would need to be recruited to the core promoter region from the silencer (NRSE) site. Therefore, we focused in this study on the interaction of NRSF with core promoter factors in NRSF-mediated transcriptional repression.
We show herein that the NRSF N- and C-terminal repression domains interact with TBP and some other factors and that inhibition of the interaction between the C-terminus of NRSF and TATA-binding protein (TBP) causes loss of the repression activity of this NRSF domain. Our results indicate that this NRSF–TBP interaction is an important mechanism for the transcriptional repression of neuronal genes via the C-terminal domain of NRSF in addition to the thus far known HDAC-dependent mechanism.
MATERIALS AND METHODS
Plasmid constructs
Plasmids encoding human TBP deletion constructs were a generous gift from P.Carlsson (Göteborg University, Sweden) (23). To prepare additional TBP deletion constructs for in vitro translation, we amplified inserts by PCR with 5′ BamHI and 3′ XhoI adapter primers and subcloned them into the unique BamHI and XhoI sites of pcDNA3. Mouse TFIIB cDNA was obtained by reverse transcription–PCR and subcloned into pcDNA3. The construction of glutathione S-transferase (GST)–NRSF fusion proteins, e.g. GST–NRSF-N (1–153) and GST–NRSF-C (989–1097), was previously described (12). Similar GST fusion proteins containing various portions of NRSF were constructed by standard recombinant DNA methodology using the pGEX-5X vector (Amersham Pharmacia). The series of Gal4 DNA-binding domain (G4 DBD) (1–147)–NRSF fusion constructs used for a luciferase assay were described previously (12). TBPAS expression vectors and a c-fos TGTA reporter plasmid were provided by W.Herr. (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) (24).
GST pull-down and in vitro binding assays
The GST–NRSF fusion proteins were expressed in Escherichia coli BL21 and purified and immobilized on glutathione–Sepharose beads (Amersham Pharmacia). 35S-radiolabeled TBP and TFIIB were synthesized in vitro using a single tube, coupled transcription–translation system (Novagen). The 35S-labeled proteins were incubated at 4°C for 1 h with a 50% slurry of immobilized GST fusion proteins in 1 ml of buffer A (phosphate-buffered saline containing 100 mM KCl and 0.25% NP40). The beads were then washed five times with 1 ml of buffer A each time. The bound proteins were eluted into Laemmli’s loading buffer, fractionated by SDS–PAGE and detected by autoradiography. For identification of the NRSF-binding region on TBP, their respective deletion constructs were radiolabeled with [35S]methionine and the GST pull-down assay was performed similarly to as described above. For analysis of the interactions of endogenous general transcription factors with NRSF-N and NRSF-C, GST fusion proteins were incubated with 100 µg of nuclear extract of NIH 3T3 cells in buffer B [20 mM Tris–HCl at pH 8.0, 150 mM NaCl, 0.5% NP40, 10% glycerol and protease inhibitor complete (Roche)]. Nuclear extracts were prepared as described earlier (25). Bound proteins were analyzed by western blotting using anti-TBP antibody N-12 and anti-TFIIB antibody C-18 (all from Santa Cruz Biotechnology). Autoradiograms were quantified using PhosphoImaging.
Immunoprecipitation and western blotting
HEK 293 cells were transfected with FLAG-tagged NRSF using Lipofectamine Plus reagent (Gibco-BRL). Nuclear extracts of HEK 293 cells transfected with FLAG-tagged NRSF were precleared by rotating at 4°C for 1 h with protein G–Sepharose beads in buffer C [20 mM Tris–HCl at pH 8.0, 150 mM NaCl, 0.1% NP40, 10% glycerol and protease inhibitor complete (Roche)]. Nuclear extracts were then recovered by centrifugation and added to 5 µg of anti-NRSF antibody (P-18). Immunoprecipitations were performed by rotation for 2 h or overnight at 4°C. Then protein G beads were added and after additional rotation for 2 h, the beads were washed three times with buffer C and bound proteins were eluted by boiling in Laemmli sample buffer. The proteins were resolved by SDS–PAGE and western blotting was performed using anti-TBP (N-12), anti-mSin3B (AK-12), anti-polymerase II (pol II) (C-21) (Santa Cruz) and anti-FLAG (M2) (Sigma) antibodies.
TBPAS transfection and reporter gene assay
Neuro2a cells were transiently transfected using Lipofectamine Plus. For luciferase assays, 0.8 × 105 cells were plated in 24-well plates and were transfected 24 h later with 200 ng of luciferase reporter, 200 ng of TBPAS expression plasmid, 1.5–22.5 ng of G4 DBD–NRSF or 1–10 ng of G4 DBD–NRSF-N or G4 DBD–NRSF-C and 50 ng of Renilla luciferase vector (pRL-TK) (Promega) as an internal control. Cells were harvested 48 h after transfection and dual luciferase activities were measured with a luminometer (Lummat LB96V; EG & G, Salem, MA). Three independent experiments were performed in duplicate or triplicate and the values are presented as means ± SE. Statistical analysis was performed using Fisher’s post hoc test (P < 0.05 versus G4 DBD).
RESULTS
Specific binding of NRSF RD-1 (N) and RD-2 (C) with TBP and other transcriptional core factors
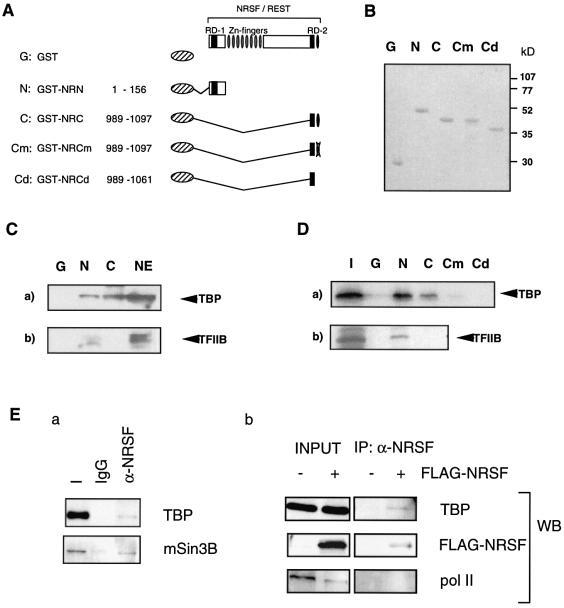
We first tested whether the N- or C-terminal repression domain of NRSF could interact with various general transcription factors (see Fig. 1). GST pull-down analysis using nuclear extracts of NIH 3T3 cells showed that GST-fused NRSF-N bound to endogenous TBP and TFIIB but that GST-fused NRSF-C bound only to TBP (Fig. 1C). We also tested possible binding to TFIIF-RAP30, TAFII250, TFIIA and pol II, however, we could not detect any significant binding with GST–NRSF-N or GST–NRSF-C, except RAP30 (data not shown). We next analyzed the direct interaction of NRSF with 35S-labeled general transcription factors (GTFs) synthesized by in vitro transcription–translation. Consistent with the former results, GST–NRSF-N bound to TBP and TFIIB, whereas GST–NRSF-C bound to TBP only (Fig. 1D). A weak interaction of NRSF-C with TFIIF-RAP30 was also detected (not shown). When the zinc finger motif in NRSF-C (RD-2) was mutated (Cm) or deleted (Cd), this specific binding of RD-2 with TBP was abolished, indicating that the interaction of RD-2 with TBP required the C-terminal zinc finger motif (Fig. 1D).
Figure 1.
Specific binding of NRSF RD-1 (N) and RD-2 (C) to transcriptional core promoter factors. (A) The series of GST-fused NRSFs used in the GST pull-down assay. Cm and Cd indicate the C1062A mutant and a deletion mutant of the C-terminal zinc finger, respectively. (B) Purity and molecular sizes of fusion proteins, i.e. N, C, Cm, Cd and GST alone, on SDS–PAGE stained with CBB. (C) Association of NRSF-N and NRSF-C with endogenous core promoter factors in cellular extracts from NIH 3T3 cells. Nuclear extracts were incubated with immobilized GST (G), GST–NRSF-N (N) or GST–NRSF-C (C) fusion protein and blotted with anti-TBP (a) and anti-TFIIB (b) antibodies. Note that NRSF-N associates with all of these core factors but that NRSF-C does so only with TBP. A 20% aliquot of nuclear extract (NE) was loaded as a control. (D) Specific binding of NRSF-N and NRSF-C with in vitro translated core factors. TBP and TFIIB bound to GST–NRSF were analyzed by SDS–PAGE followed by autoradiography. A 20% aliquot of total input protein was loaded (I) as a control. Disruption of the C-terminal zinc finger structure (as shown in Cm and Cd) abolished the NRSF-C and TBP interaction. (E) NRSF interacts with TBP and mSin3 in vivo. (a) Nuclear extracts of FLAG-tagged NRSF transfected HEK 293 cells were immunoprecipitated with anti-NRSF antibody (α-NRSF) or preimmune IgG (IgG). Bound proteins were detected with anti-TBP or anti-mSin3B antibody. (b) Nuclear extracts of HEK 293 cells transfected with FLAG-tagged NRSF or FLAG vector alone were immunoprecipitated with anti-NRSF antibody. Bound proteins were detected with anti-TBP, anti-FLAG or anti-pol II antibody.
To confirm the interaction between NRSF and TBP, we further tested whether TBP formed a complex with NRSF in vivo. Co-immunoprecipitation experiments using nuclear extracts of HEK 293 cells overexpressing FLAG-tagged NRSF revealed that TBP was present in the anti-NRSF immunoprecipitates, but not in the preimmune IgG immunoprecipitates as a control (Fig. 1E, a). To confirm the specificity of this binding, we also tested whether pol II, which should not bind, was precipitated with NRSF or not. Only TBP was precipitated by anti-NRSF antibody in the presence of FLAG–NRSF, while pol II was not (Fig. 1E, b). These results show that TBP formed a complex with NRSF, indicating that TBP is relevant, at least in part, to NRSF-mediated transcriptional repression. The NRSF–TBP complex also seems to be associated with mSin3 (see Fig. 1E, a).
Identification of TBP-binding regions on NRSF RD-1
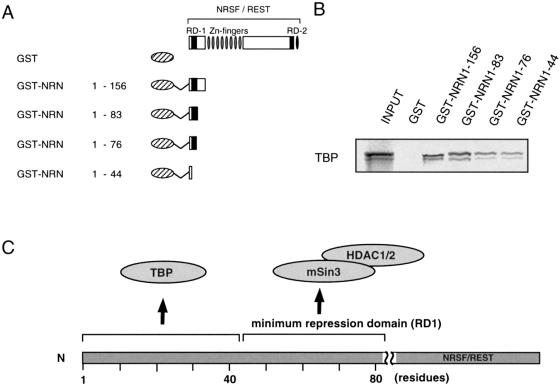
To further narrow down the TBP-binding regions on NRSF RD-1, we next performed a GST pull-down assay using the deletion forms of GST-fused NRSF RD-1 (Fig. 2A) and in vitro translated TBP. The results revealed that GST–NRSF(1–44) was sufficient for binding to TBP (Fig. 2B). In a previous study (12) we showed that amino acids 1–44 of NRSF RD-1 did not retain sufficient repression activity in themselves. Therefore, this result suggests that the interaction of TBP with the NRSF N-terminal region is not essential for the repression activity of RD-1. However, TFIIB and TFIIF-RAP30 bound to GST–NRSF(1–83), but interacted little or not at all with GST–NRSF(1–76), indicating that residues 76–83 of the RD-1 region are required for sufficient binding to TFIIB or RAP30 (Supplementary Material, Fig. 7S). These results suggest that interaction of the NRSF RD-1 with TFIIB or TFIIF-RAP30 may be involved in the repression mechanism of RD-1, because residues 76–83 of NRSF were previously shown to be necessary for the repression activity of RD-1 (12). To test for a possible direct interaction of NRSF with TFIIB, we made TFIIB deletions [the N-terminal region, TFIIB(1–121); the internal direct repeat, TFIIB(122–215); the C-terminal direct repeat, TFIIB(216–316)] (26). These constructs were radiolabeled by in vitro transcription–translation and then processed for a GST pull-down assay using GST-fused NRSF RD-1 and RD-2 (Supplementary Material, Fig. 7S). The results revealed that the internal and C-terminal domains of TFIIB bound to NRSF RD-1, whereas NRSF RD-2 interacted weakly with the TFIIB C-terminus. In contrast, the TFIIB N-terminal domain did not bind to NRSF. These results indicate that TFIIB directly interacts with the NRSF RD-1 through internal and/or C-terminal direct repeats of the TFIIB molecule.
Figure 2.
Minimum interaction regions of NRSF-N terminal domain for TBP. (A) Schematic representation of the various forms of GST-fused NRSF proteins used in (B). (B) Interaction of in vitro translated TBP-binding region with the NRSF N-terminal regions. In vitro binding, the so-called GST pull-down assay, was tested against a series of deletion constructs (NRN1–44, NRN1–76, NRN1–83 and NRN1–156) or GST as a negative control. (C) Summary of interaction of TBP and co-repressor complex with the N-terminal repression domain of NRSF.
NRSF RD-1 and RD-2 bind to the evolutionally conserved H1–H2 and H1′–H2′ regions, respectively, on TBP
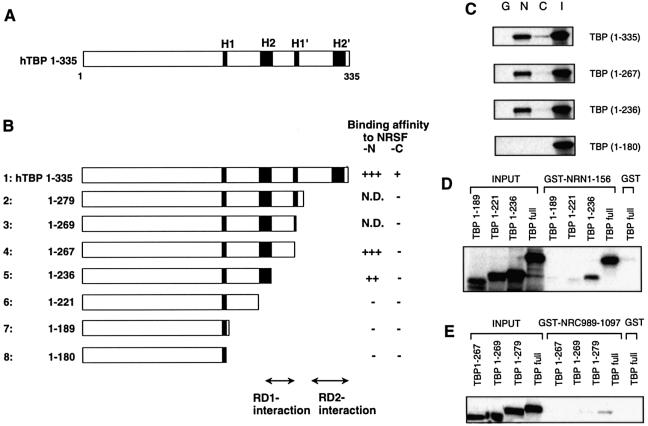
We next determined the NRSF RD-1- and RD-2-binding regions on TBP. The tertiary structure of TBP has been determined and the conserved C-terminal domain was shown to form a saddle-shaped structure with its underside containing eight antiparallel β-sheets (27) that form the DNA (TATA box)-binding surface of TBP (28,29). When TBP is bound to the TATA box, much of the opposite surface of the molecule, including its four α-helices and many of the loops connecting the β-strands, should be available for interaction with other proteins.
To test the possible direct interaction of NRSF with these regions of TBP, we made C-terminal deletion constructs of TBP radiolabeled by in vitro transcription–translation and then performed GST pull-down assays using GST-fused NRSF RD-1 and RD-2 (Fig. 3B). We first examined four TBP constructs [the full-length TBP(1–335), TBP(1–267), TBP(1–236) and TBP(1–180)] (Fig. 3C). TBP(1–267), a construct with H1′ and H2′ deleted, still retained a strong affinity for NRSF-N. TBP(1–236), containing the H1 and H2 regions, showed weak but significant binding to NRSF-N. In contrast, TBP(1–180), containing only H1, showed no interaction with NRSF-N. On the other hand, binding to NRSF-C was detected only with wild-type TBP. To identify the binding region in detail, another series of deletion constructs of TBP were examined for their interaction with NRSF-N (Fig. 3D) and NRSF-C (Fig. 3E). TBP(1–221), a construct with a deletion from the H2 region to the C-terminal end, and TBP(1–189), with a further deletion, did not bind to NRSF-N (Fig. 3D). These results indicate that amino acids 221–267 containing the H2 region of TBP are required for the interaction of NRSF RD-1 with TBP. Since no TBP deletion constructs had affinity for NRSF-C except for full-length TBP [TBP(1–335), Fig. 3E], NRSF RD-2 seemed to interact with TBP through amino acid residues 279–335 containing the H2′ region (as summarized in Fig. 3B). This binding is weaker than that of NRSF-N. However, the specificity of this binding is also shown in Figure 1D, where Cm and Cd abolished the binding to TBP that was apparent with NRSF-C. In contrast to this binding, when nuclear extracts were used the binding became more apparent compared with that of NRSF-N (Fig. 1D). This fact may suggest that other factors may also be required for more stable binding between NRSF-C and TBP.
Figure 3.
Delineation of the NRSF RD-1- and RD-2-binding regions on TBP. (A) Schematic representation of the TBP structure. The evolutionarily conserved helix regions (H1, H1′, H2 and H2′) are indicated by shaded boxes. (B) TBP deletion constructs used in the GST pull-down assay. The relative binding affinity of TBP for NRSF-N or NRSF-C was assessed by quantification of the autoradiograms and scored from background level (–) to strong interaction (+++); N.D.; not determined. The RD-1- and RD-2-interaction regions are indicated. (C) Specific binding of NRSF-RD-1, i.e. N1–156 (N), or RD-2, i.e. C989–1097 (C) to the in vitro translated TBP deletion constructs. GST alone (G) was used as a negative control and a 20% aliquot of total input protein (I) was loaded as a control. To identify the binding region in more detail, we used another series of TBP deletion constructs with NRSF-N (D) or NRSF-C (E).
NRSF RD-2 binding to TBP is necessary for repression by NRSF
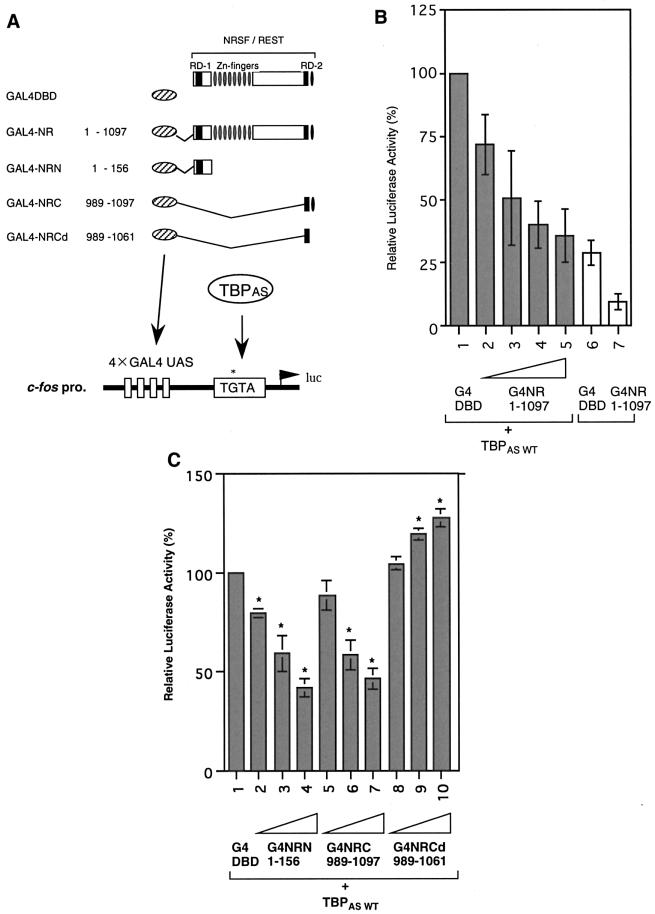
To monitor the effect of the interaction of NRSF with TBP on repression activity in vivo, we used the altered specificity TBP assay first described by Strubin and Struhl (30). This so-called TBPAS assay employs a TBP derivative with a triple amino acid substitution in its DNA-binding surface. This TBP derivative (TBPAS) has a relaxed DNA-binding specificity and recognizes an altered TATA box, i.e. TGTA, rather than the normal one. The use of this TBPAS and a TGTA promoter construct allowed us to test the effect of in vitro manipulations of TBP in vivo, as it discriminates between the activity of endogenous wild-type TBP and the manipulated one.
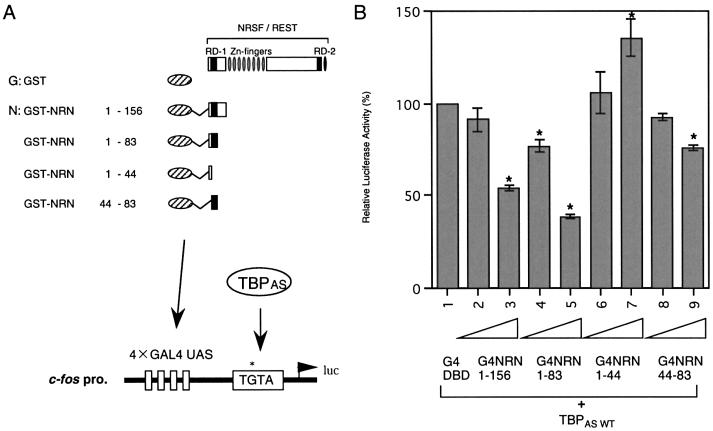
We performed a luciferase assay in Neuro2a cells using human TBPAS and a c-fos TGTA reporter linked to a GAL4–upstream activating sequence (UAS) synthetic enhancer (see Fig. 4A). At first, we examined the effect of TBPAS in transcriptional repression of a TGTA promoter, using G4 DBD-fused full-length constructs of NRSF [1.5, 7.5, 15 ng (lanes 2–4) and 22.5 ng (lanes 5 and 7]. Compared with modest repression without TBPAS (19% less activity, Fig. 4B, lanes 6 and 7), NRSF effectively repressed transcription of the TGTA promoter in the presence of TBPAS (65% less activity at best, Fig. 4B, lanes 1 and 5), suggesting that TBPAS is required for effective transcriptional repression mediated by NRSF. To further characterize the mechanism of NRSF repression, we performed this assay using NRSF RD-1 and RD-2. When G4 DBD-fused NRSF RD-1 or RD-2 was added at various concentrations (1, 5 and 10 ng), luciferase activity was decreased in a dose-dependent manner in both cases (Fig. 4C, lanes 2–4 and 5–7 compared with lane 1). Interest ingly, however, the repression activity of RD-2 was completely abolished by the use of a zinc finger deletion construct, which had lost its binding activity toward TBP (Fig. 4C, lanes 8–10). These results indicate that NRSF RD-2 binding to TBP is essential for the repression activity of RD-2.
Figure 4.
RD-2 binding to TBP is essential for NRSF-mediated transcriptional regulation via the TGTA promoter. (A) Schematic representation of reporter and effector constructs. GAL4 DBD-fused NRSF repression domain constructs used in the luciferase assay are indicated. Luciferase reporter constructs contained four synthetic GAL4 DNA-binding sites (GAL4 UAS) upstream of the c-fos promoter altered TATA box, the TGTA box, linked to a luciferase gene. (B) Reporter assay on the TGTA promoter using GAL4-fused full-length NRSF with or without TBPAS. (C) Reporter assay for the effect of NRSF N- or C-terminal repression domains on the TGTA promoter. Transcriptional activities with various amounts of GAL4-fused NRN, NRC or mutated NRCd are compared with the activity of the GAL4 DBD control.
NRSF RD-1 binding to TBP is also required for repression by NRSF
We also examined the effect of the NRSF RD-1 interaction with TBP on the repression activity of RD-1. Here we define RD-1 in a broader sense, including the very N-terminal region in addition to the previously defined RD-1 for HDAC interaction (12). Various G4 DBD-fused RD-1 deletion constructs were used for the luciferase assay (Fig. 5A). G4–NRSF-N (G4–NRN)(1–44), which contained the minimum region for binding to TBP, could not repress transcription, indicating that TBP-binding to NRSF RD-1 is not sufficient for the repression activity of RD-1 (Fig. 5B, lanes 6 and 7). G4–NRN(1–83) exhibited very efficient repression activity in this assay using TBPAS (lanes 4 and 5), compared with the weak activity of G4–NRN(44–83), which had been identified as the minimum region for NRSF RD-1 repression mediated by the mSin3–HDAC complex in our previous study (12) (lanes 8 and 9). These results indicate that the binding of TBP to the very N-terminal end of NRSF increases the repression activity of NRSF RD-1, when RD-1 forms a complex including histone deacetylases. Thus, TBP-binding via NRSF-N is required to recruit the NRSF–mSin3–HDAC complex to the core promoter region in the NRSF RD-1-mediated repression mechanism.
Figure 5.
NRSF RD-1 binding to TBP is not sufficient but is required for repression by NRSF. (A) Reporter and effector constructs used in the luciferase assay are as in Figure 4. (B) Transcriptional repression in the presence of various portions of NRSF RD-1. G4NRN1–83 could repress transcription, but G4NRN1–44 could not.
Correlation between NRSF binding to TBP and NRSF repression activity
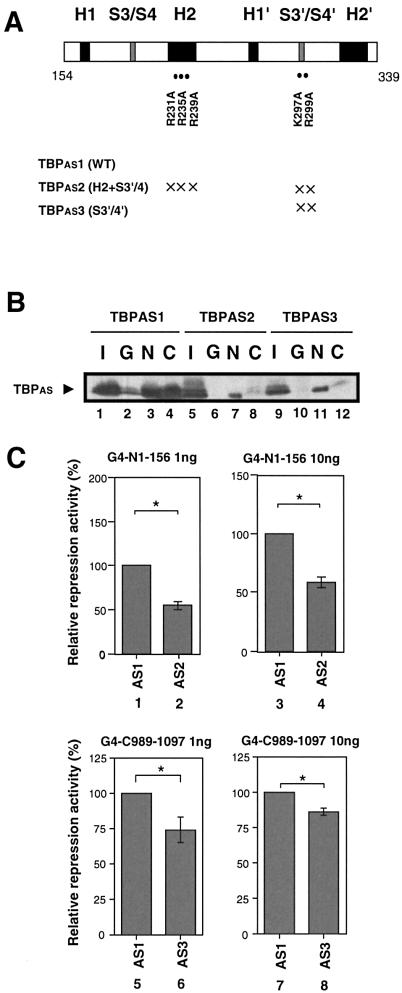
To further test the requirement for NRSF binding to TBP for NRSF-mediated transcriptional repression, we performed GST pull-down and luciferase assays using a series of TBPAS mutants (Fig. 6). Based upon the results of the aforementioned binding analysis shown in Figure 3, we used TBPAS mutants that had mutations of residues located on the surface and contained in the binding regions for NRSF-N or NRSF-C of TBP. At first, we analyzed a double substitution mutant of TBPAS, termed H2+S3′/S4′ (31). This mutant has three Arg (R)→Ala (A) substitutions at R231A, R235A and R239A on helix 2 and two additional mutations at K297A and R299A in the loop connecting sheet 3′ with sheet 4′ in the C-terminal region of TBP. The results of the pull-down assay (Fig. 6B) showed that RD-1 (N) binding to this TBPAS mutant (AS5, lane 7) was significantly decreased compared with that to wild-type TBPAS (AS1, lane 3). In addition, the repression activity of RD-1 was also decreased by this TBPAS mutant (Fig. 6C, lanes 2 and 4). Next, a TBPAS mutant termed S3′/S4′ was tested. This mutant had the same two residue amino acid replacements in the connecting loop as in the above double substitution mutant. The RD-2-binding activity of the TBPAS mutant S3′/S4′ (Fig. 6B, lane 12) was decreased compared to wild-type TBPAS (Fig. 6B, AS1, lane 4) and the repression activity of RD-2 was decreased (Fig. 6C, lanes 6 and 8). These results suggest that the repression activities of NRSF RD-1 and RD-2 correlate with their binding activity for TBP. However, we could not detect a significant correlation between the binding to TBP and repression activity with these single point mutants of TBPAS. These results suggest that the NRSF–TBP interaction occurs at multiple residues and/or induces a conformational change in the tertiary structure of the TBP-containing more global complex of the transcriptional initiation machinery.
Figure 6.
Correlation between NRSF binding to TBP and NRSF repression activity. (A) TBPAS mutants used in these NRSF binding and NRSF-mediated transcriptional repression assays. Positions of specific amino acid mutations are marked (with an ×) along with the C-terminal half domain of TBP: helix regions (H1, H2, H1′ and H2′) are indicated by filled black boxes and loop regions connecting sheet 3 and sheet 4 (S3/S4) or sheet 3′ and sheet 4′ (S3′/S4′) by filled gray boxes. TBPAS mutants at multiple positions (TBPAS 2 and 3) are shown in the panel. (B) Interactions of TBPAS mutants with NRSF as evidenced by the GST pull-down assay: GST (G), GST-NRSF-N (N) and GST-NRSF-C (C). Nuclear extracts of COS7 cells transfected with each of the TBPAS mutants (TBPAS 1, 2 and 3), as indicated, were subjected to the pull-down assay. A 10% aliquot of total input protein (I) was loaded as an internal control. (C) A series of TBPAS mutant expression plasmids were co-transfected into Neuro2a cells with GAL4 DBD–NRSF-N (G4-N1–156) (lanes 1–4) or GAL4 DBD–NRSF-C (G4-C989–1097) (lanes 5–8) constructs (1 or 10 ng) and repression activity was analyzed by the luciferase assay. Repression activities were calculated relative to the activity obtained using the GAL4 DBD plasmid as effector.
DISCUSSION
Our experiments demonstrate that NRSF interacts with core promoter factor TBP and that this interaction is required for the transcriptional repression mediated by NRSF. These results indicate that the recruitment of NRSF to the core promoter region is of importance for repression and may allow the NRSF–mSin3–HDAC complex to deacetylate histones located near the promoter region of target genes. In addition, our results suggest that a new HDAC-independent mechanism of repression mediated by a direct interaction between NRSF and TBP is part of the transcriptional regulation by NRSF.
The transcription initiation of any given gene involves an ordered assembly of pol II and other transcription core initiation factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH) to form a preinitiation complex (PIC) on the core promoter (32,33). TBP in the TFIID complex binds specifically to the TATA box to initiate a stepwise assembly of the PIC for pol II transcription. This is followed by the ordered assembly of TFIIA, TFIIB, pol II/TFIIF, TFIIE and TFIIH. The fact that both NRSF RD-1 and RD-2 could bind TBP and, more specifically, RD-1 and RD-2 bound as an ordered array along the helical stretches of the so-called ‘saddle-shaped’ structure of TBP may suggest that collaboration of RD-1 and RD-2 is necessary to form a stable inhibitory complex of NRSF–TBP at the promoters of various neuron-specific genes. On the other hand, it is also possible that these TBP–RD-1 and TBP–RD-2 interactions occur independently under various circumstances of neuronal differentiation and/or repression. Whichever the case, NRSF seems to interact with TBP at the very first step of pol II transcription.
As to repression by the N-terminal RD-1, it is worth noting that TBP binding occurred at the very N-terminal portion (1–44), whereas the ‘real’ RD-1 with repression activity resided a little downstream and included residues 44–83 (12). Since the repression activity of the latter domain was mediated by the interaction with mSin3 and HDAC (12), the N-terminal region of NRSF (RD-1 in a ‘broader’ sense) could confirm a large complex with TBP, mSin3 and HDAC.
In a previous study we reported that the repression activity of the C-terminal RD-2 was not released by TSA, suggesting the possibility of a HDAC-independent mechanism (12). A plausible mechanism would be that the NRSF–TBP interaction inhibits the formation of a transcription PIC by disrupting the interaction between TBP and other core promoter factors, such as TFIIB. This idea is supported by our results for the binding assay between NRSF and general transcription factors (Fig. 1), showing that RD-2 bound TBP but not TFIIB. In addition, the interaction of NRSF RD-1 with TFIIB may inhibit TFIIB from interacting with the TBP–DNA complex, because RD-1 interacts with the direct repeats region of TFIIB, which is also utilized for formation of the TBP–TFIIB–DNA complex (26). Recent studies by others have shown interactions between transcription factors or cofactors and general transcription factors. In the absence of its ligand, the thyroid hormone receptor makes contact with components of the general transcription machinery and interferes with formation of the transcription PIC (34). A SMRT–mSin3 complex directly interacts with TFIIB (35). In addition, Ikaros, a repressor in the lymphoid system, can repress transcription through interactions with HDAC-independent co-repressors, CtBP and CtIP, which can interact with TBP and TFIIB (36). More recently, the X-ray structure of Negative cofactor 2 (NC2), a negative regulator of TATA-dependent transcription initiation, was determined as a NC2–TBP–DNA complex, demonstrating that NC2 represses by inhibiting TFIIB from binding to the TBP–TATA element binary complex (37). In the case of NRSF, the first direct target of NRSF-N (RD-1) and NRSF-C (RD-2) seems to be TBP, and TFIIB would follow TBP and bind to the associated NRSF-N (RD-1). This scheme seems reminiscent of the aforementioned case of the NC2–TBP–TFIIB complex. Three-dimensional structural studies would allow further understanding of the molecular interaction of NRSF and core promoter factors.
So how would the NRSF–TBP interaction influence transcriptional function? One possible mechanism could be that direct interaction between NRSF and TBP prevents binding of TBP to the TATA box of the promoter, which is the first step in assembly of the PIC. A recent study revealed that Mot1, a general transcriptional repressor, prevented the binding of TBP to inactive promoters (38), whereas transcription activators enhanced TBP binding to promoters and transcriptional activity correlated strongly with the degree of TBP occupancy. In this regard, it is possible that NRSF may inhibit TBP binding. Thus, these reports support the possibility of an NRSF C-terminal repression mechanism acting via direct interaction between NRSF and TBP.
It is interesting to speculate how these dual repression domains, i.e. RD-1 and RD-2, might regulate target genes during neuronal differentiation. Since HDAC-mediated mechanisms require more complex and energy-dependent steps compared with TBP-mediated core factor inhibition, it is possible that the N-terminal domain (RD-1) is used for major regulation, such as expression of genes required for commitment from non-neuronal to neuronal cells in a chromatin-dependent fashion, whereas the C-terminal domain (RD-2) may regulate the expression of genes required for specific neuronal subsets, such as cholinergic or dorpaminergic cells. Otherwise, RD-1 and RD-2 may be utilized in a distinct spatio-temporal manner under various circumstances during the course of neural development. The expression patterns of the mSin3A and CoREST genes during mouse development support this idea. At embryonic day 8.5 (E8.5), mSin3A is expressed widely throughout the embryo, along with NRSF, whereas expression of CoREST is restricted to the head region (15). Furthermore, TSA treatment released transcription repression of the type II sodium channel gene mediated by the C-terminal domain of NRSF (21), however, TSA could not release transcriptional repression of the SCG10 and GluR2 glutamate receptor genes (12,17). These results suggest that repression of the C-terminal domain of NRSF is multiple and has a distinct promoter-specific mechanism. Further study using an in vitro transcription system may allow us to define in more detail the mechanism of NRSF repression with respect to interactions between NRSF and the general transcription factors.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr W. Herr for the series of TBPAS expression vectors and the TGTA reporter plasmid. We also thank Drs P. Carlsson, S. Kitajima and Y. Ohkuma for the gift of pET11-human TBP, pUC19-RAP30 and plasmids encoding TFIIEα and TFIIEβ, respectively. We gratefully thank Dr H. Kimura for helpful suggestions and encouragement. Additionally, the excellent technical assistance of Ms Y. Kadokawa and I.Nakano is acknowledged. This study was supported by grants from CREST, Science and Technology Corporation of Japan (JST), and the Japanese Ministry of Education, Science, Sport and Culture and also in part by the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim and funds for Comprehensive Research on Aging and Health from the Ministry of Health, Welfare and Labor (to N.M.).
REFERENCES
- 1.Fry C.J. and Peterson,C.L. (2001) Chromatin remodeling enzymes: who’s on first? Curr. Biol., 11, R185–R197. [DOI] [PubMed] [Google Scholar]
- 2.Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- 3.Emerson B.M. (2002) Specificity of gene regulation. Cell, 109, 267–270. [DOI] [PubMed] [Google Scholar]
- 4.Schoenherr C.J. and Anderson,D.J. (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science, 267, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 5.Chong J.A., Tapia-Ramirez,J., Kim,S., Toledo-Aral,J.J., Zheng,Y., Boutros,M.C., Altshuller,Y.M., Frohman,M.A., Kraner,S.D. and Mandel,G. (1995) REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell, 80, 949–957. [DOI] [PubMed] [Google Scholar]
- 6.Schoenherr C.J. and Anderson,D.J. (1995) Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr. Opin. Neurobiol., 5, 566–571. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z.F., Paquette,A.J. and Anderson,D.J. (1998) NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nature Genet., 20, 136–142. [DOI] [PubMed] [Google Scholar]
- 8.Mori N., Mizuno,T., Murai,K., Nakano,I. and Yamashita,H. (2002) Effect of age on the gene expression of neural-restrictive silencing factor NRSF/REST. Neurobiol. Aging, 23, 255–262. [DOI] [PubMed] [Google Scholar]
- 9.Schoenherr C.J., Paquette,A.J. and Anderson,D.J. (1996) Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl Acad. Sci. USA, 93, 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tapia-Ramirez J., Eggen,B.J., Peral-Rubio,M.J., Toledo-Aral,J.J. and Mandel,G. (1997) A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc. Natl Acad. Sci. USA, 94, 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiel G., Lietz,M. and Cramer,M. (1998) Biological activity and modular structure of RE-1-silencing transcription factor (REST), a repressor of neuronal genes. J. Biol. Chem., 273, 26891–26899. [DOI] [PubMed] [Google Scholar]
- 12.Naruse Y., Aoki,T., Kojima,T. and Mori,N. (1999) Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl Acad. Sci. USA, 96, 13691–13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolffe A.P. (1994) Transcription: in tune with the histones. Cell, 77, 13–16. [DOI] [PubMed] [Google Scholar]
- 14.Struhl K. (1996) Chromatin structure and RNA polymerase II connection: implications for transcription. Cell, 84, 179–182. [DOI] [PubMed] [Google Scholar]
- 15.Grimes J.A., Nielsen,S.J., Battaglioli,E., Miska,E.A., Speh,J.C., Berry,D.L., Atouf,F., Holdener,B.C., Mandel,G. and Kouzarides,T. (2000) The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem., 275, 9461–9467. [DOI] [PubMed] [Google Scholar]
- 16.Roopra A., Sharling,L., Wood,I.C., Briggs,T., Bachfischer,U., Paquette,A.J. and Buckley,N.J. (2000) Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol., 20, 2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y., Myers,S.J. and Dingledine,R. (1999) Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nature Neurosci., 2, 867–872. [DOI] [PubMed] [Google Scholar]
- 18.Kuwahara K., Saito,Y., Ogawa,E., Takahashi,N., Nakagawa,Y., Naruse,Y., Harada,M., Hamanaka,I., Izumi,T., Miyamoto,Y., Kishimoto,I., Kawakami,R., Nakanishi,M., Mori,N. and Nakao,K. (2001) The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol. Cell. Biol., 21, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey G.W., Wang,Y., Russanova,V.R., Hirai,T., Qin,J., Nakatani,Y. and Howard,B.H. (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem., 276, 6817–6824. [DOI] [PubMed] [Google Scholar]
- 20.You A., Tong,J.K., Grozinger,C.M. and Schreiber,S.L. (2001) CoREST is an integral component of the CoREST–human histone deacetylase complex. Proc. Natl Acad. Sci. USA, 98, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballas N., Battaglioli,E., Atouf,F., Andres,M.E., Chenoweth,J., Anderson,M.E., Burger,C., Moniwa,M., Davie,J.R., Bowers,W.J., Federoff,H.J., Rose,D.W., Rosenfeld,M.G., Brehm,P. and Mandel,G. (2001) Regulation of neuronal traits by a novel transcriptional complex. Neuron, 31, 353–365. [DOI] [PubMed] [Google Scholar]
- 22.Kadosh D. and Struhl,K. (1998) Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol., 18, 5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellqvist M., Mahlapuu,M., Blixt,A., Enerback,S. and Carlsson,P. (1998) The human forkhead protein FREAC-2 contains two functionally redundant activation domains and interacts with TBP and TFIIB. J. Biol. Chem., 273, 23335–233343. [DOI] [PubMed] [Google Scholar]
- 24.Haviv I., Matza,Y. and Shaul,Y. (1998) pX, the HBV-encoded coactivator, suppresses the phenotypes of TBP and TAFII250 mutants. Genes Dev., 12, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori N., Schoenherr,C., Vandenbergh,D.J. and Anderson,D.J. (1992) A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron, 9, 45–54. [DOI] [PubMed] [Google Scholar]
- 26.Hisatake K., Roeder,R.G. and Horikoshi,M. (1993) Functional dissection of TFIIB domains required for TFIIB-TFIID-promoter complex formation and basal transcription activity. Nature, 363, 744–747. [DOI] [PubMed] [Google Scholar]
- 27.Nikolov D.B., Hu,S.H., Lin,J., Gasch,A., Hoffmann,A., Horikoshi,M., Chua,N.H., Roeder,R.G. and Burley,S.K. (1992) Crystal structure of TFIID TATA-box binding protein. Nature, 360, 40–46. [DOI] [PubMed] [Google Scholar]
- 28.Kim J.L., Nikolov,D.B. and Burley,S.K. (1993) Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature, 365, 520–527. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y., Geiger,J.H., Hahn,S. and Sigler,P.B. (1993) Crystal structure of a yeast TBP/TATA-box complex. Nature, 365, 512–520. [DOI] [PubMed] [Google Scholar]
- 30.Strubin M. and Struhl,K. (1992) Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell, 68, 721–730. [DOI] [PubMed] [Google Scholar]
- 31.Tansey W.P., Ruppert,S., Tjian,R. and Herr,W. (1994) Multiple regions of TBP participate in the response to transcriptional activators in vivo. Genes Dev., 8, 2756–2769. [DOI] [PubMed] [Google Scholar]
- 32.Ranish J.A. and Hahn,S. (1996) Transcription: basal factors and activation. Curr. Opin. Genet. Dev., 6, 151–158. [DOI] [PubMed] [Google Scholar]
- 33.Roeder R.G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci., 21, 327–335. [PubMed] [Google Scholar]
- 34.Fondell J.D., Roy,A.L. and Roeder,R.G. (1993) Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev., 7, 1400–1410. [DOI] [PubMed] [Google Scholar]
- 35.Wong C.W. and Privalsky,M.L. (1998) Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms and a potential role for TFIIB. Mol. Cell. Biol., 18, 5500–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koipally J. and Georgopoulos,K. (2002) Ikaros-CtIP interactions do not require C-terminal binding protein and participate in a deacetylase-independent mode of repression. J. Biol. Chem., 277, 23143–23149. [DOI] [PubMed] [Google Scholar]
- 37.Kamada K., Shu,F., Chen,H., Malik,S., Stelzer,G., Roeder,R.G., Meisterernst,M. and Burley,S.K. (2001) Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell, 106, 71–81. [DOI] [PubMed] [Google Scholar]
- 38.Li X.Y., Virbasius,A., Zhu,X. and Green,M.R. (1999) Enhancement of TBP binding by activators and general transcription factors. Nature, 399, 605–609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.