Abstract
The epigenome is established and maintained by the site-specific recruitment of chromatin-modifying enzymes and their co-factors. Identifying the cis-elements that regulate epigenomic modification is critical to understand the regulatory mechanisms that control gene expression patterns. We present Epigram, an analysis pipeline that predicts histone modification and DNA methylation patterns from DNA motifs. The identified cis-elements represent interactions with the site-specific DNA-binding factors that establish and maintain epigenomic modifications. We catalog the cis-elements in embryonic stem cells and four derived lineages and found numerous motifs that have location preference, such as at the center of H3K27ac or at the edges of H3K4me3 and H3K9me3, which provides mechanistic insight about the shaping of the epigenome. The Epigram pipeline and predictive motifs are at http://wanglab.ucsd.edu/star/epigram.
Introduction
Epigenomic modifications, including histone modifications and DNA methylation, play critical roles in development and other key biological processes. The establishment and maintenance of specific epigenomic patterns are regulated by many factors; including: nucleosome positioning1, modifying enzymes2, transcription factors (TFs)3, non-coding RNAs4, signaling molecules5 and three-dimensional genomic organization6, 7. How exactly these mechanisms collectively regulate the epigenome remains unclear. In particular, the importance of cis-regulatory motifs, which are bound by site-specific DNA-binding factors, in regulating epigenomic modification remains unclear.
The genome sequence is unchanged between an individual's different cell-types; however, the execution of the cis-regulatory program shaping the epigenome is dynamic, as the expression and activity of chromatin-modifying enzymes and their co-factors vary between cell-types and cellular conditions (Fig. 1a). Epigenomic regulatory mechanisms use combinations of enzymes and co-factors to read a cis-regulatory code that defines locus-specific modification patterns. Therefore, given a particular epigenomic state, it is possible to identify the cis-elements that interplay with epigenomic modifications and are responsible for their establishment and/or maintenance (Fig 1b). A global picture of the cis-regulatory code that regulates the epigenome may emerge from surveying a diversity of cell-types and conditions. Indeed, evidence supporting the importance of cis-regulatory code in shaping the epigenome is rapidly accumulating8. For example, GC-rich sequences are strongly correlated with two histone modifications, H3K27me39 and H3K4me310; GC-rich motifs establish H3K27me3 by recruiting the polycomb repressive complex 2 (PRC2) through interaction with lncRNAs11, 12; the CpG-binding protein, CFP1, recruits the H3K4 methyltransferase SETD1 to GC-rich motifs10; another H3K4me3 methyltransferase, PRDM9, has a sequence-specific binding motif that directs it to meiotic recombination hotspots13. Additional examples include, TFs belonging to the PAX family that help establish H3K9me3 at pericentric heterochromatin14; and the TF NANOG that physically interacts with the methylcytosine hydroxylase, TET1, to facilitate DNA demethylation at specific loci, resulting in priming of key pluripotency genes during cellular reprogramming15. Moreover, DNA variants can cause inter-individual differences in histone modification levels by altering the binding motifs of TFs16–18.
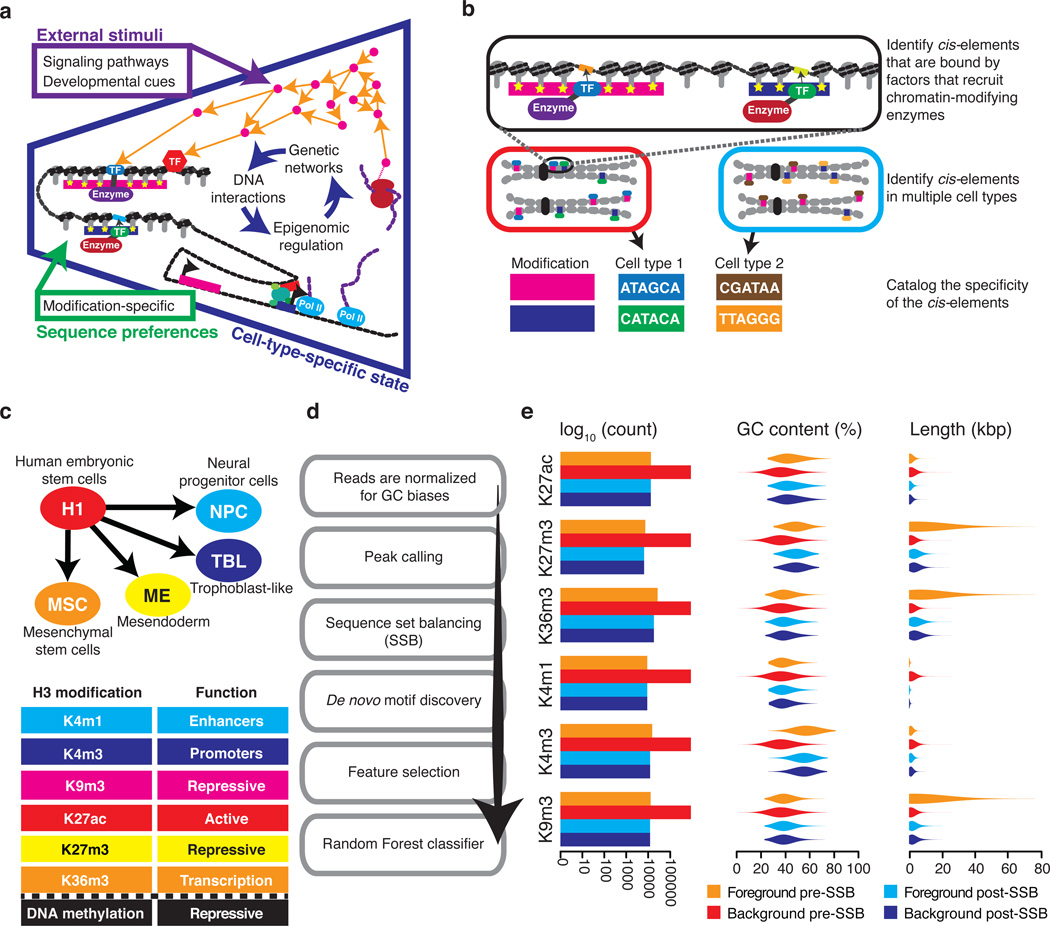
Figure 1. Identifying motifs that are predicative of epigenomic modifications.
(a) Site-specific DNA-binding factors regulate the epigenome. The blue section shows three regulatory levels of the cell-type-specific state: (i) gene regulatory network, (ii) sites-specific DNA-binding factors, and (iii) epigenomic regulation of gene expression. The green square lists non-cell-type-specific DNA sequence regulatory influences over the epigenome. The purple square lists stimuli that influence the cell-type-specific state. (b) An overview of the cis-element cataloging process. (c) A schematic showing H1 hESC and the four cell-types that were derived through in vitro differentiation. The table lists the analyzed epigenomic modifications and their roles. (d) A flow chart of the key stages in our analysis pipeline. (e) The effect of SSB on sequences sets. The bar plot shows the number of regions in a set before and after SSB. Violin plots show the distribution of region GC-content and length before and after SSB.
Despite these suggestive observations, methods to systematically catalog the epigenomes cis-regulatory program are lacking. Studies of nucleosome positioning1 have identified a ~10bp periodicity of A/T dinucleotides that oscillates out of phase with the dinucleotide GC19–21 and poly(dA:dT) tracks that inhibit nucleosome formation22–25; however, the involvement of DNA sequence in nucleosome positioning remains controversial26–28. Nevertheless, these studies did not intend to predict histone modifications from DNA sequence. Enrichment of TF binding and sequence features in various chromatin states have been examined29, but DNA motifs were not used to predict epigenomic modification. Recently, DNA 6-mers were used to predict the presence of H3K4me3 with reasonable accuracy but failed to find sequence features associated with other histone modifications30; notably this study did not focus on DNA motifs, which are recognized by DNA binding factors. These previous studies illustrate the possibility of deciphering the epigenomic cis-regulatory program; however, a predictive model that quantitatively links DNA motifs to epigenomic state has not been established, which is the major goal of this study.
Herein, we present the first comprehensive investigation of the cis-regulatory program that regulates the epigenome (Fig. 1b). We build models that use DNA motifs to predict epigenomic modifications in a cell-type-specific manner. Thus, we capture the cis-elements that interplay with the dynamic regulatory program to shape the epigenome. By surveying various cell-types we reveal mark-specific motifs, which may be universally recognized by chromatin-modifying enzymes, and motifs with cell-type-specific interplay, which may be recognized by cell-type-specific co-factors. We have successfully applied this approach to predicting the placement of six core histone modifications and DNA methylation valleys (DMV) in H1 human embryonic stem cells (hESC) and four H1-derived cell-types31 (Fig. 1c). We developed a novel analysis pipeline, Epigram, to systematically identify DNA motifs that are predictive of epigenomic modifications (Fig. 1d). To tease out the cis-elements that direct binding of epigenomic regulatory factors, such as chromatin-modifying enzymes, TFs and non-coding RNAs, we systematically removed the possible bias introduced by simple sequence patterns, such as GC-content. We observed that motifs have different location preferences within modified regions, such as the center of H3K27ac or the edge of H3K4me3 or H3K9me3. Furthermore, we demonstrate the importance of Epigram motifs in the regulation of histone modification through the significant correlation between their disruption and inter-individual H3K27ac variation. Importantly, our study provided a catalogue of cis-elements that play important roles in shaping the epigenomic modifications, which is useful for designing new epigenome editing tools.
Results
Predicting the epigenome from DNA motifs
We first examined if DNA motifs could distinguish genomic regions that possess modified histones from regions that do not possess any modified histones. For the sake of discussion, we refer to this as the ‘single mark analysis’. We started by correcting a potential bias in the ChIP-seq data that can be caused by the preferential sequencing of GC-rich genomic fragments32, 33 (Fig. 1d). To identify regions that are enriched with a histone modification from ChIP-seq, we called two types of peaks: tight for H3K27ac and H3K4me1/3; broad for H3K27me3, H3K36me3 and H3K9me3. The genome sequence of peaks from a specific modification, such as H3K27ac, formed the foreground for de novo motif finding. The background sequences consisted of genomic regions not covered by any histone modification peak (Fig. 2a). Identifying motifs that are enriched within the peaks is challenging, as methodology must be able to efficiently analyze tens of thousands of variable length regions. Thus, we employed two de novo motif-finding methods, Homer34 and Epigram’s own algorithm, as we found that the combination of both was more effective at predicting modification than either alone. In particular, Epigram is able to identify predictive motifs in very large sets of sequences. For example, Epigram could identify predictive motifs in 980,465 sequences with a mean length of 1,640 bps while Homer could not. For the purpose of feature selection, we next exploited a LASSO35 logistic regression to classify the foreground and background using the found motifs. Only the motifs with non-zero coefficients were kept to create the full set of motifs, which were then input to a Random Forest classifier. To improve interpretability, we further reduce the number of motifs by clustering the motifs by matrix similarity and from each cluster retaining a single motif, the one with the best area under the ROC curve (AUC). The reduced model motif set, was the lowest number of motifs that could achieve an AUC >95% of the full model's AUC during Random Forest prediction. We assessed our method’s performance through 5-fold cross-validation and to avoid a biased inflation of predictability we performed de novo motif discovery and feature selection using only the training data36, 37.
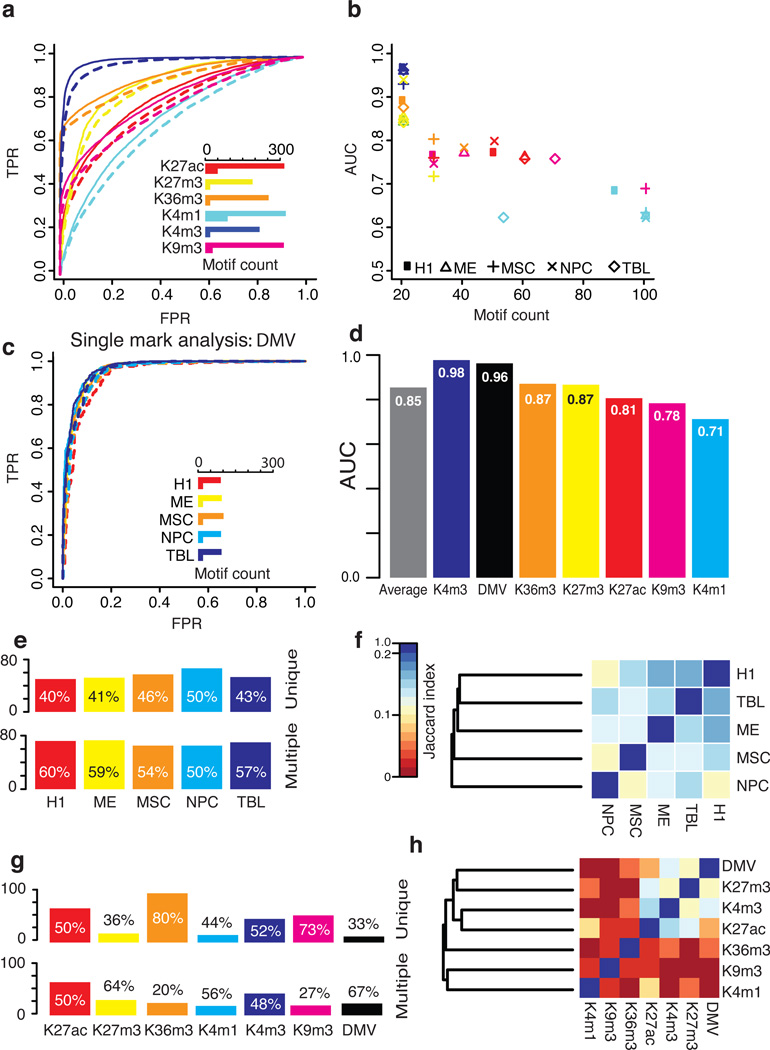
Figure 2. Predicting epigenomic modification from DNA motifs.
(a) The two sets of regions that are being distinguished. (b) A receiver-operating characteristic (ROC) curve shows the prediction performance in H1. Solid and dotted lines show the full and reduced models, respectively. The inlayed bar chart shows the number of motifs used in the full and reduced models. The same color scheme is used to represent the marks in the bar chart, the ROC curve and the scatter plot in part (c), which summarizes the performance across all cell-types. (d) ROC curves showing the DMV predictions performance. (e) The averaged results across five cell-types for the ‘single mark analysis’ (full model). (f) The number of motifs from each cell-type that are predictive of modification in only that cell-type (unique) or are also predictive of modification in other cell-types (multiple). Calculated using 589 motif groups. Motifs from the cell-type-specific comparison are excluded as H1 is featured in multiple comparisons and so are motifs enriched in the background, as they are not specifically predictive of modifications. (g) A heat map showing the proportion of shared motifs between each pair of cell-types. The Jaccard index was used to measure overlap and clustering was performed using the complete linkage method. (h;i) the same as (f) and (g) but showing modification specificity.
The selected motifs could successfully discriminate modified and unmodified regions: the average full model accuracy across all the peaks in the genome is 79%. This performance is excellent in light of the prediction challenges: (i) the large number of sequences in each set; (ii) variable region sizes; (iii) the sequence sets were greatly unbalanced for GC-content and region size; (iv) prediction requires the identification and combined predictive power of motif combinations. The excellent performance was also reflected by the average AUC in H1 of 0.85 for the full model (270 motifs) and 0.82 for the reduced (38 motifs; Fig. 2b–c). When all the five cell-types are averaged, the full model has an AUC of 0.84 (227 motifs) and reduced 0.80 (43 motifs), which shows that the total motifs can be reduced greatly while maintaining the majority of the prediction performance. Among the six marks, H3K4me3 is the most predictable in all cell-types (average AUC=0.96 for reduced models). To investigate the possible factors limiting the prediction performance, we compared the level of reads in the background for each of the modifications (Supplementary Fig. 1). The least predicable modification, H3K4me1, had the highest level of reads in its background, which reduces the distinction between foreground and background. The prediction performance for each mark is consistent across cell-types, which suggests the robustness of our model in handling possible noise in different experiments and cell-types.
It is noteworthy that the discrimination of modified regions and background is not a result of differences in GC-content or region length (Fig. 1e), which was corrected in our analysis to avoid biasing the Random Forest predictions. We refer to this step as sequence set balancing (SSB; see Methods). To demonstrate the importance of SSB, the models were tested with randomized sequences that have had their base pairs shuffled (Supplementary Fig. 2). When the shuffled sequences were used to test the dataset that had been subject to SSB, the prediction performance was destroyed as expected (Supplementary Fig. 3). However, in the dataset where the SSB step was omitted, the prediction performance remains high for all modifications except H3K27ac. This analysis clearly illustrated that SSB is critical to remove the trivial correlation between simple sequence features, such as GC-content and region size, and epigenomic modifications. Note that no similar analysis was done in the previously published work30 and the observed prediction power there may be a trivial result of GC-content.
Contributing factors in predicting histone modification
As multiple factors regulate the epigenome, we conducted additional control analyses to demonstrate that DNA motifs are predictive of histone modification. Firstly, we investigated if prediction power was affected by nucleosome-positioning related sequence features. To this end, we conducted a ‘mark-specific analysis’ by comparing regions enriched with one modification to regions with any other modification. Thus, motifs generally involved in nucleosome placement, but not histone modification per se, are present in both foreground and background, and therefore, do not contribute to discrimination. The average H1 full model accuracy was 77% with full and reduced models AUCs of 0.85 and 0.83, respectively (Supplementary Fig. 4a and 4d). The H1 full model had an average of 259 motifs while the reduced model had an average of 40. Similar results were achieved in other cell-types where the full models had an average AUC of 0.84 and 253 motifs while the reduced models with an average AUC of 0.82 and 39 motifs. Importantly, the model performance is comparable to the ‘single mark analysis’, which illustrates that mark-specific motifs, and not just sequence features involved in nucleosome positioning, can be captured by our method and are predictive of specific histone modifications.
Secondly, specific histone modifications often occur in particular genomic regions, such as H3K4me3 at promoters, and these possess characteristic sequence features, such as CpG islands. Therefore, we conducted the ‘typical background analysis’ to ensure discrimination is not coming from genomic features that are associated with regions where a modification commonly occurs. To this end, the modified regions were compared to regions that typically possess the modification but are unmodified in our comparison. For example, all H3K4me3 enriched regions were compared to annotated promoters without any H3K4me3 signal. The average accuracy for H1 was 75% with an AUC of 0.82 for the full model (237 motifs) and 0.79 (35 motifs) for the reduced (Supplementary Fig. 4b and 4d). Similar results were achieved overall where the full models had average AUC of 0.83 with 243 motifs and the reduced models average AUC of 0.80 (32 motifs). The AUC comparable to the ‘single mark analysis’, suggests that sequence features that are generally associated with regions, which histone modifications typically occupy, are generally removed from our analysis. As discussed above, 6mers have achieved comparable prediction accuracy on only (H3K4me3)30. However, while the GC-bias was recognized as being important for H3K4me3 prediction, it was not corrected during their analysis. Thus, their predictions were not based upon DNA motifs, which are recognized by DNA-binding factors; furthermore, no sequence feature was found to be predictive for other modifications. In contrast, we corrected the GC-bias in our analysis, as we are only interested in the predictive power of DNA motifs. We still identified sophisticated GC-rich motifs (see below), which suggests that specific regulatory factors have evolved to recognize these motifs in the GC-rich context of promoters.
Thirdly, we examined the impact of cell-type-specificity on the epigenomic modifications by investigating whether cell-type-specifically modified regions can be discriminated by motifs. Since four of the cell-types are derived from H1, we compared each histone modification in a H1-derived cell-type to the same modification in H1. A drop in model performance was observed; the average full model accuracy over the four cell-types was 61% (AUC=0.67). Even though we removed the small datasets (<2000 regions), the average full AUC was still only 0.69 (Supplementary Fig. 4c–d). As the similarity between the four H1-derived cell-types and H1 likely makes it difficult for this cell-type-specific analysis, we further compared H1 to eight more distant cell-types: A549, CD14+, GM12878, HeLa, HepG2, HUVEC, IMR90 and K562 (Supplementary Fig. 5). The prediction accuracy for the full model was improved to 68% (AUC=0.74) but still lower than the ‘single mark analysis’ (accuracy=79%; AUC=0.84). As the same histone modification was compared in different cell-types, the significantly decreased AUC further confirmed the existence of mark-specific motifs that direct chromatin-modifying enzymes to form specific histone modifications. The remaining discrimination may come from the binding of cell-type-specific factors, which are regulated by cell-type-specific patterns of gene expression and open chromatin29, 38.
These control analyses illustrate that our method can detect DNA motifs that are recognized by mark-specific chromatin-modifying enzymes and regulatory co-factors. These motifs are read by the genetic network to execute the cis-regulatory program that specifies the placement of the histone modifications39.
Motifs are predictive of DNA methylation
To further demonstrate the ability of DNA motifs to predict epigenomic modification, we applied the Epigram pipeline to DMVs, which are defined as large genomic domains (>5kb) that are devoid of DNA methylation31. DMVs have been shown to be enriched for early developmental regulatory genes and gain methylation in cancer cells, suggesting their biological importance. DMVs are relatively few in number (639–1004 per cell-type in: H1, ME, MSC, TBL and NPC) and show substantial overlap between cell-types (461 are common to all of these cell-types). Therefore, we conducted ‘single mark analysis’ on DMVs and not the additional control analyses. The average AUC for the DMVs was 0.96 for the full model (95 motifs; accuracy=0.91) and 0.95 for the reduced (Fig. 2d). The prediction performance remained high in all cell-types and the models all reduced down to 20 motifs, which is the lowest number of motifs assessed.
Furthermore, we report prediction of methylation status (hypo or hyper) at tissue-specific DMRs (TSDMRs) from 18 human tissues in a separate study (see details in40): the average AUC was 0.79 while an AUC of 0.85 was achieved when only 20 motifs were used to predict adrenal gland TSDMR status. Critically, the overlap between predictive motifs and SNPs that “break” motifs was compared between genotypes with DNA methylation concordance and discordance. This analysis identified a 2.6-fold enrichment of motif “breaking” SNPs, which strongly supports the association between our predictive motifs and epigenomic modifications.
Taken together, we have shown that the patterns of histone modifications and DNA methylation can be successfully predicted from DNA motifs. Considering the difficulty of such prediction, the overall full model achieved an excellent average accuracy (including DMV) of 79% for the ‘single mark analysis’ with an AUC=0.85 (Fig. 2e) and the reduced model an accuracy of 76% with an AUC=0.81.
Comparison of DNA motif specificities
The landscape of interplay between cis-elements and epigenomic modifications is complex (Supplementary Fig. 6). To pinpoint the cis-elements that are recognized by specific factors, we conducted comparative analyses to identify mark/cell-type-specific and independent DNA motifs. The five cell-types have similar proportions of cell-type-specific (unique) motifs (Fig. 2f). The degree of motif overlap between the cell-types is consistent with the known similarity between the five cell-types; for example, H1 is most related to TBL but most distinct from NPC (Fig. 2g). The number of motifs per modification varies considerably (Fig. 2h). The active enhancer mark H3K27ac41 has the most motifs, which is not unexpected as chromatin marks at enhancers have been shown to be dynamic across cell-types42. As H3K27ac is a mark of enhancer activity and the placement of H3K27ac is expected to be more cell-type-specific, it would require more cell-type-specific interactions to guide its placement. DMVs had the fewest motifs, which may reflect the stability of these large domains. The transcriptional activity mark H3K36me3, has the highest proportion of unique motifs, which suggests that their motif-based regulation is the most distinct. H3K4me3 and H3K27ac, both enriched at active promoters, share the most motifs (Fig. 2i). They form a larger cluster with H3K27me3 and DMV because H3K4me3, H3K27me3 and DMV share GC-rich motif regulation (Fig. 4 and Discussion), which is consistent with the lowest proportion of unique motifs that DMV and H3K27me3 have. Although the two active enhancer marks, H3K27ac and H3K4me1, do not cluster adjacently, their proportion of overlap is similar to that of H3K27ac and H3K4me3 (Fig. 2i).
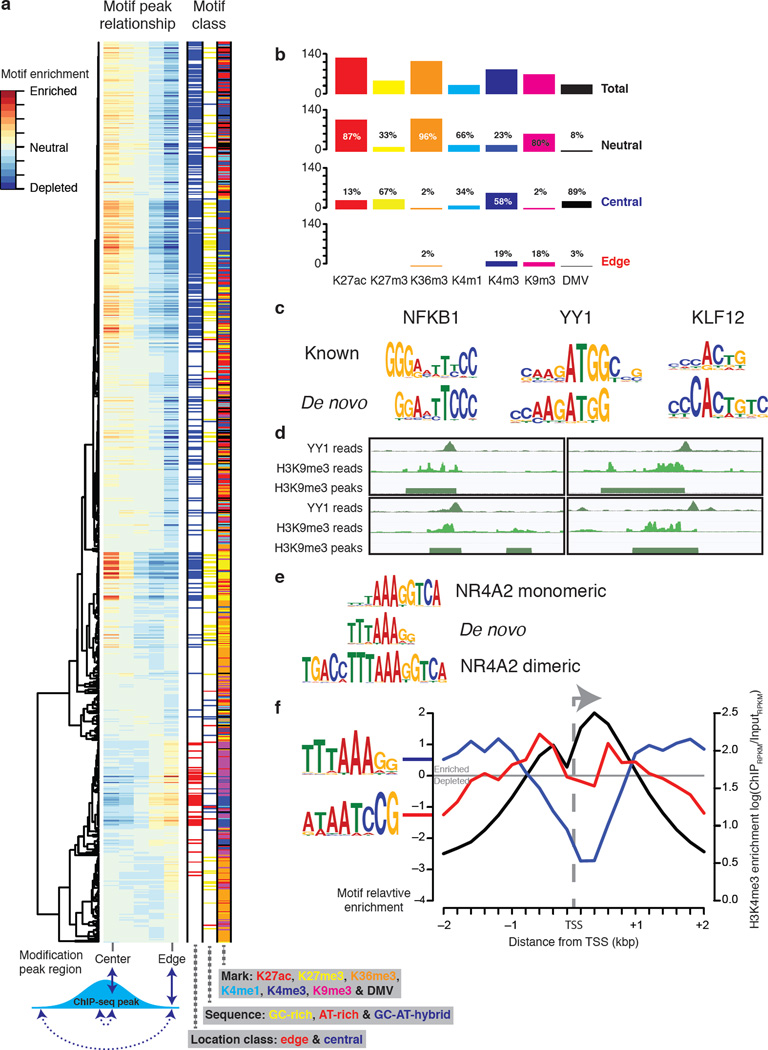
Figure 4. Predictive motifs have location preferences.
(a) Hierarchical clustering of 812 motifs showing positive interplay in the "single-mark" analysis. The motifs were scanned against their corresponding modification peaks. The scores were then summed in five bins that represent different regions of the peaks (see Supplementary Fig. 10). The bin scores for each motif were then hierarchically clustered using Ward’s method72. Motifs with edge or central preferences were classified by comparing edge and center bin scores and by using a Chi-square test P-value cut-off of <1.0e-10 (see methods for full details). (b) A summary of the location preference of motifs by mark specificity. (c) The motifs that interplay with H3K9me3 edges in H1. NFKB1 is given as an example of ‘Rel homology domain’. (d) Four screen shots showing YY1 ChIP-seq reads at the edge of a region of H3K9me3. Going clock-wise from the top left the four YY1 sites start at: chr2:17515620, chr6:16069456, chr12:14424514 and chr2:626745 (genome assembly version: hg18). (e) A de novo GC-AT-hybrid motif aligned to the NR4A2 monomer and dimeric motifs. (f) The average profile of two GC-AT-hybrid motifs and H3K4me3 at 13,962 TSS’s (see methods for full details).
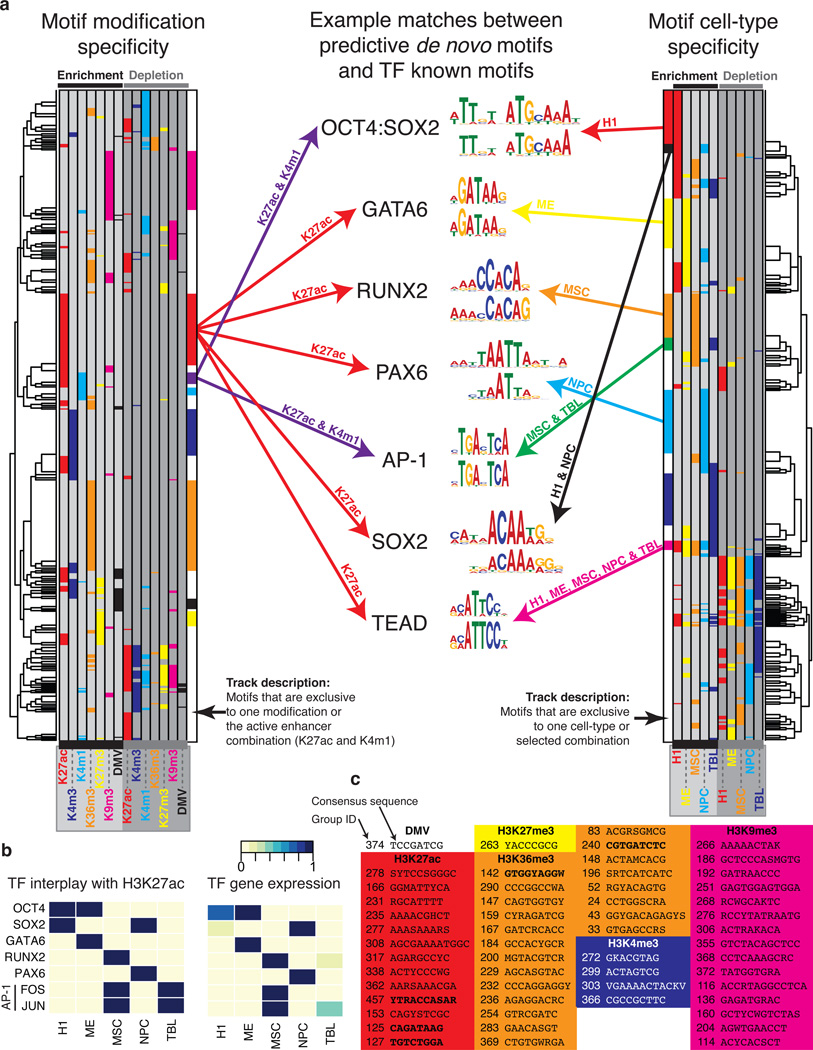
To identify cell-type or mark-specific motifs, we separately clustered the motifs by cell-type and modification specificity (Fig. 3a). The clusters contain motifs whose gene expression patterns match their interplay with H3K27ac (Fig. 3b) and that have known associations with particular epigenomic modifications and cell-types. For example, the SOX2 monomer motif is found associated with H3K27ac in H1 and NPC while the OCT4:SOX2 heterodimer motif is found only in H1. This observation is consistent with the functional roles of OCT4 in H1 and SOX2 in both H1 and NPC43. Another example is the motif recognized by the four TEAD family members, which is associated with enhancer marks H3K27ac (all cell-types) and H3K4me1 (TBL only). Remarkably, our finding is consistent with a previous study showing that deletion of a TEAD binding site from upstream of TCRJα locus resulted in a loss of H3 histone acetylation44. Furthermore, TEAD family members are known to promote cell proliferation by interacting with the Hippo signaling pathway45, which is critical for self-renewal and expansion of ESC into lineage-specific progenitors46. In mice TEAD family members establish enhancers during the initial development that occurs after the formation of the zygote47, 48. In human TEAD4 is crucial in determining the trophectoderm transcriptional program, which directs segregation from the inner cell mass49. Taken together, experimental evidence shows that TEAD family members play critical roles in directing histone acetylation to embryonic enhancers, which is consistent with our findings.
Figure 3. The specificities of interplay between DNA motifs and the epigenome.
(a) The left hand heat map shows 589 motif groups hierarchically clustered by their interplay with epigenomic modification. Each row represents a different motif and the positions are colored if the motif interplays with the modification. The first six columns show positive interplay (when a motif is enriched within a modification peaks) and the last six columns show negative interplay (when a motif is depleted in the modification peaks). The inner most bars indicate groups of motifs that are specific to certain modifications or combinations thereof. These bars follow the same color scheme as the heat map. Additionally purple represent H3K4me1 and H3K27ac, which corresponds to active enhancers. The right hand heat map shows the groups clustered by cell-type-specificity. Here additional colors represent the following combinations of cell-types: black, positive interplay with both H1 and NPC; orange, positive interplay with both MSC and TBL; magenta, positive interplay with all cell-types. In the center of the figure example motifs are shown: top, the known motif; lower, the identified de novo motif. (b) Positive interplay between H3K27ac and TFs is shown on the left. The normalized expression values of the genes are shown on the right. Gene expression values were taken from31 and normalized for each gene separately. The low expression levels of FOS in TBL can be explained by JUN can bind the AP-1 binding site as a homodimer71. (c) A table of modification specific motifs. The motif group numbers and consensus sequences are given. Motifs in bold match known motifs (see text).
To systematically identify motifs that may be involved in the placement of specific epigenomic modifications, we identified those that were selected in more than one analysis but associated with only one modification. We found 56 of these motifs (Fig. 3c) that may represent the binding preferences of modification-specific chromatin-modifying enzymes or their cofactors. These motifs include matches to three known TF motifs that interplay with H3K27ac (groups 457, 125 and 127 respectively match RUNX, GATA and HNRNPH3) and two that interplay with H3K36me3 (groups 142 and 240 respectively match ELSPBP1;MYOD1;MYOG and PSMD9). Two motifs match to families of known TFs (RUNX50 and GATA51) that are known to be involved in embryonic development.
Predictive motifs have location preferences
The identified cis-elements may play various roles in shaping the epigenome, such as setting the boundary of a histone modification domain or opening chromatin to allow remodeling enzymes to bind DNA. These roles may restrain the relative location (edge or center) of a motif within the modified regions (Fig. 4a). While the majority of the motifs fall into the neutral category, numerous motifs showed biased location distributions (Fig. 4b). The heterochromatin mark H3K9me3 is associated with edge and neutral motifs but not with any central motifs. This observation is consistent with the large domain of H3K9me3 and the edge motifs may help set the boundary. Consistently, concentrated marks of H3K4me1/3 and H3K27ac are associated with central motifs, which may guide the recruitment of the chromatin-modifying enzymes to initiate or other factors to maintain the modifications38, 52. Interestingly, while the enhancer marks H3K4me1 and H3K27ac have no edge motifs, the promoter marker H3K4me3 is associated with several edge motifs, which may help define the promoter boundary. Contradictory to H3K9me3, the widespread histone mark H3K27me3 and the DMV are largely associated with central motifs, which suggests different regulatory mechanisms. The transcriptional activity mark, H3K36me3, almost exclusively associates with neutral motifs.
The majority (81%) of the H3K9me3 edge motifs were found in H1 and these match the known motifs of YY1, KLF12 and the ‘Rel homology domain’ (RHD), (Fig. 4c). Multiple lines of evidence support these associations. KLF12 mediates transcriptional repression through interaction with phosphoprotein CtBP53, which forms a complex with histone methyltransferase and DNA-binding proteins to target H3K9 for methylation54. NFKB1, a member of RHD TFs, is known to function with deacetylase SIRT6 to repress gene expression via H3K9 deacetylation55, which clears the site for methylation. YY1 is a transcriptional regulator that directs localization of histone acetyltransferases, deacetylases and members of the PRC2 complex56, which directs the placement of H3K9me3 and H3K37me357. Furthermore, YY1 knockdown during mouse spermatogenesis resulted in global decrease of H3K9me358. Given the available genome-wide binding data of YY1, we found that the motif and ChIP-seq binding profiles of YY1 are highly correlated (R2=0.86) in H3K9me3 domains that overlap with YY1 ChIP-seq peaks.
Interestingly, YY1 also marks the center of H3K4me3 peaks. In total there are 35,393 YY1 binding peaks in H1 (union of two replicates) of which 874 are within 200bps of H3K9me3 peak start/end boundaries (Fig. 4d) whereas 14,587 are within 200bps of an H3K4me3 peak. This suggests that the primary role of YY1 in H1 is binding at promoters with a secondary role binding at edges of H3K9me3 peaks. The dual activating and repressing classes of YY1 binding sites have previously been observed59. To identify co-factors that differentiate YY1 sites that exclusively overlap with H3K9me3 edges and those with H3K4me3 peaks, we examined the differential overlap of these regions with ChIP-seq data in H1 for 60 different factors (Supplementary Data 1). H3K9me3 edges were found to have modestly higher levels of overlap with OCT4, SUZ12, NANOG and BCL11A while the H3K9me3 demethylase, KDM4A (also known as JMJD2A), was found to have the lowest relative degree of overlap with H3K9me3 compared to H3K4me3. Interestingly, we found that the YY1 known motif scored higher at YY1 sites that overlap with H3K9me3 edges than YY1 sites that overlap with H3K4me3 peaks (Supplementary Fig. 7), suggesting that YY1 ChIP-seq sites at the edge of H3K9me3 are more likely to be stronger binding sites or that YY1 binds the H3K4me3 sites with a different motif. Next, we compared the YY1 sites located at H3K9me3 edges and H3K4me3 peaks in H1 to YY1 ChIP-seq peaks from other cell-types (A549, GM12878, HepG2 and K562) and found that the H3K9me3 sites were less conserved (H3K9me3 11–32%; H3K4me3 62–81%). Furthermore, the majority of YY1/H3K9me3 edge sites in H1 were H1-specific as only 9–37% were within 200bps of a peak and 8–29% were within 200bps of a peak start/end in any of the four H1-derived cells. Finally, the expression levels of the nearest gene (with 20kb) to the H1 YY1/H3K9me3 edge sites were significantly lower than the four H1-derived cells (P-value < 0.05; paired Students t-test). Taken together, these suggest that the H1 H3K9me3 edge motifs may represent a regulatory system present in hESCs for establishing regions of heterochromatin and repressing gene expression. In light of recent findings that show H3K9me3 as a primary epigenomic determinant during iPS60 cell reprogramming, we speculate that these interactions may be important in establishing and maintaining the pluripotent state.
We also observed that many of the H3K4me3, H3K27me3 and DMV central motifs have high GC content. We defined GC-rich motifs as those containing >80% of positions where: (i) high probability positions (>0.5) must be G or C; (ii) if A and T, or G and C, at a single position, are >0.75, they must be G and C. In total, we identified 150 such motifs (Fig. 4a), which were found in all cell-types and are enriched in all modifications. The association of TF binding and GC-rich regions may explain the general abundance of GC-rich motifs61. However, the GC-rich motifs never negatively interplay (depleted from the modification peaks) with H3K4me3, H3K27me3 and DMV suggesting a more specific association. In mESCs, an artificial, promoterless and CpG-rich sequence bound by CFP1 results in H3K4me3 establishment10. Promoters with high GC-content tend to be repressed by H3K27me3 whereas other promoters tend to be repressed by DNA methylation31. Our results are consistent with these previous reports and systematically pinpoint the DNA motifs in these GC-rich sequences that are responsible for forming the specific modifications. If the same criteria were reversed to identify AT-rich motifs (Fig. 4a), only 22 motifs were found and no overall trend is observed.
When examining the GC-AT-hybrid motifs (motifs made up of a continuous stretch of G/C followed by a continuous stretch of A/T, or vice versa), we found no overall trend of their interplay with the epigenomic modifications. However, GC-AT-hybrid motifs whose GC portion occupies three or less positions are found to prefer the edge of H3K4me3 and DMV (Fig. 4a). One of the GC-AT-hybrid motifs matches the motif of the nuclear receptor NR4A2 (Fig. 4e). The NR4A family contains two members (NR4A1 and NR4A3) that have highly similar DNA-binding domains and are constitutively active62. Moreover, NR4A2 has been shown to mediate gene expression by inducing H3K4me3 and histone acetylation at the promoter of FOXP363. The pattern of GC-AT-hybrid motif enrichment and H3K4me3 at transcription start sites (TSS) (Fig. 4f) shows two roles: (i) TTTAAAGG enriched ~1kb either side of the TSS, (ii) ATAATCCG is enriched ~0.5kb either side of the TSS. Consecutive 3–5 pyrimidines are believed to narrow the minor groove of DNA64 and have been shown to flank the binding sties of TFs65. Furthermore, poly(dA:dT) control nucleosome positioning by forming nucleosome-depleted regions (because of their structural stiffness) around which nucleosomes are positioned22, 23, 25. Moreover, poly(dA:dT) tracks capped with a single G residue on the same strand as the poly(dA) have been shown to flank well positioned nucleosomes at promoters in yeast24. The adjacency of the G and A nucleotides is consistent with our findings in human. Taken together, GC-AT-hybrid motifs may define the boundary of H3K4me3 and DMV modified regions through a combination of several possible mechanisms: (i) by creating a nucleosome-free region around the TSS, (ii) by providing a stretch of G/C-free sequence that cannot be bound by the factors preferring GC-regions, (iii) by being bound by TFs, such as NR4A2, that in turn recruit chromatin-modifying enzymes.
Motif disruption is correlated with H3K27ac variation
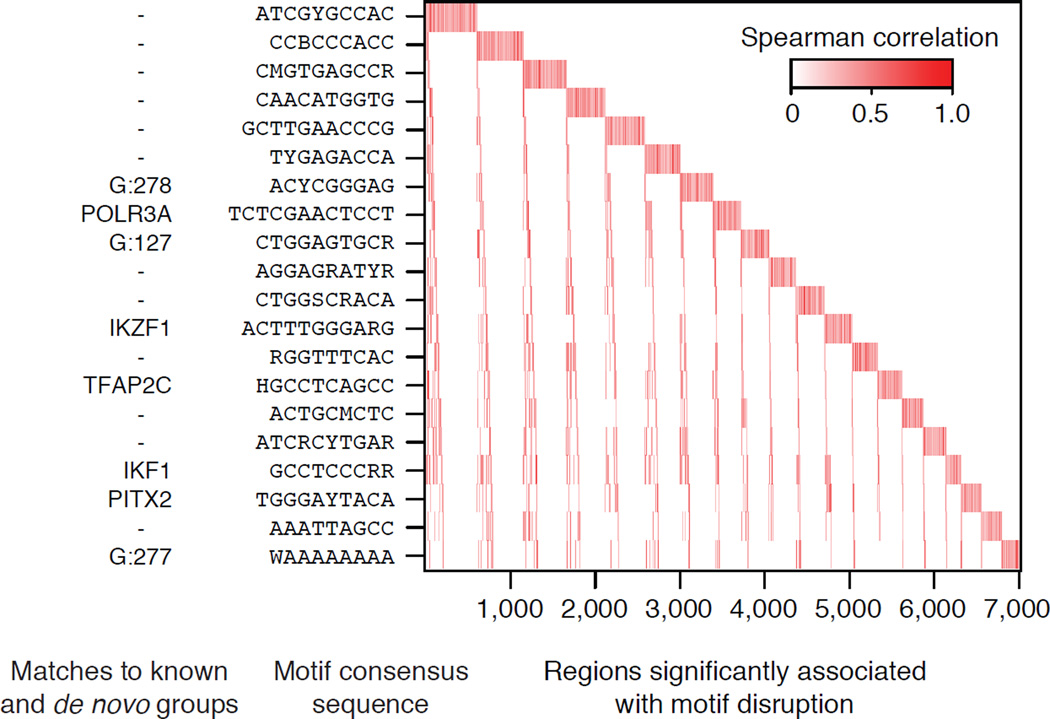
A recent study of 19 individuals correlated sequence variation at known TF motif sites with variation in H3K27ac levels at overlapping peaks16. Kasowski et al. found that H3K27ac variation in 32,886 peaks correlated with disruption of 662 known motifs by SNPs among the 19 individuals and significant association was found in 32% of regions (significance determined using Spearman’s rank and label permutation16). To demonstrate the power of the Epigram pipeline, we repeated the analyses done by Kosowski et al. by first running Epigram on the H3K27ac peaks, resulting in a full model featuring 133 motifs that are predictive of H3K27ac. Epigrams motifs were significantly correlated in 62% of regions using a motif set that is ~20% the size of those used by Kasowski et al. (662 known motifs). Thus, Epigram discovers motifs that are significantly correlated with H3K27ac variation in 30% more regions and represent the novel binding patterns for regulators of H3K27ac. Furthermore, Kasowski et al.16 showed 20 TFs that are significantly correlated within ~4,500 variable regions; whereas the motifs from Epigram’s 20 motif model are significantly correlated within 7,006 variable regions (Fig. 5). One of the Epigram’s 20 motifs matches the known IKZF1 motif, which has been shown to target chromatin remodeling and deacetylation complexes during lymphocyte differentiation66. In addition, we also found that three of these 20 motifs match motif groups identified to be associated with H3K27ac in H1, NPC, MSC and TBL. Taken together, Epigram is able to explain significantly more variants while using fewer motifs than the Kasowski et al. analysis.
Figure 5. De novo motif disruption and H3K27ac levels are correlated.
The disruption of de novo motifs was correlated with variation in H3K27ac levels.16. Motifs are sorted by their number of significantly correlated peaks; peaks are sorted by their associated motifs. Matches with known TFs and motif groups (from the analysis of H1 and the fo ur derived cell-types) are shown on the left. Motif groups start with - ‘G:’.
Discussion
Herein we present the Epigram pipeline, which is the first quantitative model to predict epigenomic modifications from combinations of sophisticated DNA motifs. This in turn reveals the cis-regulatory program that is read by the dynamic genetic network to shape the epigenome (Fig. 1a). We demonstrated the success of Epigram in hESCs and four derived lineages. Furthermore, we generated the first systematic cataloguing of mark-specific cis-elements that are predictive of epigenomic modifications. Prediction power was demonstrated by distinguishing epigenomically modified regions from non-modified regions with an accuracy of 79% across all peaks in the genome. We further demonstrated that prediction power is not the consequence of trivial correlations between epigenomic modifications and simple sequence features, such as GC-content. Indeed, our method removed many sequence features that are associated with classes of genomic regions but not epigenomic modifications. Moreover, we observed significantly reduced performance in predicting same histone modifications in different cell-types, which indicates that histone modifications are significantly decided by mark-specific cis-elements and our model can successfully detect them.
In particular, we removed biases introduced by simple sequence features, such as GC-content, and focused on identifying the cis-regulatory program that directs locus-specific epigenomic modification This was further enhanced by the comparative analyses to tease out the motifs that are directly associated with epigenomic modifications. Furthermore, regulatory connections between Epigram motifs and epigenomic modification are confirmed using inter-individual correlation between DNA motifs and: (i) H3K27ac, (ii) tissue DNA methylation40. In particular, Epigram’s motifs are significantly correlated with almost double the number of H3K27ac regions when compared to five times the number of known TF motifs. Thus, the Epigram pipeline discovers previously unknown binding motifs that are involved in regulating the placement of epigenomic modification. Recent studies have illustrated the power of editing epigenomes by changing the DNA sequences67, 68. Equipped with new genome editing technologies, such as TALEN69 and CRISPR70, our study provides the first comprehensive catalogue of DNA motifs to guide locus-specific epigenome editing through alteration of regulatory cis-elements.
Especially interesting, we found motifs that have location preference within the modified regions, which suggests possible functions in setting the boundary of modified regions or opening chromatin to establish modification. Several of these motifs match with the cis-elements recognized by TFs; such as TEAD family TFs that tend to bind at the center of H3K27ac; YY1, whose ChIP-seq peaks consistently overlap with H3K4me3 peaks and the edge of H3K9me3 regions. We also observed that H3K4me3 is consistently the most predictable with GC-rich motifs at the center of its peaks and GC-AT-hybrid motifs at the boundaries of the peaks (Supplementary Fig. 11). The role of the GC-AT-hybrid motifs is unclear but they likely play roles in nucleosome positioning or recruiting specific TFs, such as NR4A2. Although the mechanisms by which the identified motifs orchestrate the epigenome are largely unknown, these mechanisms are ultimately mediated by DNA-specific factor binding to establish locus-specific modifications and our study represents the first step towards unveiling the enigmatic cis-regulation of the human epigenome.
Supplementary Material
Acknowledgement
This work was partially supported by NIH (U01 ES017166 to W.W., PI, B. Ren at Ludwig Institute and UCSD). The authors wish to thank B. Ren, D. R. Westhead and M. H. Sherman for discussion of this work.
Footnotes
Contributions
JWW and WW conceived and designed the project, JWW performed all the analyses, ZC contributed to data analysis, WW analyzed the data, JWW and WW wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan GC. Linking genome to epigenome. Wiley Interdiscip Rev Syst Biol Med. 2012;4:297–309. doi: 10.1002/wsbm.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendenhall EM, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klattenhoff CA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulut-Karslioglu A, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol. 2012;19:1023–1030. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 15.Costa Y, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342:744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McVicker G, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342:747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. Embo J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu R, Li H. Positioned and G/C-capped poly(dA:dT) tracts associate with the centers of nucleosome-free regions in yeast promoters. Genome Res. 2010;20:473–484. doi: 10.1101/gr.103226.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raveh-Sadka T, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat Genet. 2012;44:743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan N, et al. Nucleosome sequence preferences influence in vivo nucleosome organization. Nat Struct Mol Biol. 2010;17:918–920. doi: 10.1038/nsmb0810-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pugh BF. A preoccupied position on nucleosomes. Nat Struct Mol Biol. 2010;17:923. doi: 10.1038/nsmb0810-923. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Evidence against a genomic code for nucleosome positioning Reply to "Nucleosome sequence preferences influence in vivo nucleosome organization". Nat Struct Mol Biol. 2010;17:920–923. doi: 10.1038/nsmb0810-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst J, Kellis M. Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome research. 2013;23:1142–1154. doi: 10.1101/gr.144840.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ha M, Hong S, Li WH. Predicting the probability of H3K4me3 occupation at a base pair from the genome sequence context. Bioinformatics. 2013;29:1199–1205. doi: 10.1093/bioinformatics/btt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W, et al. Epigenomic Analysis of Multi-lineage Differentiation of Human Embryonic Stem Cell. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung MS, Down TA, Latorre I, Ahringer J. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res. 2011;39:e103. doi: 10.1093/nar/gkr425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 36.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 37.Yuan Y, Guo L, Shen L, Liu JS. Predicting gene expression from sequence: a reexamination. PLoS Comput Biol. 2007;3:e243. doi: 10.1371/journal.pcbi.0030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz MD, et al. Human Body Epigenome Maps Reveal Noncanonical DNA Methylation Variation. 2013 doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 44.Mauvieux L, Villey I, de Villartay JP. TEA regulates local TCR-Jalpha accessibility through histone acetylation. Eur J Immunol. 2003;33:2216–2222. doi: 10.1002/eji.200323867. [DOI] [PubMed] [Google Scholar]
- 45.Badouel C, Garg A, McNeill H. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol. 2009;21:837–843. doi: 10.1016/j.ceb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 46.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melin F, Miranda M, Montreau N, DePamphilis ML, Blangy D. Transcription enhancer factor-1 (TEF-1) DNA binding sites can specifically enhance gene expression at the beginning of mouse development. Embo J. 1993;12:4657–4666. doi: 10.1002/j.1460-2075.1993.tb06154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev Genet. 1998;22:43–55. doi: 10.1002/(SICI)1520-6408(1998)22:1<43::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 49.Home P, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7362–7367. doi: 10.1073/pnas.1201595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi JY, et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci U S A. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 52.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuierer M, et al. Induction of AP-2alpha expression by adenoviral infection involves inactivation of the AP-2rep transcriptional corepressor CtBP1. J Biol Chem. 2001;276:27944–27949. doi: 10.1074/jbc.M100070200. [DOI] [PubMed] [Google Scholar]
- 54.Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 55.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. Variable requirements for DNA-binding proteins at polycomb-dependent repressive regions in human HOX clusters. Mol Cell Biol. 2013;33:3274–3285. doi: 10.1128/MCB.00275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de la Cruz CC, et al. The polycomb group protein SUZ12 regulates histone H3 lysine 9 methylation and HP1 alpha distribution. Chromosome Res. 2007;15:299–314. doi: 10.1007/s10577-007-1126-1. [DOI] [PubMed] [Google Scholar]
- 58.Wu S, Hu YC, Liu H, Shi Y. Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol Cell Biol. 2009;29:6245–6256. doi: 10.1128/MCB.00679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitfield TW, et al. Functional analysis of transcription factor binding sites in human promoters. Genome Biol. 2012;13:R50. doi: 10.1186/gb-2012-13-9-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome research. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 63.Sekiya T, et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun. 2011;2:269. doi: 10.1038/ncomms1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jolma A, et al. DNA-Binding Specificities of Human Transcription Factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 66.Kim J, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 67.Carone BR, Rando OJ. Rewriting the epigenome. Cell. 2012;149:1422–1423. doi: 10.1016/j.cell.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 68.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 70.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 72.Ward JH. Hierarchical Grouping to Optimize an Objective Function. Journal of the American Statistical Association. 1963;58:236–244. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.