Cytochrome c peroxidase (CcP) consumes hydrogen peroxide in mitochondria, using electrons derived from reduced cytochrome c. This and a related enzyme, horseradish peroxidase (HRP), have played key roles in the development of structural and mechanistic biochemistry and are used in biocatalysis and chemiluminescent bioassays (1). On page 193 of this issue, Casadei et al. (2) use neutron diffraction to reveal the role and origin of protons in heme oxidation by hydrogen peroxide, a key step in this essential enzymatic reaction.
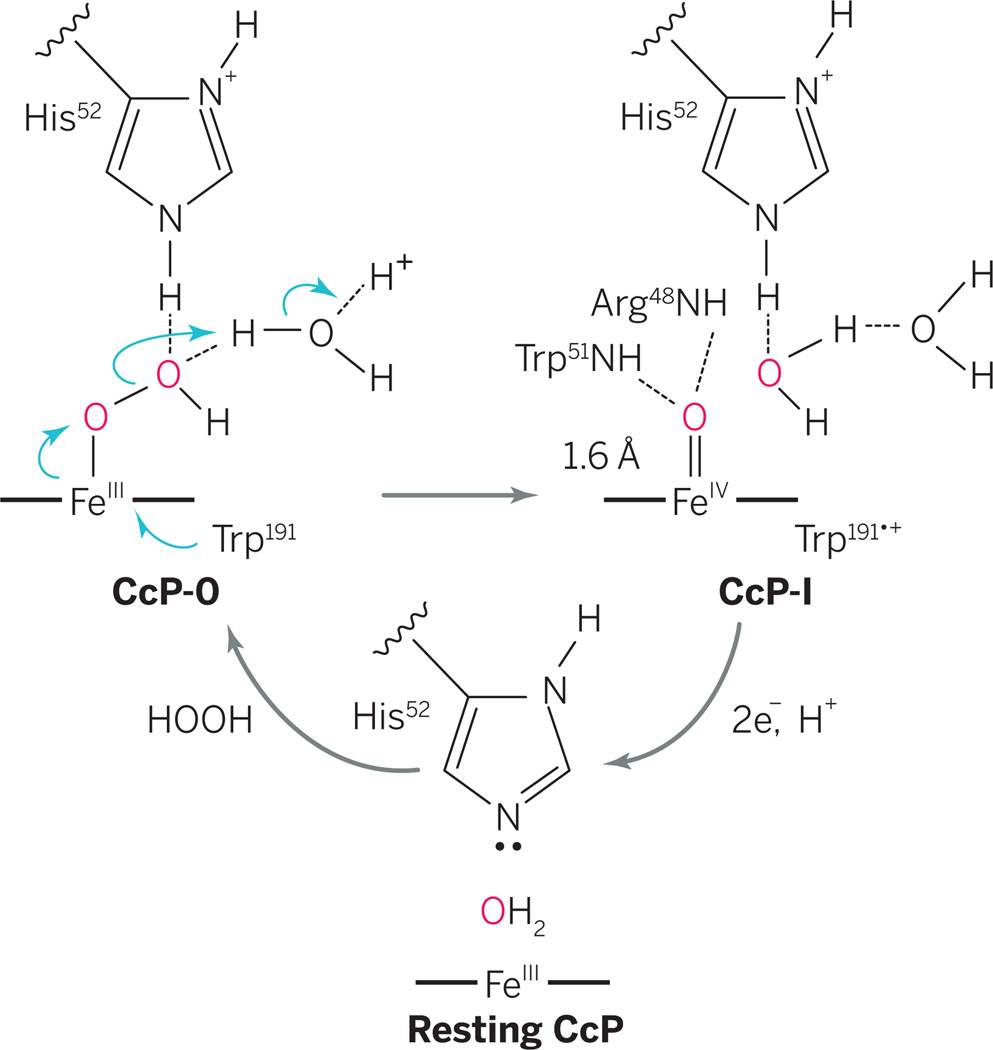
CcP and HRP were the first heme enzymes for which oxidized intermediates were observed (1). In the textbook mechanism for heme oxidation, protonated histidine N-H assists O-O bond heterolysis in an Fe(III)-OOH intermediate (CcP-0), producing CcP compound I (CcP-I) and water. The overall course of this reaction was established long ago. But where are the protons? Casadei et al. use neutron diffraction to reveal the positions of protons in resting CcP and CcP-I. They show that the iron(IV) of CcP-I is an unprotonated ferryl, Fe(IV)=O. The results bring new clarity to heme oxidation by hydrogen peroxide (see the figure).
Proton-mediated mechanism.
Reaction of ferric CcP with H2O2 first gives CcP-0, followed by O-O bond scission driven by external protonation to afford CcP-I. Casadei et al. now report neutron diffraction data that pinpoint the locations of the protons and elucidate the catalytic mechanism.
Neutron diffraction has distinct advantages over x-ray diffraction techniques for the structural characterization of enzymes that contain redoxactive metals. Non-ionizing neutron beams avoid the photoreduction that often plagues structural analysis with x-rays and that also occurs in the laser beams used for resonance Raman spectroscopy. Laser and x-radiation lead to ambiguities in the oxidation states of redox-active metals such as iron or manganese. By contrast, neutrons interact only with atomic nuclei and scatter much more effectively from hydrogen and, especially, deuterium atoms. Catalytic proton networks and even deuterated hydronium ions (D3O+) have been observed in proteins by means of neutron diffraction (3, 4).
Efforts to understand the atomic and electronic structure of the oxidized intermediates in the CcP catalytic cycle have been hampered by the fact that Fe(III)/Fe(IV) redox potentials in heme proteins lie in the same range as those of the porphyrin ring and those of tryptophan and tyrosine. This “redox non-innocence” greatly increases the complexity of these systems because it increases the number of plausible sites of oxidation. In HRP-I and in model porphyrin complexes, ferryl states, Fe(IV)=O, with very short Fe-O bond lengths have been reported (5, 6). The distinction between Fe(IV)=O species and their hydroxylated equivalents, Fe(IV)-OH, has taken on considerable importance with recent evidence that cytochrome P450 compound II is protonated and that the basicity of ferryl oxygen strongly affects heme protein reactivity (7).
To identify the positions of active-site protons in CcP and CcP-I, Casadei et al. have compared neutron and x-ray diffraction data from both species. The authors first replaced exchangeable protons with deuterons in large (1 mm) crystals of CcP, and then treated a CcP crystal with hydrogen peroxide to produce CcP-I. The results show that the catalytic imidazole of His52 is not protonated in resting CcP, as expected (see the figure). In CcP-I, both nitrogen atoms in His52 are protonated (deuterated), which was unexpected according to the generally accepted mechanism. This means that the protons required for O-O bond cleavage in CcP-0 (see the figure) must have come from another source, such as an adjacent water molecule. In this snapshot of the catalytic cycle, the ferryl oxygen, Fe(IV)=O, of CcP-I is not protonated, and the short iron-oxygen distance expected for the ferryl is confirmed.
Visualizing the mechanistically pertinent protons has important implications for the mechanism of O-O bond scission mediated by CcP and other heme proteins. A “wet” version of the peroxidase mechanism has been proposed, in which a water molecule adjacent to His52 mediates O-O bond cleavage (1, 8, 9). Retention of the His52 proton adjacent to the ferryl heme after O-O bond heterolysis, as revealed by Casadei et al., suggests that another proton, likely traveling through an aqueduct of water molecules leading to the active-site cavity, is also necessary (see the figure).
CcP-catalyzed peroxide bond heterolysis thus seems to occur via a proton relay mechanism similar to that of cytochrome P450 (10), with electrons arriving through Trp191 (see the figure). In this scenario, deprotonation of His52 would occur during subsequent reduction of CcP-I by another enzyme, ferrocytochrome c. This realization points to a water-mediated, acid-catalyzed process for O-O bond heterolysis, which is mechanistically satisfying because of its analogies to other proton relay mechanisms, such as that of cytochrome P450. Further, the need for a water channel and an external proton in peroxidase catalysis are highly informative for the design and construction of new hemeiron biocatalysts.
REFERENCES
- 1.Poulos TL. Chem. Rev. 2014;114:3919. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadei CM, et al. Science. 2014;345:193. doi: 10.1126/science.1254398. [DOI] [PubMed] [Google Scholar]
- 3.Cuypers MG, et al. Angew. Chem. Int. Ed. 2013;52:1022. doi: 10.1002/anie.201207071. [DOI] [PubMed] [Google Scholar]
- 4.Tomanicek SJ, et al. J. Biol. Chem. 2013;288:4715. doi: 10.1074/jbc.M112.436238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groves JT. J. Inorg. Biochem. 2006;100:434. doi: 10.1016/j.jinorgbio.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Penner-Hahn JE, et al. J. Am. Chem. Soc. 1986;108:7819. doi: 10.1021/ja00284a054. [DOI] [PubMed] [Google Scholar]
- 7.Yosca TH, et al. Science. 2013;342:825. doi: 10.1126/science.1244373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidossich P, et al. J. Phys. Chem. B. 2010;114:5161. doi: 10.1021/jp911170b. [DOI] [PubMed] [Google Scholar]
- 9.Sen K, Thiel W. J. Phys. Chem. B. 2014;118:2810. doi: 10.1021/jp411272h. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Peter S, Kinne M, Hofrichter M, Groves JT. J. Am. Chem. Soc. 2012;134:12897. doi: 10.1021/ja3049223. [DOI] [PMC free article] [PubMed] [Google Scholar]