Abstract
Sequence analysis of the group of proteins known to be associated with hereditary diseases allows the detection of key distinctive features shared within this group. The disease proteins are characterized by greater length of their amino acid sequence, a broader phylogenetic extent, and specific conservation and paralogy profiles compared with all human proteins. This unique property pattern provides insights into the global nature of hereditary diseases and moreover can be used to predict novel disease genes. We have developed a computational method that allows the detection of genes likely to be involved in hereditary disease in the human genome. The probability score assignments for the human genome are accessible at http://maine.ebi.ac.uk:8000/services/dgp.
INTRODUCTION
The identification of genes involved in inherited human disease requires a large effort to collect inheritance patterns from families with the disease and to perform linkage analysis and/or mutation analysis for the candidate genes in order to identify the gene(s) involved in a particular hereditary disorder (1). The mutation analysis of candidate genes is a tedious, labour-intensive activity and sometimes requires the analysis of a large number of genes before finding the causative mutation of the disease under consideration.
The genes known to be implicated in human disease and the mutations causing these disorders are collected in several databases such as OMIM (Online Mendelian Inheritance in Man) (2), LocusLink (3) and The Human Gene Mutation Database (4). This amount of information, together with the sequence data derived from the human genome (5) and other organisms (6,7), provides a unique opportunity to identify intrinsic attributes of disease-associated genes, leading to a deeper understanding of the causes of human hereditary disease.
Despite the significance of these resources for human welfare, only limited work has been carried out on the global analysis of disease genes as a group. Recently, a report has explored the functional classification of disease genes and their products, and described a correlation between the function of the gene product and general features of the disease, such as the age of onset and the mode of inheritance (8). Furthermore, sequence analysis for several eukaryotes revealed that human proteins with multiple long amino acid runs are often associated with disease (9).
A protein is involved in a hereditary disease when its corresponding gene has suffered a mutation that impairs its function or expression strongly enough to produce a certain phenotype that is classified as disease. The likelihood of a protein being involved in disease should scale with the probability of its gene to suffer mutations with large (but non-lethal) fitness effects. We can expect that long proteins with highly conserved amino acid sequences would be more likely to exhibit disease mutations. Moreover, proteins with similar paralogues would be less likely to be involved in disease since they could rescue the mutant phenotype.
We thus hypothesized that human genes involved in hereditary disease have some distinct sequence properties in common which render them more susceptible to mutations causing genetic disorders. To test this hypothesis, we compiled and analysed a set of 1567 proteins encoded by genes known to be involved in disease from the OMIM database (2). We then compared them with the rest of the human proteins for some selected properties.
MATERIALS AND METHODS
We obtained a list of genes reported to cause a disease when mutated from the OMIM database (2). The OMIM identification numbers of genes involved in hereditary disease were selected from the ‘morbid map’ table in the OMIM database. Using the National Center for Biotechnology Information (NCBI) LocusLink (10) database (from tables mim2loc and loc2ref) and the Ensembl database (11), we located the corresponding protein sequence records. The result was a list of 1567 genes associated with human diseases and their protein sequences. Each of the protein sequence entries were compared against a dataset containing all the protein sequences from complete genomes (15 Archaea, 61 Bacteria and seven Eukarya: Encephalitozoon cuniculi, Arabidopsis thaliana, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Caenorhabditis elegans and Mus musculus) obtained from CoGenT (12), and all protein sequences from SwissProt-TrEmbl (7) (January 2003) using BlastP (13). These sequences were divided into nine taxonomic groups: viruses, archaea, bacteria, protista, fungi, plants, (while metazoa were further divided in) invertebrates, vertebrates (excluding mammals) and mammals (excluding humans), as described elsewhere (14). This taxonomic partition is not designed as an evolutionary tree, but rather to serve as a landmark of phylogenetic distance of these taxa from humans. We computed phylogenetic profiles (15) for all proteins. Sequence comparisons were performed using BlastP (version 2.09) and the BLOSUM62 matrix with an E-value threshold of 10–6; sequences were filtered for compositional bias with CAST (complexity analysis of sequence tracts) (16).
To assess the statistical significance of the findings, 10 000 protein sets of the same size were randomly selected from all the human genome proteins (22638 protein sequences in total; Ensembl version 15.33.1) (11) and used as control sets. An identical BlastP analysis was performed for each of the 10 000 control sets. The number of proteins in each of the 10 001 sets (10 000 randomly selected sets and the disease set) that detected at least one homologue in each taxonomic group was considered for the analysis of phylogenetic extent. To assess the degree of conservation of disease proteins compared with the rest of the human proteins, we computed for each protein in each taxonomic group what we define as the conservation score (CS). This is the BlastP score of the closest homologue in that taxonomic group divided by the BlastP score of the protein against itself, ranging from 0 to 1 (when the closest homologue is 100% identical). This measure gives an estimation of the mutation rate that the protein has been subjected to during evolution, which is independent of the length of the protein.
To examine the degree of paralogy of disease proteins, a BlastP search was performed for each protein in the group against the longest protein sequence of each gene in the human genome (22 638 protein sequences in total), to exclude alternatively spliced forms. The best BlastP hit found in the human genome (excluding the hit against itself) was obtained for each protein and conservation scores between paralogues were calculated. In the analysis of each feature, the Z-score [Zx = (X – µx)/σx] and P-value [Px = Σ(nx ≥ X)/N] were calculated to statistically assess the results observed in the disease set (X) against the 10 000 randomly selected sets (where mean = µx, standard deviation = σx, and nx = the number of sets that satisfy the condition for the calculation of the P-value).
To test the differences in the distributions of conservation scores between disease and human proteins, the Kolmogorov–Smirnov test was applied, which provides the probability (P-value) that the two distributions are equal, and also the maximum distance between them. This analysis was also applied for distributions of length (Table 1).
Table 1. Statistical analysis.
| Phylogenetic extent Z-scorea | KS test: conservation scoreb D (%) | P-value | |
|---|---|---|---|
| Viruses | 7.2 | 5.5 | 2.8e-04 |
| Archaea | 12.8 | 11 | 7.8e-16 |
| Bacteria | 16.7 | 17.5 | <2.2e-16 |
| Protista | 6.5 | 8 | 1.1e-08 |
| Fungi | 4.9 | 6.2 | 2.2e-05 |
| Plants | 3.9 | 5 | 1.1e-03 |
| Invertebrates | 12.9 | 15.1 | 2.2e-16 |
| Vertebrates (no mammals) | 17.5 | 21.8 | <2.2e-16 |
| Mammals | 12.5 | 23 | <2.2e-16 |
| Paralogues | - | 10.5 | 2.2e-14 |
| KS test: length distributionc D (%) | P-value | ||
| Length | 24.5 | <2.2e-16 |
aZ-score of the number of proteins conserved in each taxonomic group between 10 000 randomly selected sets and disease set [Zx = (X – µx)/σx] and P-value [Px = Σ(nx > X)/N]. The P-value for all taxonomic groups is <1e-04.
bKolmogorov–Smirnov (KS) test of the conservation score between disease proteins and all the human proteins. The KS test analyses how different two distributions are, and computes a probability (P-value) that the two distributions are equal to well as the maximum distance (D) between them.
cKS test of the distribution of length between disease proteins and all human proteins.
The features used to build the decision tree-based model (17) correspond to protein length, phylogenetic extent, degree of conservation and paralogy. To represent phylogenetic extent and conservation, we computed the conservation score for each protein in each taxonomic group. To quantify paralogy, the conservation score with the closest paralogue was used as a parameter.
We searched for all these parameters in the 1567 proteins known to be involved in disease and the 1567 other proteins that were taken randomly from the human genome and were not known to be involved in any disease. The result is 3134 vectors, one for each protein, with 11 dimensions [CS viruses, CS archaea, CS bacteria, CS protista, CS plants, CS fungi, CS invertebrates, CS vertebrates (no mammals), CS mammals, CS paralogues and protein length]. These vectors were used to generate a decision tree using the ‘Machine Learning in C++’ (MLC++) library (17), through SGI’s Mineset™ machine-learning software suite (version 2.5) (18). The derived model can then be applied to other proteins in order to obtain a probability score for these proteins being involved in human disease.
To cross-validate our prediction method, we executed two widely used tests: self-consistency and jack-knife. The self-consistency test consists of estimating the probability score for each protein that has been used to build the model, called the learning set. To obtain a global estimate, this test examines how well the model can predict the entire learning set. Our model assigned a probability score of >0.5 to all the disease proteins in the learning set (100%) (Table 2).
Table 2. Validation of the prediction model.
| Probability score | Total of predicted genes | Total of known disease genes | Number of new genes | ||
|---|---|---|---|---|---|
| >0.5 | 7770 | 1567 | 6203 | ||
| >0.55 | 5795 | 1567 | 4228 | ||
| >0.60 | 4247 | 1567 | 2680 | ||
| >0.65 | 3031 | 1556 | 1475 | ||
| >0.70 | 1734 | 1073 | 661 | ||
| >0.75 | 507 | 284 | 223 | ||
| >0.80 | 101 | 52 | 49 | ||
| >0.85 |
14 |
8 |
6 |
|
|
| Actual | Predicted | Self-consistency test | Jack-knife test | ||
| n | % | n | |||
| A | Disease | Disease | 1567 | 100 | 274 |
| B | Disease | Non-disease | 0 | 0 | 115 |
| C | Non-disease | Disease | 0 | 0 | 136 |
| D |
Non-disease |
Non-disease |
1567 |
100 |
259 |
| Formula | Decision tree | ||||
| Accuracy | (A+D)/(A+B+C+D) | 68% | |||
| Sensitivity | A/(A+B) | 70% | |||
| Precision | A/(A+C) | 67% | |||
To cross-validate the disease gene prediction method, we executed two widely used tests: self-consistency and jack-knife. The self-consistency test consists of estimating the probability score of being involved in disease for each protein that has been used to build the model, called the learning set. To obtain a global estimate, this test examines how well the model can predict the entire learning set. Our model assigned a probability score of >0.5 to all the disease proteins in the learning set (100%).
The jack-knife test consists of building the model with a fraction of the data (learning set; in this case, 75% of the total) and checking how well the model is able to predict the remaining fraction that has not been seen before (test set; 25% of the total). This test was performed 10 times: on average, 70% of the disease genes in the test set were predicted correctly. We want to point out here that the values of accuracy, sensitivity and precision shown in Table 2 have been calculated for a fraction of the genome (the test set), including 392 genes involved in disease and 392 genes not known to be involved in disease. These values are calculated for a cut-off probability score of 0.5. These assessments give us an estimation of the performance of our method. It should be noted that for higher cut-off values (i.e. 0.6 or 0.7) the precision increases and the sensitivity decreases, meaning that the model predicts less real disease genes but with fewer false-positives.
Although the positive set of proteins obtained from OMIM can generally be trusted, producing negative sets for proteins that are known not to be involved in disease is not possible. We thus faced the problem that our negative examples were provided by randomly selected proteins from the human genome and presumably were not known to be involved in disease. However, some of these proteins may well be involved in disease, although this property has not been detected yet. By implication, some of the false-positive predictions might represent true positives; indeed, this is the predictive power of our current inductive approach.
Finally, a matrix of 22 638 proteins [all the genes in the human genome (Ensembl version 15.33.1) (11)] with the 11 dimensions was generated and the decision tree model was applied to all of them; the probability score for possible involvement in a hereditary disease was then calculated for each gene.
A relational database has been set up to allow queries on these results and is available via the Internet at http://maine.ebi.ac.uk:8000/services/dgp, using the mySQL relational database management system and a set of PERL scripts using the DBI package. The probability scores for genes being involved in disease can be consulted via the Internet, querying for any gene of interest or a defined chromosomal region. It can also be searched for any of the diseases that have been mapped to a chromosomal region, but the disease-causing gene has not yet been found.
RESULTS
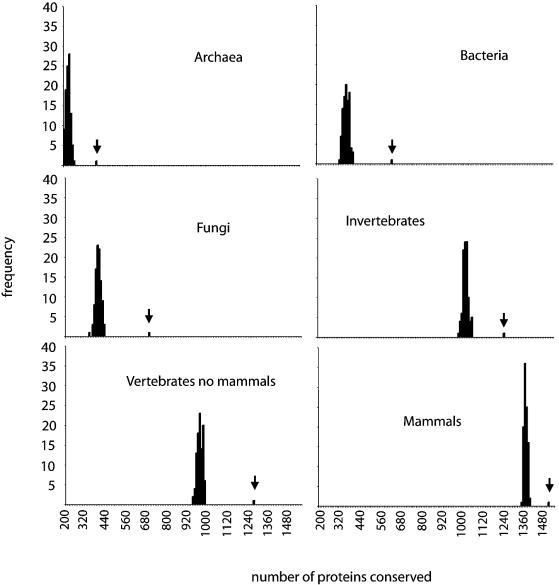
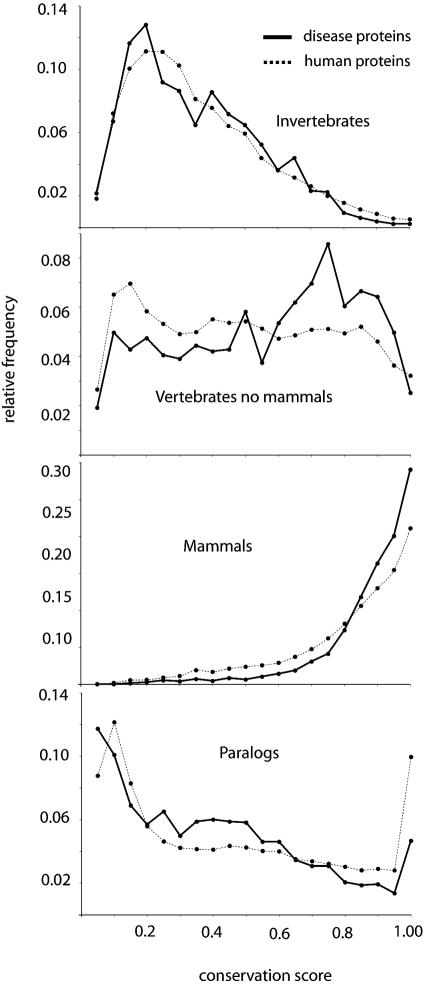
We first analysed protein phylogenetic extent and sequence conservation. We found that disease proteins exhibit a wider phylogenetic extent (Fig. 1; for statistical analysis see Table 1). Almost all disease proteins (99.5%) have homologues in mammals, and the majority of disease proteins (73%) have homologues both in vertebrates (mammals and no mammals) and invertebrates, while only 55% of human proteins have homologues in these three taxonomic groups (see Table 1 of Supplementary Material for additional information). Also, many more proteins in the disease group have homologues in bacteria (41%) or archaea (25%) compared with all human proteins (23% and 14%, respectively). Disease proteins are also more conserved than the rest of the human proteins, meaning that they tend to have larger conservation scores (Fig. 2; Table 1; see Materials and methods). On the other hand, we have also observed that extremely conserved proteins with conservation scores of ∼1 in vertebrates excluding mammals, >0.8 in invertebrates, or >0.6 in plants, fungi or protista, are less frequently found to be involved in disease (Fig. 2).
Figure 1.
Phylogenetic extent of human disease proteins. Frequency distributions of disease proteins (bars indicated by vertical arrows) with homologues in archaea, bacteria, fungi, invertebrates, vertebrates (excluding mammals) and mammals, versus 100 control sets of equal size containing randomly selected human proteins. Notice that less human proteins appear to have homologues in vertebrates (not mammals) than in invertebrates, yet this effect is only due to the fact that there are more sequences available from invertebrates (largely contributed by the two completed genomes of D.melanogaster and C.elegans). Number of proteins with homologues (from a maximum of 1567) is shown on the x-axis and the frequency of the sets on the y-axis.
Figure 2.
Conservation and paralogy of disease proteins. Distribution of conservation score of disease (solid line) and all human proteins (dotted line) against their closest homologue in invertebrates, vertebrates (not mammals) and mammals, and between paralogues. The conservation score gives an estimation of the mutation rate that the protein has been subjected to during evolution that is independent of the length of the protein: it is calculated as the BLAST score of the closest homologue in one taxonomic group, or the closest paralogue divided by the BLAST score of the protein against itself, ranging from 0 to 1 (when the closest homologue is 100% identical).
This wider phylogenetic extent could be partly due to historical biases. It is possible that genes found to be involved in disease in humans might have been preferentially sequenced in other species. Alternatively, the analysis of genes involved in disease could have been biased towards the most well studied genes, which may correspond to the most conserved ones. In order to remove these biases, we have performed an analysis of protein phylogenetic extent and conservation using data exclusively from complete genome sequences (see Table 2, and figs 1 and 2 of Supplementary Material). The results obtained using all available sequences (including SP-TrEMBL and complete genomes) or just sequences from complete genomes are very similar, indicating that the above-mentioned biases are negligible.
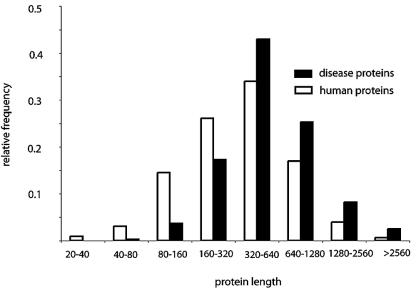
We then tested if protein sequence length is another shared property within disease genes, reasoning that the longer the coding sequence is, the more likely it is that the gene will suffer a disease-causing mutation. As predicted, we have found that proteins involved in human diseases are longer than the rest of the proteins encoded in the human genome. The average length of disease proteins is 699 amino acids, while the average length of the sequences in each randomly selected set ranges from 460 to 508 (Z-score = 19.8, P-value <1e–04, see Materials and methods). Furthermore, a comparison of the length distributions for the disease and all the human proteins shows a clear trend for the disease proteins to be longer than the rest of the proteins in the human genome (Fig. 3; Table 1). Paralogy is another salient feature of genes that causes human disease. We have found that genes with highly conserved paralogues are less frequently involved in disease (Fig. 2; Table 1).
Figure 3.
Protein length, calculated as number of amino acids, distribution of disease proteins (black) and all human proteins (white).
Taking into account that the above-mentioned features follow different trends in the proteins known to be involved in hereditary diseases compared with the rest of the human proteins, we also have the opportunity to identify which other proteins in the human genome follow this trend and thus are more likely to be involved in disease. We have developed a method based on a decision tree algorithm (17) that is able to predict whether a gene is associated with hereditary disease with 70% sensitivity and 67% precision (see Materials and methods; Table 2). This model has been applied to all the genes in the human genome and a probability score for possible involvement in a hereditary disease has been calculated for each gene. The probability score assignment for the entire human genome can be accessed online (http://maine.ebi.ac.uk:8000/services/dgp).
DISCUSSION
The proteins involved in hereditary diseases follow a specific property pattern. They tend to be long, conserved, phylogenetically extended, and without close paralogues.
Conserved proteins correspond to segments of the genome that have been exposed to strong evolutionary constraints and have not had the opportunity to accumulate many variations. These selective constraints can still be seen today in the sense that some variations in those genes have severe phenotypic consequences, thus bringing the patients in question to the attention of clinicians. Conversely, other proteins, under less selective pressure, have been able to vary among species, and are able to tolerate variations without causing a major phenotypic effect that would be classified as disease. On the other hand, we have also observed that extremely conserved proteins, with conservation scores of ∼1 in vertebrates (not mammals), >0.8 in invertebrates or >0.6 in plants, fungi or protista, are less frequently found to be involved in disease (Fig. 2). One likely explanation is that variations in this group of extremely conserved genes are mostly lethal.
These results on the conservation of proteins coded for genes that cause hereditary diseases are in agreement with the idea that genes with strong fitness effects should evolve more slowly than other genes (19,20). One analysis of the evolutionary distances between S.cerevisiae and C.elegans proteins indicates that the fitness effect of a protein influences its rate of evolution (21). More recently, another report suggested that essential genes are more evolutionarily conserved than non-essential genes in bacteria (22). Here, we demonstrate that genes with fitness effects in humans are also more evolutionarily conserved than other genes.
Other reports on the analysis of disease-causing mutations have shown that the most conserved residues of disease proteins are frequently found to be mutated in patients with the disease (23,24). Linked to our conclusions that disease proteins are mostly conserved, it is possible to infer that the mutations causing hereditary diseases in humans mostly occur in conserved residues of conserved proteins. It would be interesting to test this hypothesis in the future, provided that a reliable method for the detection of conserved residues is developed.
Genes with highly conserved paralogues are less likely to be involved in disease. This can be explained by the fact that conserved paralogues might be able to complement the function of a mutated gene, while non-conserved paralogues may have acquired a different function. Recently, it has been shown that in S.cerevisiae, there is a strong anti-correlation between the fitness effect of a deleted gene and the sequence similarity of its closest paralogue (25), suggesting that genes with highly similar paralogues are compensated for mutations more often than genes with distant paralogues.
Using the specific property pattern followed by genes involved in disease, we have developed a model to predict which genes in the human genome are more likely to be involved in disease. We have assigned a probability score of being involved in disease to all genes in the human genome. Two new disease genes have been described in the recent literature: CXCR4 as the causative gene of WHIM syndrome (26) and the GARS gene involved in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V (CMT2D/dSMA-V) (27). Both genes are predicted by our method to probably to be involved in disease, with probabilities of 0.688 and 0.695, respectively. Furthermore, CMT2D/dSMA-V diseases were mapped in a region of ∼980 kb (27), which contains 15 genes according to the Ensembl database (11), and the gene with the highest probability score in the region was the GARS gene. As a further example, the 20 highest-scoring proteins in the human genome not known to be involved in disease are shown in Table 3: these include enzymes, DNA-binding proteins, receptors, channels and proteins of unknown function.
Table 3. The top 20 predicted high-scoring genes not known to be involved in disease.
| Ensembl identifier | Name | Position | P-value |
|---|---|---|---|
| ENSG00000182175 | Repulsive guidance molecule A (RGMA) | 15q26 | 0.87 |
| ENSG00000034827 | Calcium channel alpha 1E subunit (CACNA1E) | 1q25 | 0.87 |
| ENSG00000118432 | Cannabinoid receptor 1 (brain) (CNR1) | 6q15 | 0.87 |
| ENSG00000184349 | Homologue to ephrin-A5 precusor (Dario rerio) | 5q21 | 0.86 |
| ENSG00000129348 | tRNA guanine transglycosylase (TGT) | 19p13 | 0.85 |
| ENSG00000108309 | RaP2 interacting protein 8 (RPIP8) | 17q21 | 0.85 |
| ENSG00000130702 | Laminin alpha 5 (LAMA5) | 20q13 | 0.85 |
| ENSG00000159964 | Selenoprotein SelM (SELM) | 22q12 | 0.84 |
| ENSG00000111432 | Frizzled homologue 10 (Drosophila) (FZD10) | 12q24 | 0.84 |
| ENSG00000135862 | Laminin gamma 1 (LAMC1) | 1q25 | 0.83 |
| ENSG00000155886 | Solute carrier family 24 member 2 (SLC24A2) | 9p22 | 0.83 |
| ENSG00000151498 | Acyl-coenzyme A dehydrogenase 8 (ACAD8) | 11q25 | 0.83 |
| ENSG00000082269 | KIAA1411_protein (KIAA1411) | 6q13 | 0.83 |
| ENSG00000180914 | Oxytocin receptor (OXTR) | 2p25 | 0.83 |
| ENSG00000168772 | Dvl binding protein (IDAX) | 4p24 | 0.83 |
| ENSG00000133026 | Myosin heavy chain, non-muscle type B (MYH10) | 17p13 | 0.83 |
| ENSG00000085741 | Wingless type MMTV 11 (WNT11) | 11q13 | 0.83 |
| ENSG00000110536 | NADH-ubiquinone oxidoreductase (NDUFS3) | 11p11 | 0.83 |
| ENSG00000162383 | Excitatory amino acid transporter 5 (SLC1A7) | 1p32 | 0.83 |
| ENSG00000168610 | Signal transducer and activator of transcription 3 (STAT3) | 17q21 | 0.82 |
Recently, three different computational methods for the prediction of disease-related genes have been reported (28–30). These methods score potential disease genes based on functional similarities to known disease genes that cause similar phenotypes, and/or use expression data that correlate with the phenotype of the disease. The approach of our model is entirely different to these other methods in that it is based on readily computable sequence properties, having the advantage of assigning a probability score of involvement in a disease for all proteins in the human genome, even without having information about their function or expression profiles.
This work provides further insights into the general nature of human hereditary diseases and the genes that (when mutated) cause them. We have shown that long genes that are conserved, have a wide phylogenetic extent and do not have highly similar paralogues are more likely to cause a disease due to mutations in their protein sequence. Moreover, the capability to assign probabilities to human genes with respect to whether they are likely to be involved in diseases will be highly useful in identifying new candidate genes. Researchers working on one particular disease may want to consult the probability scores of the proteins in the region linked to the disease of interest as well as obtain additional information for that gene, before starting any mutation analysis. The identification of genes with a high probability of causing hereditary diseases should provide the basis for future associations of these genes with the diseases in which they may be involved.
SUPPLEMENTARY MATERIAL
Supplementary Material are available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Benjamin Audit and José Pereira-Leal for comments, and other members of the Computational Genomics Group (EBI) for discussions. We also wish to thank Anton Enright (Memorial Sloan-Kettering Cancer Center, NY) for the pairwise similarity data from COGENT. N.L.-B. is supported by a long-term post-doctoral fellowship from the Human Frontiers Science Program. C.A.O. acknowledges support from the European Molecular Biology Laboratory, the UK Medical Research Council and IBM Research.
REFERENCES
- 1.Botstein D. and Risch,N. (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nature Genet., S33, 228–237. [DOI] [PubMed] [Google Scholar]
- 2.Hamosh A., Scott,A.F., Amberger,J., Valle,D. and McKusick,V.A. (2000) Online Mendelian Inheritance in Man (OMIM). Hum. Mutat., 15, 57–61. [DOI] [PubMed] [Google Scholar]
- 3.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 4.Krawczak M., Ball,E.V., Fenton,I., Stenson,P.D., Abeysinghe,S., Thomas,N. and Cooper,D.N. (2000) Human gene mutation database-a biomedical information and research resource. Hum. Mutat., 15, 45–51. [DOI] [PubMed] [Google Scholar]
- 5.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 6.Bernal A., Ear,U. and Kyrpides,N. (2001) Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res., 29, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimenez-Sanchez G., Childs,B. and Valle,D. (2001) Human disease genes. Nature, 409, 853–855. [DOI] [PubMed] [Google Scholar]
- 9.Karlin S., Brocchieri,L., Bergman,A., Mrazek,J. and Gentles,A.J. (2002) Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl Acad. Sci. USA, 99, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T. et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen P.J., Enright,A.J., Audit,B., Cases,I., Goldovsky,L., Harte,N., Kunin,V. and Ouzounis,C.A. (2003) COmplete GENome Tracking (COGENT): a flexible data enviroment for computationa genomics. Bioinformatics, 19, 1451–1452. [DOI] [PubMed] [Google Scholar]
- 13.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peregrin-Alvarez J.M., Tsoka,S. and Ouzounis,C.A. (2003) The phylogenetic extent of metabolic enzymes and pathways. Genome Res., 13, 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrini M., Marcotte,E.M., Thompson,M.J., Eisenberg,D. and Yeates,T.O. (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA, 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Promponas V.J., Enright,A.J., Tsoka,S., Kreil,D.P., Leroy,C., Hamodrakas,S., Sander,C. and Ouzounis,C.A. (2000) CAST: an iterative algorithm for the complexity analysis of sequence tracts. Complexity analysis of sequence tracts. Bioinformatics, 16, 915–922. [DOI] [PubMed] [Google Scholar]
- 17.Kohavi R., Sommereld,D. and Dougherty,J. (1997) Data mining using MLC++: A machine learning library in C++. Int. J. Artif. Intell. Tools, 6, 537–566. [Google Scholar]
- 18.Brunk C., Kelly,J. and Kohavi,R. (1997) MineSet: an integrated system for data mining. In Heckerman,D., Mannila,H., Pregibon,D. and Uthurusamy,R. (eds) Proceedings of the Third International Conference on Knowledge Discovery and Data Mining. AAAI Press, CA. [Google Scholar]
- 19.Wilson A.C., Carlson,S.S. and White,T.J. (1977) Biochemical evolution. Annu. Rev. Biochem., 46, 573–639. [DOI] [PubMed] [Google Scholar]
- 20.Hurst L.D. and Smith,N.G. (1999) Do essential genes evolve slowly? Curr. Biol., 9, 747–750. [DOI] [PubMed] [Google Scholar]
- 21.Hirsh A.E. and Fraser,H.B. (2001) Protein dispensability and rate of evolution. Nature, 411, 1046–1049. [DOI] [PubMed] [Google Scholar]
- 22.Jordan I.K., Rogozin,I.B., Wolf,Y.I. and Koonin,E.V. (2002) Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res., 12, 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller M.P. and Kumar,S. (2001) Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet., 10, 2319–2328. [DOI] [PubMed] [Google Scholar]
- 24.Mooney S.D. and Klein,T.E. (2002) The functional importance of disease-associated mutation. BMC Bioinformatics, 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Z., Steinmetz,L.M., Gu,X., Scharfe,C., Davis,R.W. and Li,W.H. (2003) Role of duplicate genes in genetic robustness against null mutations. Nature, 421, 63–66. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez P.A., Gorlin,R.J., Lukens,J.N., Taniuchi,S., Bohinjec,J., Francois,F., Klotman,M.E. and Diaz,G.A. (2003) Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nature Genet., 34, 70–74. [DOI] [PubMed] [Google Scholar]
- 27.Antonellis A., Ellsworth,R.E., Sambuughin,N., Puls,I., Abel,A., Lee-Lin,S.Q., Jordanova,A., Kremensky,I., Christodoulou,K., Middleton,L.T. et al. (2003) Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet., 72, 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Iratxeta C., Bork,P. and Andrade,M.A. (2002) Association of genes to genetically inherited diseases using data mining. Nature Genet., 31, 316–319. [DOI] [PubMed] [Google Scholar]
- 29.Freudenberg J. and Propping,P. (2002) A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics, 18 (Suppl. 2), S110–S115. [DOI] [PubMed] [Google Scholar]
- 30.Van Driel M.A., Cuelenaere,K., Kemmeren,P.P., Leunissen,J.A. and Brunner,H.G. (2003) A new web-based data mining tool for the identification of candidate genes for human genetic disorders. Eur. J. Hum. Genet., 11, 57–63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.