Abstract
The Q-system is a repressible binary expression system for transgenic manipulations in living organisms. Through protein engineering and in vivo functional tests, we report here new variants of the Q-system transcriptional activator, including QF2, that allows the Q-system to drive strong and ubiquitous expression for the first time in all tissues. Our new QF2, GAL4QF and LexAQF chimeric transcriptional activators substantially enrich the toolkit available for transgenic regulation in Drosophila.
The characterization and manipulation of complex biological systems requires sophisticated genetic tools. Binary expression systems are powerful tools to direct transgenic expression of effector genes. In Drosophila, the GAL4-UAS1 system has been widely adopted, but this system has its limitations. Existing GAL4 expression patterns are often too broad and require refinement, and GAL4-UAS alone is insufficient for independent manipulation of two distinct populations of cells. To extend transgenic manipulations, two additional binary expression systems have been developed: the λ phage LexA-LexAop2 and Q-system, derived from the qa gene cluster of the fungus Neurospora crassa3. The Q-system comprises the transcriptional activator QF, the QF effector QUAS, the QF suppressor QS, and the non-toxic drug quinic acid, which inhibits QS. Thus, in addition to QF being repressible like GAL4, the Q system has the additional advantage that expression can be temporally regulated by quinic acid.
Despite its wide application4–8, the original QF was lethal when expressed broadly in vivo3, which made it impossible to obtain flies that expressed QF either under the control of strong pan-neuronal or ubiquitous promoters. The cause of this toxicity was unknown. To address this problem, we aimed to identify the region of QF that was responsible for toxicity, and to generate a QF variant that retained full activity yet could be broadly expressed with no adverse effects. For our experimental approach, we created chimeric proteins between QF, GAL4 and/or LexA2. Our bioinformatic alignments of QF and GAL4 and previous studies9,10 predicted that QF is structurally similar to GAL411–14. We hypothesized that, like GAL415, QF can be subdivided into 3 domains (Fig. 1a) that perform specific and independent functions: the DNA binding and dimerization domain (DBD) containing a Zn2-Cys6 motif that recognizes and binds to UAS or QUAS sites; a middle domain (MD) that has no clear function but might be involved in endogenous regulation or stability; and a transcriptional activation domain (AD) that recruits molecular machinery necessary for transcription, and which also binds the GAL80 or QS suppressor.
Figure 1.
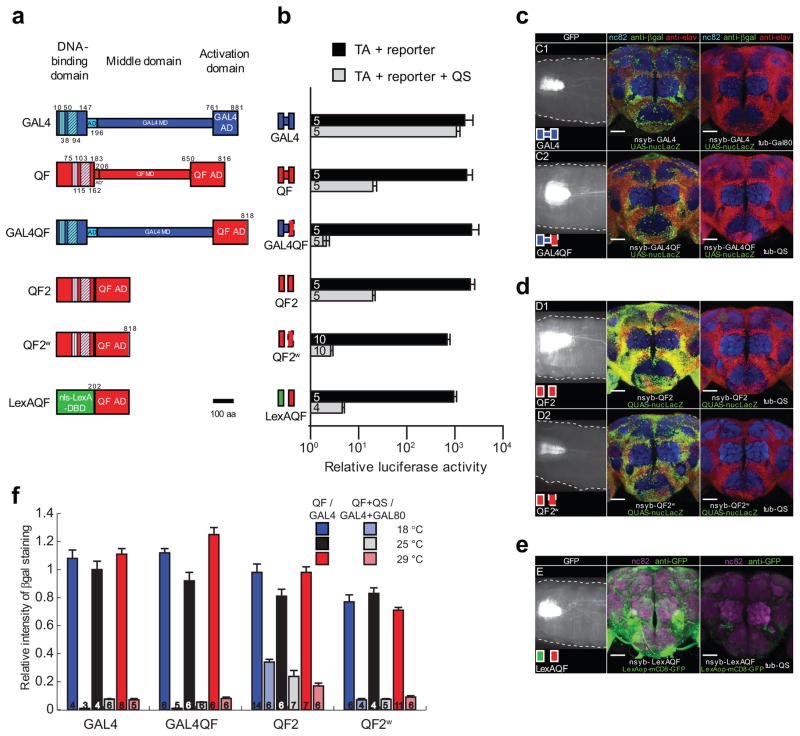
Activity of modified QF transcriptional activators in vitro and in vivo in neuronal tissue. (a) Schematics of GAL4, original QF3 and four new transcriptional activators. DBD, DNA binding domain; MD, middle domain; AD, activation domain. Vertical hatching indicate Zn2-Cys6 zinc finger motifs, diagonal hatchings mark dimerization domains. Numbers indicate amino acid position. Constructs are drawn to scale. (b) The transcriptional activity (black bars) of QF transcriptional activators (TA) was investigated by transfecting S2 cells with actin-QFX plasmid, firefly luciferase reporter plasmid (pLexAop-luc2, pQUAS-luc2 or pUAS-luc2), Rhenilla luciferase plasmid (pAC-hRluc) for normalization, and pBluescript (pBS-KS) plasmid for equalizing DNA amount. For QS repression assays (grey bars), pBluescript plasmid was replaced by pAC-QS plasmid in co-transfections. Numbers in bars indicate the number of independent repeats. Error bars are SEM. Luciferase activity is shown on a log scale. (c–e) Pan-neuronal in vivo expression of constructs driven by neuronal synaptobrevin (nsyb) promoter at 25°C. Left column shows mCD8-GFP expression in third-instar larvae (representative of n=4–6; scale bar, 100 μm), middle column shows nuclear lacZ expression in adult brain (representative of n=4–6; scale bar, 50 μm), and the right column shows the Gal80- or QS-induced suppression of lacZ expression in adult brains (representative of n=5–7). Brains were immunostained for elav (red), lacZ (green) and nc82 (blue). (e) Larval and adult expression of mCD8:GFP driven by nsyb-LexAQF construct (representative of n=5). Right panel, tubP-QS suppression of LexAQF activity (representative of n=5). (f) LacZ expression, quantified as described in Online Methods. Numbers in bars indicate the number of brains for each condition. Error bars are SEM.
To overcome limitations of the original QF, any new QF variant should be capable of generating healthy transgenic flies when broadly expressed. In addition, it should, like the original QF, exhibit strong transcriptional activity yet remain QS suppressible. We generated a series of constructs (Fig. 1a and Supplementary Fig 1a) in which certain QF domains were: 1) mutated to reduce activity by altering the charge on the C-terminus (GAL4QF, QF2w(eaker), QFe), 2) either completely (QF2, QF2w, LexAQF) or partially (QFf-i) removed, or 3) swapped for GAL4 or LexA domains (GAL4QF, LexAQF, QFa-d,j-l). To quantitatively measure activity levels, we performed luciferase assays in cultured S2 cells (Fig. 1b). To assay for QF toxicity, we attempted to generate transgenic animals expressing each construct under the pan-neuronal synaptobrevin (nsyb) promoter (Fig. 1; Supplementary Fig. 1; Supplementary Table 1). To allow direct comparison between transgenic constructs that use the same enhancer activation sequence (UAS, QUAS or LexAop), we utilized the PhiC31 integrase system to target all transgenic insertions to the same attP2 genomic landing site (3L: 68A4). Note that direct comparison between transgenic factors utilizing different activation sequences, e.g. UAS reporters (Fig. 1c) vs. QUAS reporters (Fig. 1d), cannot be made due to differing activities of the reporters.
Relative luciferase activity assays and in vivo expression analyses showed that the optimal QF variants exhibited high activity levels (Figs. 1b–f; Supplementary Table 1), were efficiently repressed by QS (Supplementary Table 1), and produced healthy transgenic animals. We initially hypothesized that a potent QF activation domain may be the source of toxicity as it may be squelching cellular transcription factor components16. Contrary to our expectations, QF variants that contained the original (QF2, LexAQF) and mutated activation domain of QF (QF2w and GAL4QF) were not toxic. Instead, constructs containing the middle domain of QF either failed to produce transgenic animals (QFd and QFg, Supplementary Fig. 1) or were extremely unhealthy (QFf). This implicated the QF middle domain as the major source of QF toxicity. Deletion of this domain yielded two smaller QF variants, QF2 and QF2w(eaker), which exhibited strong but differing QF activities in vitro (2089±477 and 685±44 times above control, respectively, P<0.001) and in vivo (Figs. 1c–e and Supplementary Fig. 1c). Both QF2 and QF2w were capable of generating healthy pan-neuronally expressing transgenic animals. Thus, the QF middle domain is dispensable for full QF activity, yet is the major source of QF toxicity in vivo.
We assessed expression patterns and strength of the transactivators at 18°C, 25°C and 29°C with both membrane tagged GFP (Supplementary Fig. 2) and nuclear lacZ reporters (Fig. 1f). Similar to the in vitro results (Fig. 1b), we found that QF2, QF2w, GAL4QF and LexAQF have activity levels comparable to GAL4 and can be robustly repressed by QS at all tested temperatures. In agreement with Mondal et al17, GAL4 activity did not vary with temperature. This contrasts with the temperature dependence often observed with many GAL4 enhancer traps18 which likely reflects the use of temperature-sensitive elements in these constructs1.
We quantified the expression level for LexABD:QFAD chimeric proteins (LexAQF and QFl) only with a GFP reporter (Fig. 1e, Supplementary Fig. 1d and 2), due to unavailability of LexAop-nucLacZ reporter lines. Qualitatively, both constructs drive strong expression in vivo, and pan-neuronally expressing transgenic animals were healthy. Quantitatively, expression levels of LexAQF-driven GFP were similar to those of GAL4-driven GFP at 18°C, 25°C and 29°C (Supplementary Fig. 2), and could be repressed by QS at all temperatures (data not shown). The LexAQF chimeras offer a useful alternative to LexA:VP16 and LexAGAL4 transcriptional activators2 in that LexAQF chimeras are independent of the GAL4-UAS system and can be reversibly suppressed by QS (Fig. 1e; Supplementary Fig. 1d).
The nuc-lacZ quantification of QS-suppressed activity of QF2 in vivo (Fig. 1f) suggested that a number of cells were still weakly labelled and detectable by our algorithm. To further validate the ability of QS to functionally inhibit QF2 and QF2w, we performed whole animal rescue experiments. We drove the expression of the temperature-sensitive endocytotic recycling protein shibire (at 29°C) with nsyb-QF2 or nsyb-QF2w, which resulted in no surviving adults as expected. This lethality was fully rescued in flies that also carried a tubulin-QS transgene (Supplementary Table 2). These results indicate that QF2 and QF2w are efficiently suppressed by QS in vivo. In addition, the activity of all QF activation domain variants (QF2, QF2w, GAL4QF, LexAQF) could be regulated by feeding larvae or adult flies with the non-toxic drug quinic acid (Supplementary Fig. 3). Quinic acid has a stronger effect on peripheral receptor neurons than on central brain neurons, presumably reflecting differential exposure of the neurons to QA.
The new transactivators (QF2, QF2w, GAL4QF and LexAQF), together with GAL4, offer a possibility to use GAL4, LexA and Q-systems simultaneously in overlapping subsets of cells. We verified that activity of any two of these transactivators in the same cell does not result in toxicity or reporter silencing effects by generating all possible binary combinations of the nsyb transactivator flies (Supplementary Fig. 4).
To test whether expression of the new transactivators might cause toxicity in non-neuronal tissues, we generated flies that express QF2, QF2w, GAL4QF and LexAQF under the control of the ubiquitous promoters tubulin or actin. These flies are viable, and activity of the transactivators was robust in late embryos (data not shown), larvae and in adult flies (Fig. 2a). All new ubiquitous drivers can be effectively suppressed by QS in the whole larva or adult flies (Fig. 2a; Supplementary Fig. 5). Examination of imaginal discs (epithelial tissue) and larval body walls (muscle) (Supplementary Fig. 5a–b) confirmed the broad transactivator expression patterns of QF2, QF2w, GAL4QF and LexAQF.
Figure 2.
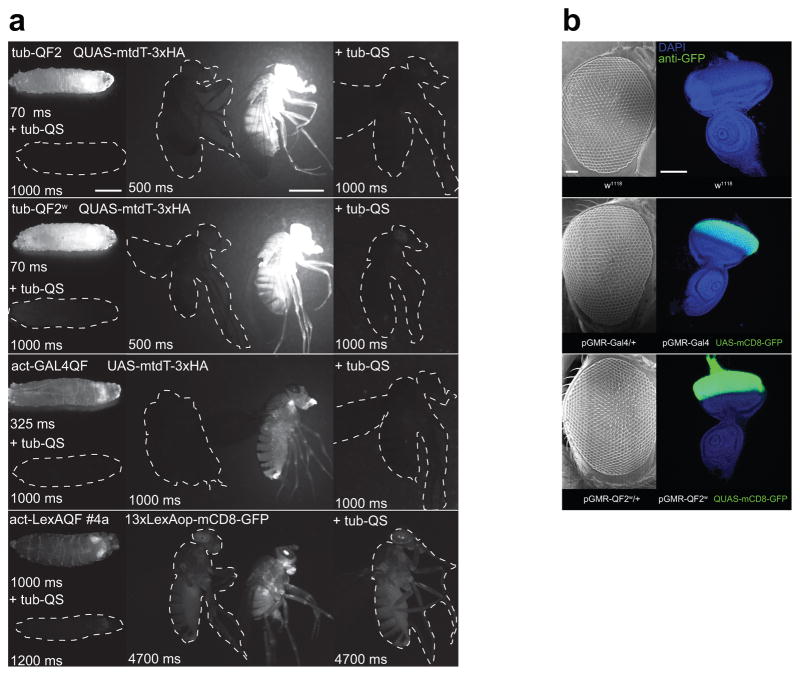
In vivo expression driven by tubulin, actin and GMR promoters. (a) Ubiquitous expression of mtdTomato or GFP reporters in third instar larvae (left column, representative of n=4–6) and in the adult flies (right column, representative of n=5). Larvae in the top row of each subpanel carry a tubulin or actin driver line and an mtdt or mCD8-GFP reporter. Larvae in the bottom row of each subpanel (marked by “+tub-QS”) also carry a tub-QS transgene. Adult flies (right column, middle) are imaged next to the controls (right column, left) that bear only the TA or only effector transgenes (dashed white outline). The rightmost subpanels (marked by “+tub-QS”) show flies that, in addition to the indicated driver and reporter transgenes, also carry a tub-QS. Expression of act-LexAQF driver is visualised with a mCD8-GFP reporter. Imaging settings were identical for all images, apart from the duration of exposure, which is indicated for each image in milliseconds. Scale bars, 1 mm. (b) Scanning electron micrograph images of the adult female eyes (left column, representative of n=10) and GFP expression in the eye-antennal imaginal disc (right column, representative of n=5) for w1118, pGMR-GAL4/+ and pGMR-QF2w/+ flies. Scale bars, 50 μm.
Experimental evidence that GAL4, when driven at very high levels, could be toxic to the fly was first determined in experiments using the very strong eye promoter pGMR19,20. pGMR-QF2w transgenic animals exhibited strong QF-induced GFP expression in the eye-antennal imaginal disc (Fig. 2b), yet had no morphological eye defects at the adult stage. These results suggest that QF2w, even when very strongly expressed, was not toxic to the cell.
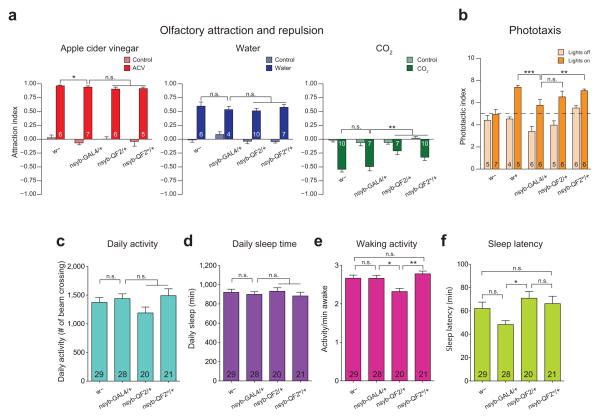
As a final readout of QF2 and QF2w effects in vivo, we examined three different behaviors in flies pan-neuronally expressing QF2, QF2w, or GAL4 in the same w1118 genetic background (Fig. 3 and Supplementary Table 3). Behaviors are a sensitive measure of in vivo health requiring many key processes to be un-affected, such as development, neuronal wiring, and neuronal function. The nsyb-QF2 and nsyb-QF2w flies were indistinguishable from nsyb-GAL4 controls in olfactory attraction to apple cider vinegar and humidified air, but were slightly but significantly (P<0.01) less repelled by CO2 gas than the controls (Fig. 3a). nsyb-QF2 and nsyb-QF2w flies exhibited phototactic responses comparable to nsyb-GAL4 flies and wild-type controls (Fig. 3b). In locomotor activity assays (Figs. 3c–f), daily activity and daily sleep amounts were not significantly different (P>0.05) between nsyb-QF2, nsyb-QF2w and control flies. Both nsyb-QF2 and nsyb-QF2w flies exhibited normal circadian rhythms under constant darkness (Supplementary Table 3). Taken together, these results demonstrate that pan-neuronal expression of QF2 and QF2w is compatible with proper neuronal development and function.
Figure 3.
Olfactory, phototaxic, and activity/sleep behavioral analyses of nsyb-GAL4, nsyb-QF2 and nsyb-QF2w transgenic flies. (a) Flies were tested in groups of 50 in a 4-field olfactometer assay. Control scores are attraction index (AI), calculated for the odor quadrant over 5 mins preceding presentation of an odorant. Odorant scores are AI’s calculated for the last 5 mins of the 10 mins odor presentation. We examined a highly attractive odor, 5% apple cider vinegar in water (left); a mildly attractive stimulus, pure water (middle) and a repulsive stimulus, 5% CO2 gas (right). Numbers in bars indicate number of independent experiments for each condition. Error bars are SEM. (b) Visually-induced responses of two wild-type strains (w− and w+), nsyb-Gal4/+, nsyb-QF2/+ and nsyb-QF2w/+ were investigated in a 10-step countercurrent phototaxis assay (see Online Methods for details). Phototactic index (PI) is the average number of approaches to the light that each fly made. The PI scores of the nsyb-QF2 and nsyb-QF2w flies were comparable with those of nsyb-Gal4/+ and w+ flies. Numbers in bars indicate number of independent experiments for each condition. Error bars are SEM. (c) Daily Activity, (d) daily sleep time, (e) waking activity, and (f) sleep latency for w− (n=29 flies), nsyb-Gal4/+ (n=28), nsyb-QF2/+ (n=20), and nsyb-QF2w/+ (n=21) flies, assayed as outlined in Online Methods. Error bars represent SEM. “*”, “**”, “***”, and “n.s.” denote P<0.05, P<0.01, P<0.001, and not significant, respectively, for all panels, as determined by the non-parametric Kolmogorov-Smirnov test.
In summary, we have developed two next-generation versions of QF, QF2 and QF2w, which have dramatically reduced toxicity and can be expressed broadly in vivo. QF2 is best suited when strong transcriptional activity is required in subsets of cells. QF2w is optimal for broad expression patterns or strong promoters. We have also developed chimeric GAL4QF and LexAQF transactivators which, while still activating UAS-geneX and LexAop-geneX effectors respectively, are QS-suppressible, quinic acid regulatable, and GAL80-insensitive. These new transactivators substantially expand the range of possible applications of the Q-system by itself, and also in combination with GAL4-UAS and LexA-LexAop for intersectional targeting.
Online Methods
Bioinformatics of QF
Phyre221 was used for bioinformatic prediction of QF secondary protein structures and disordered regions to guide domain borders for deletion or chimeric protein constructs. The QF dimerization domain at amino acids 115 to 162 was predicted based on protein alignments with GAL422 and Phyre2 predictions of a coiled-coil secondary structure. An internal QF activation domain was predicted based on protein alignments with GAL4, and charge plots15 which indicate a negatively charged basic region at aa 182–206. The QF Zn(II)2Cys6 binuclear DNA binding domain at amino acids 75–103 was as predicted in Reference #9. The QF activation domain was predicted as in Wei et al7. An InterProScan23 of QF predicted a conserved fungal transcription factor domain at amino acids 372–465, which was the basis for construct QFi.
Toxicity of transgenic QF constructs
The following constructs were generated, but failed to produce transgenic animals after multiple attempts (>1000 embryo injections per construct):
neuronal synaptobrevin promoter (nsyb) constructs:
nsyb-QF, in a piggyBac transformation vector for random genomic insertions (pXL-BACII-nsyb-QF-hsp70)
nsyb-QF, in an attB vector directed to attP2 (pattB-nsyb-QF-hsp70)
tubulin promoter (tubP) constructs:
tubulinP-QFcodon_deoptimzed (cdo), in a piggyBac transformation vector (pXL-BAC-tubulinP-QFcdo).
tubulinP-QF::QF2ADweak, in a piggyBac transformation vector; this contains the same activation domain mutant as in QF2w but with full length QF (pXL-BAC-tubulinP-QF2M1).
tubulinP-QF::QFeAD, in a piggyBac transformation vector; this contains the same activation domain mutant as in QFe but with full length QF (pXL-BAC-tubulinP-QF2M2).
Note that nsyb-QFcdo, in a piggyBac transformation vector for random genomic insertions, was able to generate transgenic animals. Of the 10 original lines, induced QUAS-mCD8GFP activity was weak, and none of the lines exhibited pan-neuronal expression. In addition, since we were unable to generate tubulinP-QFcdo transgenic animals, QFcdo constructs were not characterized further.
Our initial hypothesis was that the QF activation domain was the major source of QF toxicity. To circumvent this toxicity when generating QF transgenic animals, we injected constructs into flies containing a tubP-QS transgenic background. However, this did not help yield transgenic animals. Our recent findings suggest this is likely due to the middle domain of QF being the major source of toxicity, which would not be attenuated by QS expression.
Nonetheless, transgenic animals containing the GAL4 binding domain and QF activation domain (QFb, Supplementary Fig. 1) demonstrated the strongest activity of all the constructs, and were not as healthy as the same QF chimera (GAL4QF, Fig. 1) containing the QF2w activation domain. This suggests that a fully potent QF activation domain, in some instances, might contribute towards in vivo toxicity. We note that even GAL4 can be toxic when expressed at high levels16,19.
Recombinant DNA construction
Plasmids were constructed by standard procedures including enzyme digestions, PCR and subcloning. Some of the plasmids were manufactured using the In-Fusion® HD Cloning System (Clontech, Cat #639645). Plasmid inserts were verified by DNA sequencing. Apart from the constructs used in the reported experiments, we describe four additional constructs (pPT-QF2-hsp70, pattB-DSCP-QF2-hsp70, pattB-hsp70-QF2-hsp70, pCasper-QF2-hsp70) that may be useful for creating new QF2 lines. Primer sequences are shown in Supplementary Table 4.
QF codon variants
QFrco, QF recodonized
The original QF sequence from Neurospora often yielded tracheal expression in enhancer trap constructs3. This was likely due to a cryptic tracheal enhancer in the QF gene sequence. To eliminate this tracheal enhancer, the entire coding region of QF was re-codonized (DNA 2.0, Menlo Park, CA) by manually choosing codons expected to yield average expression (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=7227). Transgenic flies expressing QFrco enhancer traps no longer exhibited background tracheal expression (C. Potter, data not shown).
QFcdo, QF codon-deoptimized
A Drosophila codon de-optimized variant of QF led to reduced expression levels in transgenic constructs (C. Potter, data not shown). QFcdo enhancer trap flies induced weak reporter activity, and also no longer exhibited tracheal expression.
Chimeric and deletion cloning strategy
Chimeric constructs and deletions were generated by a multi-step PCR process using a high fidelity Taq polymerase (Phusion Taq, NEB). PCR fragments were generated that had terminal regions of sequence overlap (typically 17–25 base pairs to achieve a predicted Tm of 62–65°C) to other PCR fragments. The overlapping PCR fragments were used in a second round of PCR in which the overlapping PCR fragments each acted as primers for PCR amplification. After 5 cycles, additional oligos were included to selectively amplify full-length PCR products. All constructs were sequence verified before generating transgenic animals. All pattB-nsyb-geneX-hsp70 constructs were generated by inserting an EcoRI/AatII digested PCR product (GAL4, QFa-QFe, GAL4QF) or In-Fusion compatible PCR product (QF2, QF2w, QFf-QFk, LexAQF, LexAG4QF) into the EcoRI/AatII site of QF excised pattB-nsyb-QF-hsp70.
pattB-synaptobrevin-GAL4-hsp70 (GAL4) (Addgene#46107)
PCR template DNA: pGAWB1; Primers: RI-G4BD-FOR, AATII-G4AD-REV
pattB-synaptobrevin-1-QFBDAD-G4DM-hsp70 (QFa) (Addgene#46109)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: RI-QFBD-FOR, G4-QFDB-REV
PCR fragment 2- Template DNA: pGAWB; primers: QF-G4DM-FOR, QF-GFDM-REV
PCR fragment 3- PCR template DNA: pattB-tubP-QFrco; primers: G4-QFAD-FOR, AAtII-QFAD-REV
PCR fragments 1 and 2 were incubated in a PCR reaction for 5 cycles. In a separate PCR reaction, PCR fragments 2 and 3 were incubated together for 5 cycles. These two reactions were then mixed, cycled 5 times, and external oligos included for another 25 cycles.
Full length product- primers: RI-QFDB-FOR, AatII-QFAD-REV
pattB-synaptobrevin-2-G4BDDM-QFAD-hsp70 (QFb) (Addgene#46110)
PCR fragment 1- Template DNA: pGAWB; primers: RI-G4BD-FOR, QF-G4DM-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: G4-QFAD-FOR, AatII-QFAD-REV
Full length product- primers: RI-G4BD-FOR, AatII-QFAD-REV
pattB-synaptobrevin-3-QFBD-G4DMAD-hsp70 (QFc) (Addgene#46111)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: RI-QFBD-FOR, G4-QFBD-REV
PCR fragment 2- Template DNA: pGAWB; primers: QF-G4DM-FOR, AatII-G4AD-REV
Full length product- primers: RI-QFBD-FOR, AatII-G4AD-REV
pattB-synaptobrevin-4-QFBDDM-G4AD-hsp70 (QFd) (Addgene#46112)
PCR fragment 1- Template DNA: pGAWB; primers: QF-G4AD-FOR, AatII-G4AD-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: RI-QFBD-FOR, G4-QFDM-REV
Full length product- primers: RI-QFBD-FOR, AatII-G4AD-REV
pattB-synaptobrevin-5-G4BDDM-QFADM1-hsp70 (GAL4QF) (Addgene#46113)
PCR fragment 1- Template DNA: pGAWB; primers: RI-G4BD-FOR, QF-G4DM-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrcoM1; primers: G4-QFAD-FOR, AatII-QFADM1-REV
Full length product- primers: RI-G4BD-FOR, AatII-QFADM1-REV
pattB-synaptobrevin-6-G4BDDM-QFADM2-hsp70 (QFe) (Addgene#46114)
PCR fragment 1- Template DNA: pGAWB; primers: RI-G4BD-FOR, QF-G4DM-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: G4-QFAD-FOR, AatII-QFADM2-REV
Full length product- primers: RI-G4BD-FOR, AatII-QFADM2-REV
pattB-synaptobrevin-7-QFBDAD-hsp70 (QF2) (Addgene#46115)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-FOR, IF-QFAD-QFBD-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-QFAD-FOR, IF-QFAD-REV
Full length product- primers: IF-QFBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-8-QFAD184-214-hsp70 (QFf) (Addgene#46117)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-FOR, IF-QFAD-QFAD-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: IF-QFAD-QFAD-FOR, IF-QFAD-REV
Full length product- primers: IF-QFBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-9-QFMD184-475-hsp70 (QFg) (Addgene#46118)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-FOR, IF-QFAD-QFMD-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: IF-QFMD-QFAD-FOR, IF-QFAD-REV
Full length product- primers: IF-QFBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-10-QFMD475-650-hsp70 (QFh) (Addgene#46119)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-FOR, IF-QFMD475-QFBD-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-QFMD-FOR, IF-QFAD-REV
Full length product- primers: IF-QFBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-11-QFMD262-475-hsp70 (QFi) (Addgene#46120)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-FOR, 11L_QF262MD_QFBD_REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: 11M_QFBD_QF262MD_FOR, 11M-QFAD_QFMD475_REV
PCR fragment 3- Template DNA: pattB-tubP-QFrco; primers: 11R_QF475MD_QFAD_FOR, IF-QFAD-REV
Full length product- primers: IF-QFBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-12-QFBDAD-G4DM50-761-hsp70 (QFj) (Addgene#46121)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-FOR, 12L-G4MD50-QFBD-REV
PCR fragment 2- Template DNA: pGaWB; primers: 12M-QFBD-G4MD50-FOR, 12M_13M-QFAD-G4MD-REV
PCR fragment 3- Template DNA: pattB-tubP-QFrco; primers: 12R-G4MD-QFAD-FOR, IF-QFAD-REV
Full length product- primers: IF-QFBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-13-QFBDAD-G4MD257-761-hsp70 (QFk) (Addgene#46122)
PCR fragment 1- Template DNA: pattB-tubP-QFrco; primers: IF-QFBD-FOR, 13L-G4MD-QFAD-REV
PCR fragment 2- Template DNA: pGaWB; primers: 13M-QFAD-G4MD-FOR, 12M_13M-QFAD-G4MD-REV
PCR fragment 3- Template DNA: pattB-tubP-QFrco; primers: 13R-G4MD-QFAD-FOR, IF-QFAD-REV
Full length product- primers: IF-QFBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-14-LexA-QF-hsp70 (LexAQF) (Addgene#46123)
PCR fragment 1- Template DNA: pBPnlsLexA::GADflUw24 ; Addgene#26232>; primers: IF-LEXBD-FOR, 14L-QFAD-LEXBD-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: 14R-LEXBD-QFAD-FOR, IF-QFAD-REV
Full length product- primers: IF-LEXBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-15-LexABD-G4MD-QFAD-hsp70 (LexAG4QF) (Addgene#46124)
PCR fragment 1- Template DNA: pBPnlsLexA::GADflUw; primers: IF-LEXBD-FOR, 15L-QFAD-G4MD-REV
PCR fragment 2- Template DNA: pattB-tubP-QFrco; primers: 15R-G4LXMD-QFAD-FOR, IF-QFAD-REV
Full length product- primers: IF-LEXBD-FOR, IF-QFAD-REV
pattB-synaptobrevin-QF2w-hsp70 (QF2w) (Addgene#46116)
PCR template DNA: pAC-QF2w; primers: RI-QFBD-FOR, AatII-QFADM1-REV
S2 cell culture constructs
pAC-QFx (Addgene # 46089 - 46105)
These plasmids contain QFx variants under the control of the actin5c promoter for expression in S2 cell culture. The vector backbone for these constructs was obtained by digesting the pAC-QF plasmid3 with BamHI and NotI to remove QF gene. New QF variants were PCR-amplified from corresponding pattB-QFx plasmids and ligated into the vector backbone by an In-Fusion reaction. For PCR amplification, the same forward primer was used for all QF variants in combination with GAL4 DNA binding domain (IF_FOR_GAL4DBD), QF DNA binding domain (IF_FOR_QFDBD) and LexA binding domain (IF_FOR_LEXADBD) primers. Likewise, the same reverse primer was used to PCR amplify constructs with a GAL4 activation domain (IF_REV_GAL4AD) or an original QF activation domain (IF_REV_QFAD). Constructs with the LexA binding domain were amplified with reverse primer IF_REV_LEXADBD. The reverse primers for the following were: GAL4QF, IF_REV_GAL4QF; QF2w, IF_REV_QF2W; QFf, IF_REV_QF_F.
p-LexAop-Luc2
This construct allows expression of the firefly luciferase reporter under the control of LexAop in S2 cell culture experiments. The vector backbone was obtained by cutting pQUAS-luc23 with HindIII to remove QUAS. 5x LexAop was PCR-amplified from pJFRC19-13XLexAop2-IVS-myr::GFP24 (Addgene Plasmid #26224) with forward primer IF_FOR_LexAOP_LUC and reverse primer IF_REV_LexAOP_LUC, and sub-cloned by an In-Fusion reaction.
Additional constructs used for generating transgenic animals
pCasper4-tubP-QF2-hsp70 (Addgene #46127)
This construct was used to generate fly lines with random genomic insertions of the tubulinP-QF2 transgene. The vector backbone was obtained by cutting pCasper4-tubP-GAL8025 with NotI and XhoI. The QF2-hsp70_terminator insert was PCR-amplified from the pattB-QF2 plasmid with forward primer IF_FOR_TUB_QF2and reverse primer IF_REV_TUB_QF2, and cloned into the digested vector by an In-Fusion reaction.
pCasper-act(B)-QF2w-hsp70
This plasmid was used to generate fly lines with random genomic insertions of the actin-QF2w transgene. Transgenic flies are not described in this paper due to the availability of a stronger ubiquitous driver line, obtained with pCasper-tubP-QF2w-hsp70, but are available upon request. pCasper-act(B) (DGRC stock#1068) was digested with EcoRI and PstI, QF2-hsp70 was PCR-amplified from pattB-nsyb-QF2 with forward primer IF_FOR_ACT_QF2W and reverse primer IF_REV_ACT_QF2W, and cloned into the digested vector by an InFusion reaction.
pCasper-tubP-QF2w-hsp70 (Addgene #46128)
The 371 bp terminus of QF2w was excised from pattB-nsyb-QF2w-hsp70 by digestion with NheI/XhoI, and cloned into pCasper4-tubulinP-QF2-hsp70 in which the QF2 C-terminus had been excised by NheI/XhoI digestion.
pCasper-act-GAL4QF
This construct was used to generate transgenic flies with random genomic insertions of the actin-GAL4QF transgene. pCasper-act(B)-QF2w-hsp70 was digested with BamHI and NotI to remove QF2w-hsp70_terminator fragment. The GAL4QF-hsp70_terminator fragment was PCR-amplified from 5-pattB-synaptobrevin-G4BDDM-QFADM1-hsp70 with the forward primer IF_FOR_ACT_GAL4QF and reverse primer IF_REV_ACT_GAL4QF, and subcloned into the cut pCasper-act vector by an In-Fusion reaction.
pCasper-act-LexAQF
This construct was used to generate transgenic flies with random genomic insertions of the actin-GAL4QF transgene. pCasper-act(B)-QF2w-hsp70 was digested with BamHI and NotI to remove QF2w-hsp70_terminator fragment. The LexAQF-hsp70_terminator fragment was PCR-amplified from pattB-synaptobrevin-14-LexA-QF-hsp70 with the forward primer IF_FOR_ACT_LEXAQF and reverse primer IF_REV_ACT_LEXAQF, and subcloned into the cut pCasper-act vector by an In-Fusion reaction.
pGMR-QF2w (Addgene # 46130)
This plasmid was used to generate random genomic insertions of QF2w, driven by the strong GMR eye-specific promoter19,20. Vector pGMR-GAL4 was digested with EcoRI to remove the GAL4 gene, QF2w was PCR-amplified from pAC-QF2w with forward primer IF_FOR_GMR_QF2W and reverse primer IF_REV_GMR_QF2W, and subcloned into the digested pGMR vector by an In-Fusion reaction.
Additional plasmids generated for QF2
pCasper-act(B)-QF2w-act_term (Addgene # 46126)
QF2w was excised from pAC-QF2w by digestion with BamHI/NotI and ligated into pCasper4-actin5cB-QF2 digested with BamHI/NotI to excise QF2.
pPT-QF2-hsp70 (Addgene # 46136)
Vector pPTGAL26 was digested with PstI, QF2-hsp70 PCR-amplified from pattB-hsp70-QF2-hsp70 with forward primer IF_FOR_PPT_QF2 and reverse primer IF_REV_PPT_QF2, and subcloned into the digested vector by an In-Fusion reaction. This construct contains a minimal hsp70 promoter and allows for convenient subcloning of enhancers upstream of QF2.
pattB-DSCP-QF2-hsp70 (Addgene # 46133)
The pattb-QF-hsp70 plasmid3 and pattb-syb-QF2 were both cut with EcoRI and ZraI, and the isolated QF2 insert was ligated into the digested pattb-hsp70 vector using the Rapid DNA Ligation Kit (Roche). The pattb-QF2-hsp70 plasmid was digested with EcoRI and BamHI, the DSCP promoter PCR-amplified from pattB-DSCP-QF-SV403 with forward primer IF_FOR_DSCP_QF2 and reverse primer IF_REV_DSCP_QF2, cloned into the digest vector by an In-Fusion reaction. This PhiC31 integrase compatible plasmid utilizes the DSCP promoter27 to allow for the cloning of enhancer regions to drive QF2 expression.
pattB-hsp70P-QF2-hsp70T (Addgene # 46134)
The pattb-QF2-hsp70 plasmid was digested with EcoRI and BamHI, the hsp70 promoter PCR-amplified from pUAST1 with forward primer IF_FOR_ATTB_QF2 and reverse primer IF_REV_ATTB_QF2, and subcloned into the digested vector by an In-Fusion reaction. This PhiC31 integrase compatible plasmid utilizes the hsp70 minimal promoter to allow for the cloning of enhancer regions to drive QF2 expression.
pCasper-act(B)-QF2-act_term (Addgene #46125)
QF2 was PCR amplified from pattB-nsyb-QF2 using oligos IF-FOR-pCasper-ActB-QF7 and IF-REV-pCaspActB-QF7 and In-Fusion (Clontech, California) cloned into pCasper-act(B) digested with BamHI.
pCasper4-QF2-hsp70 (Addgene #46135)
The QF2-hsp70 cassette was PCR amplified from pattB-nsyb-QF2-hsp70 to include flanking XbaI restriction sites, and inserted into the XbaI site of pCasper4 (DGRC stock# 1213).
Progenitor plasmids for constructs described in this paper
pattB-synaptobrevin-QFcdo-hsp70
The synaptobrevin promoter was PCR amplified from pattB-nsyb-DSCP-QF-SV40 to include flanking BamHI and EcoRI restriction sites. The tubulin promoter from pattB-tubulinP-QFcdo-hsp70 was excised by digestion with BamHI/EcoRI, and replaced with the digested n-synaptobrevin PCR product.
pattB-nsyb-DSCP-QF-SV40
The n-synaptobrevin promoter was PCR amplified from the plasmid pPTGAL4+n-syb (a vector containing the n-synaptobrevin promoter upstream of the CMV minimal promoter, kindly provided by Julie Simpson, Janelia Farm Research Campus) to include flanking EcoRI restriction sites. The EcoRI digested PCR product was ligated into the EcoRI restriction site of pattB-DSCP-QF3.
pattB-tubulinP-QFcdo-hsp70
The hsp70 terminator from pXN-QF-hsp70 was PCR amplified to include AscI/NotI restriction sites (hsp70-AscI-FOR, hsp70-NotI-REV), and ligated into the pattB-tubulinP-QFcdo-SV40 vector digested with AscI/NotI to excise the SV40 terminator.
pattB-tubulinP-QFcdo-SV40
QFcdo was excised from p35030-QFcdo (synthesized by DNA2.0, Menlo Park, CA) by digestion with EcoRI/AscI, and replaced by ligation the QF in plasmid pattB-tubulinP-QF+AscI digested with EcoRI/AscI.
pattB-tubulinP-QF+AscI
The tubulin promoter from pCasper-tubulinP-GAL8025 was excised by digestion with BamHI/EcoRI and ligated into the BamHI/EcoRI site of the pattB-QF+AscI plasmid.
pattB-QF+AscI
An AscI restriction site between QF and the SV40 terminator was introduced into pattB-QF-SV403 by digestion with AatII and ligation of a compatible sticky ended annealed double stranded DNA of target sequence TGGCGCGCCA.
pattB-synaptobrevin-QF-hsp70
The n-synaptobrevin promoter was PCR amplified from pattB-nsyb-QF-SV40 to include BamHI and EcoRI restriction sites, and ligated into the BamHI/EcoRI multi-cloning site of pattB-QF-hsp703.
pattB-tubulinP-QFrco-hsp70
The QFcdo-hsp70 fragment from pattB-tubP-QFcdo-hsp70 was excised by EcoRI/XhoI digestion, and replaced with an EcoRI/XhoI QFrco-hsp70 cassette digested from pXL-BAC-attP-Ppromoter-QFrco-hsp70.
pXL-BAC-attP-Promoter-QFrco-hsp70
pJ241-QFrco (synthesized by DNA2.0, Menlo Park, CA) was digested with SnaBI/BglII to excise QFrco, and ligated into the SnaBI/BglII site of pXL-BAC-LoxP-DsRed-LoxP-attP-Promoter-QF-hsp70 in which QF had been excised by digestion with SnaBI/BglII.
pXL-BAC-tubulinP-QFrco-M1(EQ->KK)
The C-terminus of QFrco was mutated by PCR amplifying QFrco from pXL-BAC-tubulinP-QFrco-hsp70 using primers QFrco-NheI-FOR, QFrcoM1-REV, digestion with NheI/BglII, and ligation into NheI/BglII digested pXL-BAC-tubulinP-QFrco-hsp70.
pXL-BAC-tubulin-QFrco-M2(E->K)
The C-terminus of QFrco was mutated by PCR amplifying QFrco from pXL-BAC-tubulinP-QFrco-hsp70 using primers QFrco-NheI-FOR, QFrcoM2-REV, digestion with NheI/BglII, and ligation into NheI/BglII digested pXL-BAC-tubulinP-QFrco-hsp70.
Drosophila genetics
Flies were kept on a standard fly medium with a 12h:12h light:dark cycle in a 25 °C incubator, unless indicated otherwise. UAS, QUAS, and LexAop reporter lines were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537). Reporter transgenes were integrated at various genomic positions, but where possible, we utilized the same reporter line with new transactivator driver lines to compare activation strengths. Standard procedures were used to generate transgenic Drosophila by either P-element or piggyBac transgenesis, or by PhiC31 integration3.
Quinic acid experiments
To investigate the effect of Quinic acid (QA), we used 2–3 d.o. female flies of one of the following genotypes:
tub-QS/+; nsyb-QF2, QUAS-mCD8:GFP/+
tub-QS/+; nsyb-QF2w, QUAS-mCD8:GFP/+
tub-QS/+; nsyb-GAL4QF, UAS-mCD8:GFP/+
tub-QS/+; nsyb-LexAQF, LexAop-IVS-myr:GFP/+.
These flies were raised on standard fly medium until they were 2–3 day old adults, at which point they were transferred into vials containing 10 ml of 1% agarose (Denville Scientific Inc, Cat #CA3510-8) supplemented with 0.1g of sucrose (Sigma, Cat # S0389) and 0.6 grams of QA (Sigma, Cat # 138622). The vials also contained yeast paste made from dry yeast and QA solution (3 grams of QA per 10 ml of H2O, neutralized to pH 6.5 by 10 mM NaOH solution). The same QA solution was used to moisten a kimwipe that was embedded into the agarose gel. Flies were kept in these QA-containing vials for 3 days, after which brains were dissected and immunostained as described below.
S2 cell transfections and luciferase assays
S2 cells (Life Technologies, Cat #R690-07) were cultured in Express Five ® SFM (Serum Free Medium, Gibco®, Cat #10486-025), supplemented with 18mM L-Glutamine (Gibco®, Cat # 25030-081) and Pennicillin/Streptomycin/L-Glutamine mixture (25K/25K/200mM, Lonza, Cat #17-718R, 4.5 ml per 1L of SFM). The cells were maintained in 75cm2 tissue culture flasks (Sarstedt, Cat #83.1811.002) at room temperature and atmospheric CO2, and passaged every 4–6 days for no more than 26 generations. Cells were tested for mycoplasma infection by a PCR reaction using primers specific to 16S mycoplasma ribosomal RNA coding regions. For transfections, 0.3ml/well of cell-containing medium and 0.3ml/well of fresh medium were placed into 24-well plates (Corning, Cat #3524) 24h prior to transfection. All transfections were performed using Effectene Transfection Reagent (Qiagen, Cat #301425). 200ng of DNA (per well) was mixed with Effectene reagent, enhancer, and buffer according to manufacturer’s instructions, supplemented with 0.4ml (per well) of fresh medium and carefully pipetted into the wells. For QFx activity assays, each well was transfected with 12.5ng of a transcription factor pAC-QFx plasmid, 50ng of firefly luciferase reporter plasmid (pLexAop-luc2, pQUAS-luc2, or pUAS-luc2), 50ng of Rhenilla luciferase plasmid (pAC-hRluc) for normalization, and 87.5ng of pBluescript (pBS-KS) plasmid. In the controls, 12.5ng of the transcription factor plasmid was replaced by 12.5ng of pBluescript. We always transfected one of the wells in each 24-well plate with 200ng of pBluescript and left one well untransfected for control purposes. For QS repression assays, 87.5ng of pBluescript were replaced by 87.5ng of pAC-QS plasmid and co-transfected together with pAC-QFx, firefly and Rhenilla luciferase plasmids. Controls for QS assays were the same as for QF activity assays (firefly and Rhenilla reporters, and pBluescript plasmid). Cells were lysed 48h after transfection by replacing the medium in the wells with 0.1ml of Passive Lysis Buffer (PLB) from the Dual Luciferase Reporter Assay System (Promega, Cat # E1980) and shaking the plates at room temperature for 10 min. For luciferase activity measurements, the original lysates were diluted 10000 times in PLB, and analysed by a Fluorostar Optima (BMG Labtech) plate reader immediately after lysation. Each lysate sample was placed into 3 different wells in a 96-well plate; from each well the luminescence was measured automatically 6 times (once per second) after the addition of Firefly luciferase substrate, and 6 times (once per second) after the addition of Rhenilla luciferase substrate. The relative luminescence (RL) of each well was calculated as:
where is the average luminescence signal in response to luciferase X substrate. The average was calculated for measurements 3 to 6 because the first two measurements were often substantially different from the following four. To obtain the relative luciferase activity (RLA, Fig. 1b), the RL was averaged between the three wells that contained the same lysate. Next, this average RL, calculated for wells with transcription factor, was divided by a control RL, obtained from a corresponding control wells (only reporter plasmids without transcription factor).
For example, for pAC-GAL4 plasmid
and for pAC-QF plasmid
Thus, one RLA measurement was obtained for each of the wells from the original 24-well plate that contained a transcription factor plasmid. Fig. 1b shows the results of 4–5 RLA measurements for each construct, apart from pAC-QF2w, that was measured 10 times. Each RLA measurement was obtained from independent transfections performed on different days.
Immunohistochemistry
Dissection of larval imaginal discs and adult brains, immunostaining, and confocal imaging were done as described previously28. In short, brains of 3rd instar larvae or 4–5 days old adult flies were dissected in PBS, fixed for 20 mins at room temperature, washed at room temperature in PBT for 5–6 hours, blocked in 5% NGS in PBT, and placed in primary antibody mixes for 3 nights at 4°C. Next, the brains were washed for several hours in PBT at room temperature, and placed in secondary antibodies mix for 2 nights at 4°C. The following day the brains were washed in PBT and placed in mounting solution (Slow Fade Gold) overnight at 4°C, and mounted on a microscope slide the next day. To visualise GFP expression, we used rabbit anti-GFP (Life Technologies #A11122, 1:100), chicken anti-GFP (Aves Labs Inc, #GFP1020, 1:250) and mouse nc82 (DSHB, 1:25, not used for larval brains) primary antibodies; for lacZ experiments we used pre-absorbed rabbit anti-β-galactosidaze (MP Biomedicals #08559762, 1:50), Rat-ELAV-7E8A10 anti-ELAV (DSHB, 1:50) and mouse nc82 (1:25) primary antibodies; to visualise mtdt-3HA we used rat anti-HA (Roche #11867423001) primary antibody (1:100). Secondary antibodies used for GFP expression were Alexa-488 anti-Rabbit (Invitrogen #A11034, 1:200) and Cy3 anti-Mouse (Jackson ImmunoResearch #115-165-062, 1:200). For lacZ experiments: Cy3 anti-Rabbit (Jackson ImmunoResearch #111-165-144, 1:200), 633 anti-Rat (Invitrogen #A21094, 1:200) and Alexa488 anti-Mouse (Invitrogen #A11029, 1:200). For mtdt-3HA experiments: Cy3 anti-rat (Jackson Immuno Research #112-165-167, 1:200). Larval imaginal discs were stained in DAPI (1:100) for 10 min during one of the PBT washes after secondary antibody incubation.
Whole-animal imaging
Third instar larvae were placed on a small metal plate on top of crushed ice or on a temperature-controlled plate, and imaged by a Zeiss SteREO DiscoveryV8 microscope equipped with a GFP-470 and ds-Red filters and a Jenoptik ProgRes ® MF cool CCD camera. Monochrome images were acquired in ProgRes ®Mac Capture Pro 2.7 software and stored in *.tif format. Adult flies (3–5 d.o.) were anaesthetised on a CO2 pad and imaged as described for larvae. Images that are compared to each other were obtained under identical hardware and software settings.
Confocal imaging and image processing
Brains were imaged on a LSM 700 Zeiss confocal microscope, equipped with a LCI Plan-Neofluar 25x/0.8 Imm Korr DIC M27 water immersion objective, at 512*512 pixel resolution, with 1 μm or 2.37 μm Z-steps. Zen 2012 Release Version 8 software was used for image acquisition. Microscope settings were kept the same for the genotypes that were later compared to each other, i.e. all nsyb-QFx/(Q)UAS-mcd8-GFP brains, all nsyb-QFx/(Q)UAS-nucLacZ brains, etc.
For illustration purposes, confocal images were processed in ImageJ to collapse Z-stacks into a single image using maximum intensity projection, and to pseudo-colour different acquisition channels using a RGB Merge plugin. No other image processing was performed on the confocal data.
To quantify LacZ expression, we used a custom-written Matlab (Mathworks, MA, USA) script. The script (Fig. 1f) identified cells in the elav channel, and used the outlines of these cells as a mask to select the corresponding pixels in the lacZ channel. Then it calculated the average intensity of these pixels in the lacZ channel, and normalized it by the average intensity of initially selected elav cells. The algorithm for identifying cells was adapted from a script by Thomas Kuo and Jiyun Buyn (Center for Bioimage Informatics, also used in Ref. 3). The cells were identified for every image in a Z-stack, and the intensity measures of each image were averaged to produce one number per brain.
To quantify GFP expression (Supplementary Fig. 2), we identified pixels with above-threshold intensity in the GFP channel on each image of a z-stack. Next, we calculated the average intensity of the identified pixels, producing one number per imaged brain. Finally, we averaged the intensity measures of separate brains.
Scanning electron microscopy imaging
Heads of 3–5 day old female flies were mounted without any processing onto aluminium stubs with double stick carbon tape (Ted Pella) or Blu-Tack® (Bostik). Images were acquired at 200x magnification with a Leo 1530 Field Emission Scanning Electron Microscope operating at 1kV.
Behavioral tests
All flies used in behavioral tests were outcrossed to the same wild-type isoD1 white- background for 5 generations. Control and experimental datasets were compared using the non-parametric Kolmogorov-Smirnov test.
Phototactic behavior
Experiments were conducted in a photography dark room using overhead infrared lights for illumination. The F15T8/WW fluorescent lamp light source was switched off during control experiments. 50 male and 50 female 5 days old flies were used for each experiment. Prior to the assay, flies were kept in vials with regular fly medium at room temperature. The experimental setup was as described previously29 and consisted of 21 cell culture tubes (14 ml, BD Falcon, REF 352059), arranged in two rows of 10 and 11 tubes so that the open ends of the tubes are facing each other. For example, tube 0 is opposite of tube 0′, tube 1 is opposite of tube 1′, and so on. Flies were initially placed in tube 0 and given 2 minutes to walk towards the light source and into tube 0′. Next, tube 0′ was shifted into register with tube 1, the flies were tapped down from tube 0′ into tube 1, and again given 2 minutes to walk towards the light and into tube 1′, and so on. In total, each fly had 10 chances to walk towards the light source in the course on an experiment. The Phototaxis Index (PI) was calculated as:
where Ni is the number of flies in tube i at the end of the experiment. PI equals to the average number of times a fly walked towards the light source, with a PI=10 indicating that all flies always walked towards the light, and PI=0 meaning that no flies walked towards the light. We repeated the experiment and the lights-off control 4–7 times for each genotype. Fig. 3b represents the data as an average PI for each genotype and experimental condition; error bars show standard error of the mean.
Activity and sleep assays
For activity/sleep measurements, flies were outcrossed 5 times into iso31 background (Bloomington # 5905). Flies were entrained to a 12 hr:12 hr light:dark (LD) cycle for at least two days before being assayed. Flies were kept in glass tubes containing 5% sucrose and 2% agar and monitored using the Drosophila Activity Monitoring System (Trikinetics). Activity counts from 4–7 day old female flies were collected in 1 min bins in LD at 25 °C for 2 days. Activity/Sleep parameters were computed using MATLAB-based (MathWorks) custom software. Sleep was identified as periods of inactivity lasting at least 5 minutes. For circadian behavior measurement, activity counts were recorded in 30-min bins in constant darkness over a 6-day period and analyzed using ClockLab (Actimetrics). Period length (tau) was determined by X2 periodogram analysis, and rhythm strength was measured by relative FFT, calculated by fast Fourier transform analysis.
Olfactory behaviors in the 4-field assay
These olfactory experiments were conducted as described previously30. The experimental setup consisted of a temperature-controlled light-proof chamber (45 cm * 27 cm * 49 cm) that was equipped with 4 air inlets, CCD camera (Sony CCD IR XC-E150 with Pentax 12.5mm 1:1.4 TV lens) and two arrays of IR LEDs. The chamber was designed to accommodate a rectangular arena (23 cm* 23 cm * 3 cm), the corners of which could be connected to the four air inlets. The arena consisted of a Teflon base sandwiched between two glass plates. The bottom glass plate had a hole (D=6mm) in the middle to let out the air that was pumped into the arena from the corners. The arena was placed horizontally inside the chamber and filmed by a CCD camera from above. The video data was acquired at 30 fps, 640*480 pixels, by a custom-written GUI. Immediately after acquisition, the data was processed by custom-written Matlab scripts and stored as a *.mat file. The data structure contained information about coordinates of each detected fly at each point in time, and also about trajectories of individual flies, whenever the trajectories could be resolved unequivocally.
25 female and 25 male flies were starved for 41–43 hours before each experiment and were 5 days old when tested. The flies were transferred without anaesthesia into the 4-field arena that was immediately placed into the experimental setup and flushed with clean dry air (DA) at 0.1 l/min from each corner for 20 min. We recorded the flies’ activity for 10 mins in DA and for the following 10 mins with 5% CO2, water vapor, or 5% apple cider vinegar in water blown into one quadrant of the arena. Three other quadrants were flushed with DA at all times. Experiments were conducted in the dark, at 25°C maintained in the experimental chamber. Flies’ activities were quantified as an Attraction index (AI) calculated for the odorant quadrant. The 10 min DA recording served as a control for the odor experiment. If the flies’ activity was too low or they were distributed unevenly in the arena (|AI|>0.15) during the 10 min DA recording, this group of flies was not tested with an odorant. The AI was calculated as:
where Nodorant is the number of datapoints in the odorant quadrant during 10 min, and is the average number of data points in the other three quadrants that were always flushed with DA. Each walking fly generated 30 datapoints per second. A fly was deemed stationary if its speed was consistently below 4.5 pixel/s for 3.3 s. The datapoints from stationary flies were discarded. AI=−1 corresponds to complete repulsion from the odor quadrant, and AI=1 corresponds to complete attraction towards the odor quadrant.
Code availability
Custom-written Matlab scripts, used for quantifying confocal imaging data and for behavioral analyses, are available upon request.
Supplementary Material
Acknowledgments
We thank S. Djuranovic, A. Radhakrishnan and A. Schuller for S2 cells and training in S2 cell culturing and luciferase assays, B. Smith for help with SEM, T. Shelley for manufacturing the olfactometer setup, J. Simpson (Janelia Farm Research Campus) for a n-synaptobrevin promoter containing construct, and C.-C. Lin, B. Akitake, R. Reed and Center for Sensory Biology members for discussions and suggestions. This work was supported by grants from the Whitehall Foundation (C.J.P), US National Institutes of Health (R01DC013070, C.J.P.; R01NS079584, M.N.W., R01 DC005982, L.L.), and Howard Hughes Medical Institute (L.L.).
Footnotes
Note: Supplementary Information and Source Data files are available in the online version of the paper.
Author contributions
C.J.P and O.R. conceived the project and designed most of the experiments. S.L. and M.N.W designed the daily activity and sleep tests. O.R., E.M., S.L. and C.J.P. performed the experiments. D.L. performed embryo injections and generated most of the transgenic animals. L.L. provided reagents and suggestions. O.R. and C.J.P. wrote the manuscript with feedback from all authors.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Brand AH, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 2.Lai SL, Lee T. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 3.Potter CJ, Tasic B, Russler EV, Liang L, Luo L. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silies M, Gohl DM, Fisher YE, Freifeld L, Clark DA, Clandinin TR. Neuron. 2013;79:111–127. doi: 10.1016/j.neuron.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Garijo A, Fuchs Y, Steller H. eLife. 2013;2:e01004. doi: 10.7554/eLife.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann K, Gordon MD, Scott K. Neuron. 2013;79:754–765. doi: 10.1016/j.neuron.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei X, Potter CJ, Luo L, Shen K. Nature Methods. 2012;9:391–395. doi: 10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, Lin CC, et al. Nat Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuji G, Kenmochi Y, Takano Y, Sweigard J, Farrall L, Furusawa I, et al. Mol Microbiol. 2000;38:940–954. doi: 10.1046/j.1365-2958.2000.02181.x. [DOI] [PubMed] [Google Scholar]
- 10.Baum JA, Geever R, Giles NH. Mol Cell Biol. 1987;7:1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidalgo P, Ansari AZ, Schmidt P, Hare B, Simkovich N, Farrell S, et al. Genes Dev. 2001;15:1007–1020. doi: 10.1101/gad.873901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters KJ, Dayie KT, Reece RJ, Ptashne M, Wagner G. Nat Struct Mol Biol. 1997;4:744–750. doi: 10.1038/nsb0997-744. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis PJ, Raine ARC, Gadhavi PL, Laue ED. Nature. 1992;356:448–450. doi: 10.1038/356448a0. [DOI] [PubMed] [Google Scholar]
- 14.Marmorstein R, Carey M, Ptashne M, Harrison SC. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 15.Ma J, Ptashne M. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 16.Gill G, Ptashne M. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 17.Mondal K, Dastidar AG, Singh G, Madhusudhanan S, Gande SL, VijayRaghavan K, et al. J Mol Biol. 2007;370:939–950. doi: 10.1016/j.jmb.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 18.Wilder EL, Perrimon N. Development. 1995;121:477–488. doi: 10.1242/dev.121.2.477. [DOI] [PubMed] [Google Scholar]
- 19.Freeman M. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 20.Kramer JM, Staveley BE. Genet Mol Res. 2003;2:43–47. [PubMed] [Google Scholar]
- 21.Kelley LA, Sternberg MJE. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Bermingham-McDonogh O, Turcotte B, Guarente L. Proc Natl Acad Sci USA. 1993;90:2851–2855. doi: 10.1073/pnas.90.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zdobnov EM, Apweiler R. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 24.Pfeiffer BD, Ngo TTB, Hibbard KL, Murphy C, Jenett A, Truman JW, et al. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee T, Luo L. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 26.Sharma Y, Cheung U, Larsen EW, Eberl DF. Genesis (New York, NY : 2000) 2002;34:115–118. doi: 10.1002/gene.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeiffer BD, Jenett A, Hammonds AS, Ngo TTB, Misra S, Murphy C, et al. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu JS, Luo L. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- 29.Benzer S. Proc Natl Acad Sci USA. 1967;58:1112. doi: 10.1073/pnas.58.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao XJ, Potter CJ, Gohl DM, Silies M, Katsov AY, Clandinin TR, et al. Curr Biol. 2013;23:1163–1172. doi: 10.1016/j.cub.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.