Abstract
DNAzymes (synthetic catalytic DNA) have emerged as a new class of nucleic acid-based gene silencing agent. Using DNAzymes targeting the human mRNA of the immediate-early gene and C2H2-class zinc finger transcription factor early growth response-1 (EGR-1), we demonstrate here that EGR-1 plays an indispensable role in breast cancer proliferation, migration, chemoinvasion and xenograft growth in nude mice. DNAzyme inhibition of these tumorigenic processes and EGR-1 protein expression in breast carcinoma cells is sequence-specific and EGR-1 transcription-independent. These agents inhibit breast carcinoma cell migration and chemoinvasion in microchemotaxis chambers and solid tumour growth in athymic nude mice. Thus, DNAzymes targeting specific genes can inhibit multiple key tumorigenic processes in vitro and in vivo and may serve as useful anti-cancer agents.
INTRODUCTION
Human early growth response-1 (EGR-1, also known as NGFI-A, Zif268, Krox24 and Tis8) is a nuclear phosphoprotein that contains a highly conserved DNA-binding domain which binds to GC-rich recognition motifs (1). It is the prototypical member of a family of zinc finger transcription factors (that belong to the C2H2 subclass) that includes at least three other members (EGR-2, -3 and -4). EGR-1 also contains a nuclear localization signal, two activator domains and a repressor domain (2). EGR-1 function is negatively regulated by NAB-1 and NAB-2 through specific protein–protein interactions via its repressor domain (3–5). EGR-1 is induced by a range of molecular and environmental stimuli including growth factors, cytokines, ultraviolet light, ionizing radiation, mechanical injury and fluid biomechanical forces (1,6).
EGR-1 plays numerous regulatory roles in various breast cancer cell lines. EGR-1 is expressed in human MCF7 and MDA-MB-231 breast carcinoma cells (7). EGR-1 has been linked with transformed growth (8), multidrug resistance (MDR)-1 gene transcription (9) and anti-oestrogen-responsiveness (10) in MCF7 human breast carcinoma cells. It is activated by oestrogen in association with Raf-1 kinase activity in this cell type (11).
DNA enzymes, or DNAzymes, of the ‘10–23’ subclass have emerged as a new class of nucleic acid-based gene targeting tools that can be used to cleave its target mRNA in a gene-specific manner (12). These agents have been used in a number of in vivo applications to inhibit the expression of their target gene (13) and their dependent genes (14). Their capacity to block the development of a diverse range of pathologies in animal models (13–18) suggests a potential use for DNAzymes as therapeutic tools. These agents are usually stabilized against nucleolytic attack by incorporation of modified nucleotides that do not inadvertently obstruct cleavage activity (13).
Here, we demonstrate that DNAzymes targeting EGR-1 block proliferation, migration and chemoinvasion of human breast carcinoma cells and inhibit solid tumour xenograft growth in nude mice in a sequence-specific manner.
MATERIALS AND METHODS
Cell culture and DNAzyme transfections
Human MCF7 and MDA-MB-231 breast carcinoma cells were obtained from American Type Culture Collection (ATCC) and cultured in RPMI (Gibco-BRL) pH 7.4, supplemented with 10% fetal calf serum (FCS). All media were supplemented with 10 µg/ml streptomycin and 10 IU/ml penicillin and all cultures were incubated in a humidified atmosphere of 5% CO2 at 37°C. Cells were passaged routinely by washing with phosphate-buffered saline (PBS) pH 7.4 and trypsinization. Subconfluent MCF7 cells (60–80%) were incubated for 6 h in serum-free medium prior to transfection with DNAzyme (0.4 µM, unless indicated otherwise) using FuGENE6 according to the manufacturer’s instructions (Roche). Twenty-four hours after the medium change to serum-free, the cells were washed with PBS and transfected again as described, in medium supplemented with 5% FCS.
DNAzymes
Oligonucleotides were commercially synthesized (Tri Link Biotechnologies) with an inverted thymidine (Ti) at the 3′ position [to provide resistance to nucleolytic degradation (19)] and purified by HPLC. DzF (5′-GCGGGGACAGGCT AGCTACAACGACAGCTGCATTi-3′) targets human EGR-1 mRNA and DzFscr (5′-GGAGCTGACGGCTAGCTA CAACGAGATCGACGCTi-3′) is the size-matched counterpart of DzF, containing an active catalytic (15 nt) domain but scrambled arms (9 nt each) (15). The scrambled DNAzyme (DzFscr) served as a control for sequence specificity and equal loading as indicated.
Western blot analysis
Subconfluent MCF7 cells grown in 100 mm plates were transfected as described above and incubated for 1.5 h at 37°C in medium supplemented with 5% FCS. Cells were then washed with cold PBS and lysed in RIPA buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 5 mM EDTA, 50 µg/ml leupeptin, 1% Trazylol and 2 mM PMSF). Total protein (5 µg of extract from mock-transfected, DNAzyme or scrambled DNAzyme groups) was electrophoresed through 10% denaturing SDS–polyacrylamide gels and electroblotted onto PVDF nylon membranes (Millipore). The membranes were air-dried and blocked in PBS containing 5% skim milk and 0.05% Tween 20. Rabbit polyclonal antibodies (Santa Cruz Biotechnology Inc.) to Egr-1 were used as the primary incubate before washing, incubation with HRP-linked Ig secondary antiserum (Dako) and chemiluminescence detection (Perkin-Elmer).
Cell proliferation assays
Subconfluent MCF7 cells grown in 96-well micro-titre plates were growth-arrested and mock-transfected or transfected with 0.1 µM of DNAzyme or scrambled DNAzyme as above. A second transfection was performed as above in 5% FCS before incubation for 48 h at 37°C. Cells were then washed with PBS, trypsinized and resuspended in 10 ml of Isoton II for counting. Cells were counted using an automated Coulter counter (Coulter Z series).
Migration assay
MCF7 cells were grown in 100 mm plates and mock-transfected or transfected with 0.3 µM DNAzyme or scrambled DNAzyme in serum-free medium as described above. Twenty-four hours following medium change to serum-free, cells were harvested for migration assay by exposure to trypsin, suspended in RPMI supplemented with 10% FCS, washed with RPMI supplemented with 1% bovine serum albumin and 25 mM HEPES and subsequently resuspended in this medium. This medium was placed in the lower wells of 48-well microchemotaxis chambers (Neuroprobe, Gaithersburg, MD) before seeding 2 × 104 cells/upper well in triplicate for each treatment. The upper and lower chambers were separated by a 12 µm pore polycarbonate membrane (Neuroprobe, Gaithersburg, MD) that was coated with human Collagen IV (Sigma). Chambers were incubated for 24 h at 37°C, before fixing membranes in methanol and staining in Harris’ haematoxylin. Cells in 3–6 microscopic fields were counted, by microscopy, to measure transmigration in each well.
Chemoinvasion assays
Chemotaxis chambers and 12 µm pore polycarbonate membranes coated with 500 µg Matrigel were used in chemoinvasion experiments. DNAzyme/scrambled DNAzyme transfections and chemotaxis assays were carried out as described for migration assays, except conditioned medium produced separately with NIH 3T3 cells was placed in the lower wells and chambers were incubated for 5 h at 37°C in 5% CO2.
Transient transfections and CAT activity assays
A plasmid construct containing a 5′-deletion fragment of the human EGR-1 promoter fused to chloramphenicol acetyltransferase (CAT) cDNA was used in transient transfections for subsequent CAT activity assays. MCF7 cells were co-transfected with 15 µg of the plasmid construct and 0.1–0.4 µM DNAzyme or scrambled DNAzyme in serum-free medium, as described above. Following a 10 h incubation at 37°C in 5% CO2, cells were harvested using trypsin, suspended in cold RPMI containing 10% FCS, washed with cold PBS and resuspended in 0.3 M Tris–HCl pH 7.8, containing 10–15 µg/ml aprotinin. Cells were lysed by probe sonication and frozen at –80°C for at least 30 min. Endogenous deacetylases were inactivated by incubation at 65°C for 10 min. The lysate was clarified by centrifugation followed by incubation at 37°C with 3H-labelled acetyl-CoA and chloramphenicol as previously described (20). CAT activity was measured using a liquid scintillation analyser (Packard).
Tumour xenograft mouse models
Balb/c nude mice (5 weeks of age) were injected s.c. in the dorsal midback region with 1 × 106 logarithmically growing MDA-MB-231 human breast cancer cells in 100 µl of 50% (v/v) Matrigel. The mice were checked daily, and left with food and water ad libitum in a 12 h/12 h day/night cycle. After 21 days, when tumours were palpable, the animals were injected intra-tumourally with 10 µg of DzF, DzFscr or saline twice/week in an injectate volume of 10 µl. Mice were weighed and tumour volumes assessed using digital Vernier callipers prior to injections in a blinded fashion. Throughout the study, animals did not exhibit signs of discomfort due to tumour growth or oligonucleotide administration. The studies were approved by the UNSW Animal Care and Ethics Committee.
RESULTS AND DISCUSSION
Blockade of EGR-1 protein expression in breast carcinoma cells
DNAzymes are enzymatic molecules composed entirely of DNA that can be made to cleave their target mRNA in a gene-specific manner (12). We have previously demonstrated that DNAzymes targeting Egr-1 (rodent Egr-1 mRNA) block intimal thickening after arterial balloon injury (13) and angiogenesis (14). We also showed that DNAzymes targeting human EGR-1 inhibit in-stent restenosis after coronary stenting in pigs (15). DzF, a human EGR-1 DNAzyme, bears a 15 nt catalytic domain flanked by two 9+9 nt arms which direct the DNAzyme to the G198U site in the mRNA (15). Despite this progress using DNAzymes to implicate a regulatory role for EGR-1 in vascular growth, it remains unclear whether human EGR-1 DNAzymes have any effect on key tumorigenic processes such as migration, invasion and solid human tumour growth. DzF (0.4 µM) strongly inhibited EGR-1 protein expression in MCF7 human breast carcinoma cells (Fig. 1). In contrast, the scrambled arm counterpart of DzF, which retains the catalytic core, had only a modest effect on EGR-1 protein levels (Fig. 1).
Figure 1.
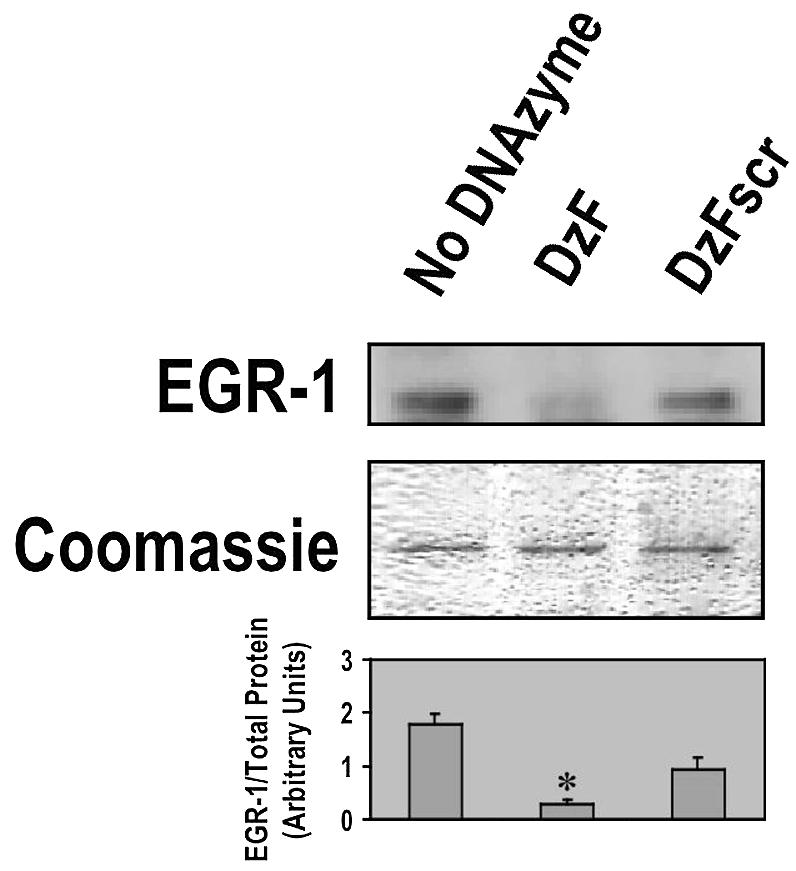
Inhibition of EGR-1 protein expression in breast carcinoma cells. DzF suppresses EGR-1 protein expression in MCF7 cells. Subconfluent growth-quiescent MCF7 cells grown in 100 mm plates were transfected twice with DNAzyme (DzF or DzFscr, 0.4 µM), or mock-transfected and incubated in medium containing 5% serum at 37°C for 1.5 h. Total cell lysates were analysed by western blot using rabbit polyclonal antibodies targeting human EGR-1 and chemiluminescence detection. The figure shows the corresponding Coomassie Blue-stained protein loading control. The histogram shows the mean ± standard error of the mean (SEM) of EGR-1 protein levels relative to the amount of protein loaded. The data are representative of at least two independent experiments. Asterisk indicates P < 0.05 using Student’s t-test relative to the mock-transfected control.
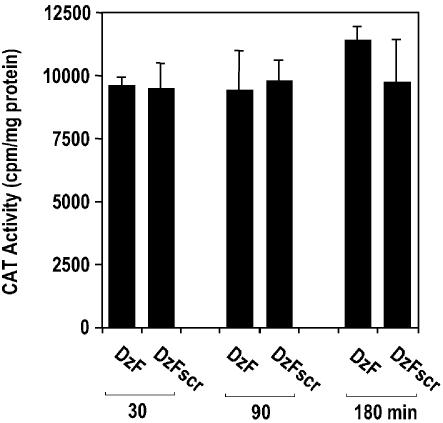
EGR-1 DNAzymes have no effect on EGR-1 promoter-dependent CAT reporter activity
The mechanism of DNAzyme-mediated gene suppression is thought to be mediated via transcription-independent degradation of the target mRNA; this to our knowledge has not yet directly been demonstrated. Therefore, to exclude the possibility that DNAzyme inhibition of EGR-1 protein was due to its suppression of EGR-1 transcription, we assessed EGR-1 promoter-dependent CAT activity in MCF7 cells treated with DzF or DzFscr. Unlike sequence-specific DzF inhibition of EGR-1 protein (Fig. 1), no such difference was observed at the level of EGR-1 transcription (Fig. 2).
Figure 2.
EGR-1 DNAzyme inhibition of MCF7 EGR-1 protein expression is independent of EGR-1 transcription. Subconfluent growth-quiescent MCF7 cells were transiently transfected with 15 µg of the construct and 0.4 µM of DNAzyme or scrambled DNAzyme (DzF or DzFscr) and incubated for 10 h at 37°C prior to harvest. CAT activity was normalized to total protein content in each sample. The data (mean ± SEM) are representative of at least two independent experiments performed in duplicate.
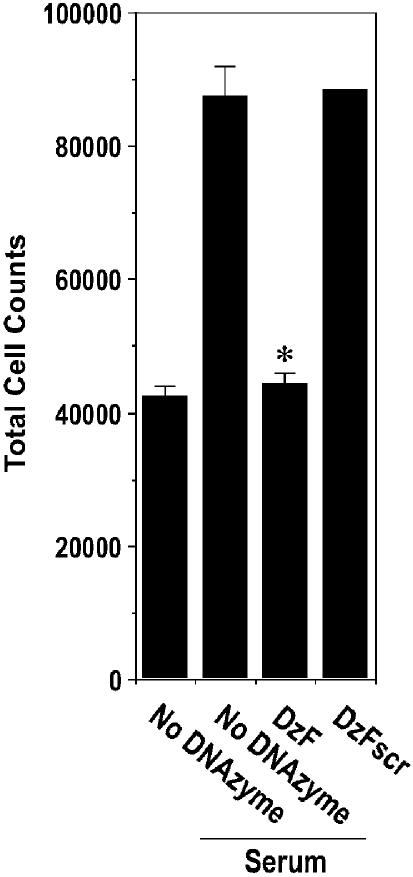
Inhibition of breast carcinoma cell growth with EGR-1 DNAzymes
Since EGR-1 levels are reduced by DzF in MCF7 cells (Fig. 1), we hypothesized that these cells would be amenable to growth inhibition by the EGR-1 DNAzyme. Serum-inducible MCF7 cell proliferation, assessed by Coulter quantitation, was blocked in the presence of DzF, whereas DzFscr had no effect (Fig. 3).
Figure 3.
EGR-1 DNAzymes inhibit breast carcinoma cell proliferation. Subconfluent growth-quiescent MCF7 cells were transfected twice with DNAzyme or scrambled DNAzyme (DzF or DzFscr, 0.1 µM), or mock-transfected and incubated in medium containing 5% serum at 37°C for 48 h. The cells were washed, trypsinized and quantitated using an automated Coulter counter. The data (mean ± SEM) are representative of at least two independent experiments performed in duplicate. Asterisk indicates P < 0.05 using Student’s t-test relative to the mock-transfected control.
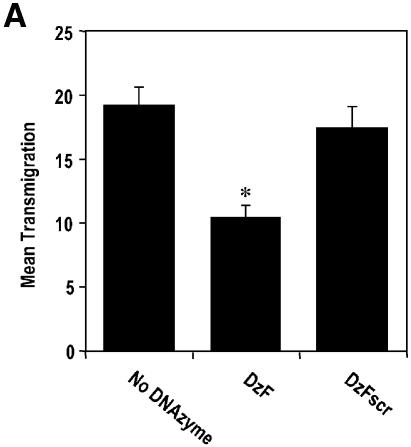
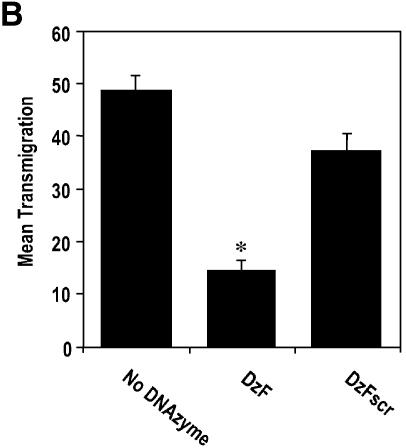
EGR-1 DNAzymes inhibit breast carcinoma cell migration and chemoinvasion
We next investigated the capacity of EGR-1 DNAzymes to influence MCF7 cell migration and chemoinvasion. We evaluated MCF7 migration from the upper to the lower chamber in microchemotaxis chambers 24 h after cell seeding. DzF inhibited transmigration, whereas the number of cells migrating to the lower chamber in the DzFscr group did not differ from DNAzyme-exempt control (Fig. 4A). When the chambers were coated with basement membrane extract (Matrigel), we observed a 2-fold increase in the number of transmigrating cells even after 5 h (Fig. 4B). DzF suppressed this chemoinvasive cellular response, whereas DzFscr had no effect (Fig. 4B).
Figure 4.
EGR-1 DNAzymes inhibit breast carcinoma cell migration and chemoinvasion. (A) MCF7 cells transfected with DNAzyme or scrambled DNAzyme (DzF or DzFscr, 0.3 µM), or mock-transfected, were seeded in the upper wells of 48-well microchemotaxis chambers and incubated at 37°C for 24 h. Cells were fixed and stained before quantitating transmigration in 3–6 microscopic fields per well. (B) MCF7 cells transfected with DNAzyme or scrambled DNAzyme (DzF or DzFscr, 0.3 µM), or mock-transfected, were seeded in the upper wells of 48-well microchemotaxis chambers containing Matrigel and incubated at 37°C for 5 h. Cells were fixed and stained before quantitating transmigration in six high-power fields per well. The data (mean ± SEM) are representative of at least two independent experiments performed in triplicate. Asterisk indicates P < 0.05 using Student’s t-test relative to the mock-transfected control.
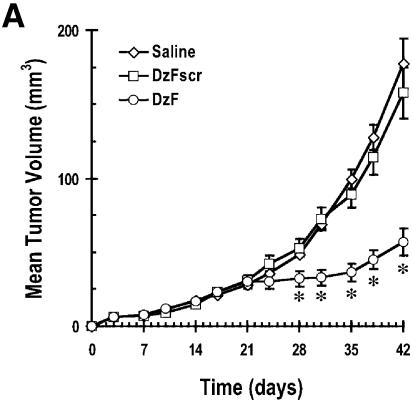
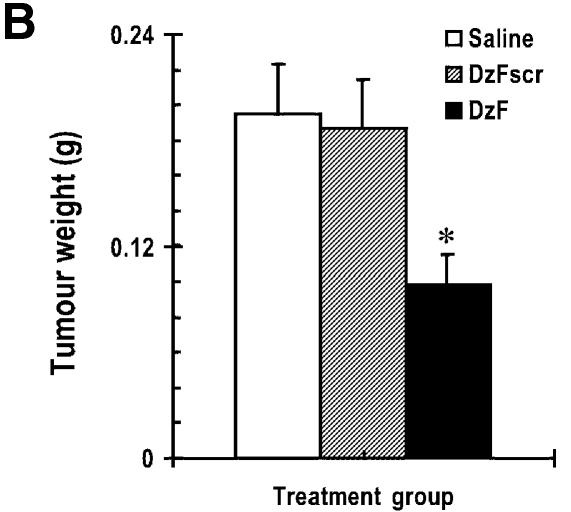
Solid breast carcinoma growth in athymic nude mice is inhibited by DzF
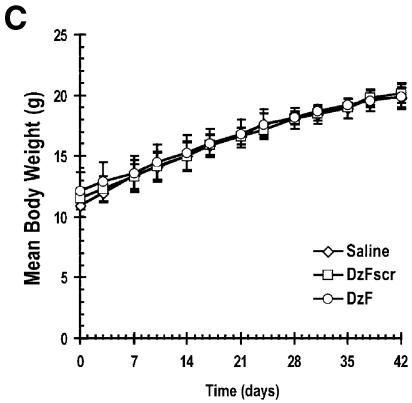
Finally we evaluated the capacity of DzF to inhibit the growth of highly aggressive human breast cancer cells as solid tumours in athymic nude mice. Palpable human MDA-MB-231 (21) xenografts were established after 21 days. DNAzymes (DzF or DzFscr, 10 µg) in saline, or saline alone, were administered intra-tumourally immediately after digital assessment of tumour volume twice a week. DzF caused profound sequence-specific inhibition of solid tumour growth (Fig. 5A and B). DzF inhibition of solid tumour growth was apparent even after the first administration of DNAzyme (Fig. 5A). Body weights of the mice were not adversely affected by DNAzyme or scrambled DNAzyme (Fig. 5C).
Figure 5.
Solid breast carcinoma growth in athymic nude mice is inhibited by DzF but not DzFscr. (A) DNAzymes (DzF or DzFscr) or saline was administered intra-tumourally (10 µg per 10 µl injection) into solid MDA-MB-231 tumours immediately after digital caliper assessment of tumour volume twice a week. Treatment was commenced 21 days after initial inoculation when the tumours were palpable. (B) Mean tumour weight immediately after resection 21 days after commencement of therapy. (C) Body weight of mice was assessed twice a week each time tumour size was evaluated. n = 5 mice per group. The error bars in all panels represent the mean value ± standard deviation. Asterisk indicates P < 0.05 by Analysis of Variance or using Student’s t-test. The data are representative of two independent experiments.
The present study demonstrates direct, sequence-specific inhibition of breast cancer cell proliferation, migration, chemoinvasion and solid tumour growth by catalytic antisense DNA molecules targeting human EGR-1. These properties, coupled with the ability of Egr-1 DNAzymes to block angiogenesis (14), suggest that these novel, small-molecule gene-targeting agents may be useful as both direct and indirect anti-cancer agents.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Mr Roger Fahmy for excellent technical assistance. This work was supported by grants from the NHMRC, Johnson and Johnson Research Pty Limited and an Infrastructure Grant from NSW Department of Health. L.M.K. is a Principal Research Fellow of the NHMRC.
REFERENCES
- 1.Gashler A. and Sukhatme,V. (1995) Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog. Nucleic Acid Res. Mol. Biol., 50, 191–224. [DOI] [PubMed] [Google Scholar]
- 2.Gashler A.L., Swaminathan,S. and Sukhatme,V.P. (1993) A novel repression module, an extensive activation domain and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol. Cell. Biol., 13, 4556–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russo M.W., Sevetson,B.R. and Milbrandt,J. (1995) Identification of NAB-1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc. Natl Acad. Sci. USA, 92, 6873–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svaren J., Sevetson,B.R., Apel,E.D., Zimonjic,D.B., Popescu,N.C. and Milbrandt,J. (1996) NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol. Cell. Biol., 16, 3545–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverman E.S., Khachigian,L.M., Santiago,F.S., Williams,A.J., Lindner,V. and Collins,T. (1999) Vascular smooth muscle cells express the transcriptional corepressor NAB2 in response to injury. Am. J. Pathol., 155, 1311–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khachigian L.M. and Collins,T. (1998) Early growth response factor 1: a pleiotropic mediator of inducible gene expression. J. Mol. Med., 76, 613–616. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y.L., Lei,Z.M., Huang,Z.H. and Rao,C.V. (1999) Determinants of transcription of the chorionic gonadotropin/luteinizing hormone receptor gene in human breast cells. Breast J., 5, 186–193. [DOI] [PubMed] [Google Scholar]
- 8.Huang R.P., Fan,Y., de Belle,I., Niemeyer,C., Gottardis,M.M., Mercola,D. and Adamson,E.D. (1997) Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int. J. Cancer, 72, 102–109. [DOI] [PubMed] [Google Scholar]
- 9.Gill P.K., Gescher,A. and Gant,T.W. (2001) Regulation of MDR1 promoter activity in human breast carcinoma cells by protein kinase C isozymes alpha and theta. Eur. J. Biochem., 268, 4151–4157. [DOI] [PubMed] [Google Scholar]
- 10.Gu Z., Lee,R.Y., Skaar,T.C., Bouker,K.B., Welch,J.N., Lu,J., Liu,A., Zhu,Y., Davis,N., Leonessa,F. et al. (2002) Association of interferon regulatory factor-1, nucleophosmin, nuclear factor-kappaB and cyclic AMP response element binding with acquired resistance to Faslodex (ICI 182 780). Cancer Res., 62, 3428–3437. [PubMed] [Google Scholar]
- 11.Pratt M.A., Satkunaratnam,A. and Novosad,D.M. (1998) Estrogen activates raf-1 kinase and induces expression of Egr-1 in MCF-7 breast cancer cells. Mol. Cell. Biochem., 189, 119–125. [DOI] [PubMed] [Google Scholar]
- 12.Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santiago F.S., Lowe,H.C., Kavurma,M.M., Chesterman,C.N., Baker,A., Atkins,D.G. and Khachigian,L.M. (1999) New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth factor injury. Nature Med., 5, 1264–1269. [DOI] [PubMed] [Google Scholar]
- 14.Fahmy R.G., Dass,C.R., Sun,L.Q., Chesterman,C.N. and Khachigian,L.M. (2003) Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nature Med., 9, 1026–1032. [DOI] [PubMed] [Google Scholar]
- 15.Lowe H.C., Fahmy,R.G., Kavurma,M.M., Baker,A., Chesterman,C.N. and Khachigian,L.M. (2001) Catalytic oligodeoxynucleotides define a key regulatory role for early growth response factor-1 in the porcine model of coronary in-stent restenosis. Circ. Res., 89, 670–677. [DOI] [PubMed] [Google Scholar]
- 16.Iversen P.O., Nicolaysen,G. and Sioud,M. (2001) DNA enzyme targeting TNF-alpha mRNA improves hemodynamic performance in rats with postinfarction heart failure. Am. J. Physiol. Heart Circ. Physiol., 281, H2211–H2217. [DOI] [PubMed] [Google Scholar]
- 17.Lowe H.C., Chesterman,C.N. and Khachigian,L.M. (2002) Catalytic antisense DNA molecules targeting Egr-1 inhibit neointima formation following permanent ligation of rat common cartoid arteries. Thromb. Haemost., 87, 134–140. [PubMed] [Google Scholar]
- 18.Zhang L., Gasper,W.J., Stass,S.A., Ioffe,O.B., Davis,M.A. and Mixson,A.J. (2002) Angiogenic inhibition mediated by a DNAzyme that targets vascular endothelial growth factor receptor 2. Cancer Res., 62, 5463–5469. [PubMed] [Google Scholar]
- 19.Santiago F.S., Atkins,D.A. and Khachigian,L.M. (1999) Vascular smooth muscle cell proliferation and regrowth after injury in vitro is dependent upon NGFI-A/Egr-1. Am. J. Pathol., 155, 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khachigian L.M., Williams,A.J. and Collins,T. (1995) Interplay of Sp1 and Egr-1 in the proximal PDGF-A promoter in cultured vascular endothelial cells. J. Biol. Chem., 270, 27679–27686. [DOI] [PubMed] [Google Scholar]
- 21.Hardwick M., Fertikh,D., Culty,M., Li,H., Vidic,B. and Papadopoulos,V. (1999) Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res., 59, 831–842. [PubMed] [Google Scholar]