Abstract
Autosomal dominant and/or recessive progressive external ophthalmoplegia (ad/arPEO) is associated with mtDNA mutagenesis. It can be caused by mutations in three nuclear genes, encoding the adenine nucleotide translocator 1, the mitochondrial helicase Twinkle or DNA polymerase γ (POLG). How mutations in these genes result in progressive accumulation of multiple mtDNA deletions in post- mitotic tissues is still unclear. A recent hypothesis suggested that mtDNA replication infidelity could promote slipped mispairing, thereby stimulating deletion formation. This hypothesis predicts that mtDNA of ad/arPEO patients will contain frequent mutations throughout; in fact, our analysis of muscle from ad/arPEO patients revealed an age-dependent, enhanced accumulation of point mutations in addition to deletions, but specifically in the mtDNA control region. Both deleted and non-deleted mtDNA molecules showed increased point mutation levels, as did mtDNAs of patients with a single mtDNA deletion, suggesting that point mutations do not cause multiple deletions. Deletion breakpoint analysis showed frequent breakpoints around homopolymeric runs, which could be a signature of replication stalling. Therefore, we propose replication stalling as the principal cause of deletion formation.
INTRODUCTION
Human mitochondrial DNA (mtDNA) is maintained in multiple copies in each mitochondrion, and is present in thousands of copies per cell in most human tissues. mtDNA is organized in protein–DNA complexes often referred to as nucleoids (1,2). Sporadic germline and maternally inherited mutations of mtDNA, including single deletions/duplications and point mutations, are a frequent cause of human disease (3).
A unique group of disorders is associated with mtDNA depletion (4,5) or multiple deletions of mtDNA in somatic cells, but is primarily caused by defects in nuclear genes. The nuclear background of these syndromes, autosomal dominant or recessive progessive external ophthalmoplegia (ad/arPEO) with multiple mtDNA deletions (6,7), and mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) (8,9), is starting to unravel. PEO in its mildest form presents as external eye muscle weakness, eye lid ptosis and skeletal muscle weakness. To date, mutations in three nuclear genes have been linked to this disorder. These include the muscle-, brain- and heart-specific isoform of the adenine nucleotide translocator (ANT1) (10), C10orf2 encoding the mitochondrial helicase Twinkle (11,12), and the mitochondrial DNA polymerase γ gene, POLG (13). ArPEO has been associated exclusively with compound heterozygous mutations in POLG (13,14). In addition, recessive POLG mutations are associated with more severe or complex disorders (15,16).
The most frequently reported mutation in POLG, Y955C, occurs, at the active site of its polymerase domain. In vitro analysis of the enzymatic properties of this mutant protein has led to a suggestion that reduced POLG fidelity could precipitate adPEO (17,18). However, POLG proofreading was also shown to be capable of counteracting infidelity resulting from the Y955C mutation in the polymerase motif B. In addition, the Y955C mutation resulted in a 45-fold decreased affinity for dNTPs (17), suggesting that under conditions of nucleotide limitation, replication stalling could occur. This effect might be enhanced at replication of short homopolymeric runs, frequently occurring in mtDNA.
MNGIE is caused by mutations in the gene ECGF1, encoding cytoplasmic thymidine phosphorylase (19). This protein is involved in the nucleoside/nucleotide salvage pathway, which is important for nucleotide homeostasis in post-mitotic cells and dividing cells outside the S phase. ECGF1 mutations in MNGIE are generally loss-of-function mutations resulting in enhanced levels of thymidine and deoxyuridine in, for example, plasma (20,21). Interestingly, in cultured skin fibroblasts, this appears to result in site-specific point mutations in mtDNA mostly affecting residues preceding runs of Ts in the de novo synthesized strand (21). Most mutations could be explained by a next nucleotide effect and a slippage mechanism in homopolymeric A–T runs. mtDNA deletions are usually not observed in cultured skin fibroblasts, but are prominent in skeletal muscle of MNGIE and PEO patients. Conversely, site-specific point mutations were demonstrated at very low levels in skeletal muscle of the MNGIE patients, while being prominent in fibroblasts (21). Recent deletion mapping of mtDNA of a MNGIE patient showed both perfect and imperfect direct repeats flanking some of the more frequent deletions (22). Microdeletions within the longer imperfect direct repeats were also observed. The data were considered to be most compatible with a homologous recombination mechanism, because that could possibly explain features, such as the microdeletions, via branch migration.
In order to study how multiple mtDNA deletions are provoked in ad/arPEO, we sequenced regions of mtDNA of PEO patients with mutations in either POLG or Twinkle, and related the findings to the pathogenic mechanism. The results do not support a generalized decrease in mtDNA replication fidelity in either POLG or Twinkle patients. Rather, deletion breakpoint mapping suggests stalling at regions of difficulty for DNA polymerases, such as homopolymeric runs and microsatellite-type repeats, and the additional involvement of the replication fork barrier at bp 16 070, as a primary cause of mtDNA deletion formation.
MATERIALS AND METHODS
Patient samples
Table 1 describes the patient material used in this study. PEO-affected individuals 1, 2, 6 and 7 and sample preparation were described previously (13,23). Patients 3–6 were described elsewhere (24). Controls Δ1–5 are sporadic patients affected by PEO (Δ1–4) or mild Kearns Sayre syndrome (KSS) (Δ5) but carrying a single mtDNA deletion; all patients were adults at the time of biopsy except for patient Δ5 who was 5 years of age. Healthy controls 1–6 are unaffected relatives of PEO patients 3–5 (Fig. 1A), controls 1–5 being brothers and sisters, thus sharing the same mitochondrial background, and control 6 being a distant paternal relative. Healthy controls, 7–10 are all direct relatives of patients 7–11 all sharing mtDNA transmitted through the same maternal line (Fig. 1B). External eye muscle from patient 6 has been treated as skeletal muscle in the statistical analysis comparing the healthy controls with PEO patients. All the samples have been taken in accordance with the Declaration of Helsinki and with informed consent.
Table 1. Patient description and materials.
| Patient | Mutation | Tissue (mtDNA) | Age | Reference |
|---|---|---|---|---|
| 1 | POLG R3P/A467T | Skeletal muscle | 60 | (13) |
| 2 | POLG Y955C | Skeletal muscle | 48 | (13) |
| 3 | POLG Y955C | Skeletal muscle | 39 | (24) |
| 4a | POLG N468D/A1105T | Skeletal muscle | 50 | (24) |
| 5a | POLG N468D/A1105T | Skeletal muscle | 44 | |
| 6a | POLG N468D/A1105T | Skeletal muscle | 34 | |
| 7a | 352–364 AA duplication Twinkle | Extraocular eye muscle | 73 | (23) |
| 8a | 352–364 AA duplication Twinkle | Skeletal muscle | 60 | |
| 8a | 352–364 AA duplication Twinkle | Frontal cortex | 60 | |
| 9a | 352–364 AA duplication Twinkle | Skeletal muscle | 33 | |
| 10a | 352–364 AA duplication Twinkle | Skeletal muscle | 31 | |
| 11a | 352–364 AA duplication Twinkle | Skeletal muscle | 20 | |
| 1L | POLG R3P/A467T | Blood lymphocytes | 60 | (13) |
| 2L | POLG Y955C | Blood lymphocytes | 48 | (13) |
| Controls | ||||
| Δ1 | Single mtDNA deletion | Skeletal muscle | 60 | |
| Δ2 | Single mtDNA deletion | Skeletal muscle | 48 | |
| Δ3 | Single mtDNA deletion | Skeletal muscle | 35 | |
| Δ4 | Single mtDNA deletion | Skeletal muscle | 40 | |
| Δ5 | Single mtDNA deletion | Skeletal muscle | 5 | |
| 1a | None | Skeletal muscle | 32 | |
| 2a | None | Skeletal muscle | 36 | |
| 3a | None | Skeletal muscle | 51 | |
| 4a | None | Skeletal muscle | 53 | |
| 5a | None | Skeletal muscle | 55 | |
| 6b | None | Skeletal muscle | 72 | |
| 7a | None | Skeletal muscle | 32 | |
| 8a | None | Skeletal muscle | 55 | |
| 9a | None | Skeletal muscle | 62 | |
| 10a | None | Skeletal muscle | 66 |
Age of patients refers to the age at the time of the biopsy.
aPatients and controls indicated in Figure 1.
bControl 6 is a distant paternal relative of the POLG N468D/A1105T family members. Patients 1–3 and all single deletion cases are unrelated.
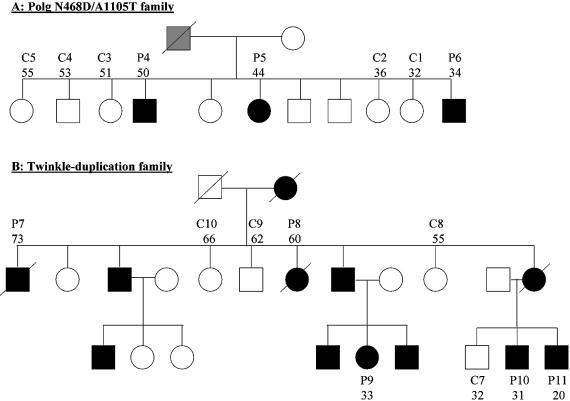
Figure 1.
Pedigrees of PEO families. (A) POLG N468D/A1105T pedigree including patients 3–5 and controls 1–5 (see also Table 1). Black symbols indicate affected family members, white symbols unaffected ones, and the gray symbol indicates unknown disease status of a subject. A slash over a symbol denotes deceased individuals. (B) Twinkle dupAA352–364 pedigree including patients 7–11 and controls 7–10. Numbers below patient numbers indicate age at the time of biopsy.
PCR amplification, cloning and sequencing
Parts of the mtDNA control region and cytochrome b gene (cyt b) were PCR amplified using the primers shown in Table 2a, yielding a 576 bp (control region) or a 654 bp (cyt b) fragment. Reactions were performed in a 50 µl volume containing 100 ng of total genomic DNA template, 1.5 U of Pfu DNA polymerase (Promega), 250 µM dNTPs, 10 µM of each primer, and the buffer supplied by the manufacturer. The amplification conditions were as follows: initial denaturation of 2 min at 95°C; 21 cycles of 30 s at 95°C, 1 min at 57°C and 2 min at 72°C; final extension step of 10 min at 72°C. mtDNA deletions were PCR amplified using various primer pairs shown in Table 2b, yielding multiple fragments according to the sizes of the deletions. Reactions were as above but with 5% dimethylsulfoxide, 20 µM of each primer, and the buffer supplied by the manufacturer. The amplification conditions were: initial denaturation of 2 min at 93°C; 29 cycles of 30 s at 93°C, 1 min at 57°C and 7 min at 72°C; final extension step of 15 min at 72°C. Following PCR amplification, control region and cyt b fragments as well as the deletion-containing PCR products were cloned using the TOPO Zero-Blunt PCR cloning kit (Invitrogen). Plasmids from individual bacterial colonies were purified using the Macherey–Nagel Nucleospin Robot-96 plasmid kit on a Tecan multipipetting robot, and verified for the presence of inserted fragments by restriction digestion and agarose gel electrophoresis. The plasmids carrying the inserts were sequenced on an ABI Prism 3100 DNA sequencer, using BigDye terminator chemistry (Applied Biosystems) and M13 forward and reverse oligonucleotides as well as mtDNA-specific oligonucleotides. Sequences were analyzed using the SeqMan™II program of DNASTAR. Differences from the Cambridge reference sequence (25) present in all clones, and for patients 1 and 2 present in leukocyte mtDNA, were discarded as polymorphisms. In addition, the control region homopolymeric D310 tract was deliberately excluded from our analysis since it is highly polymorphic (see http://www.mitomap.org/). Rare 1–2 bp insertions or deletions are included with point mutation counting, but the small duplications identified in patients 1, 3 and 4 were not. Repeated PCR cloning and sequencing of one of the samples showed good reproducibility. Full-length and deleted mtDNA sequence features such as base composition, nucleotide word counts, palindrome and direct repeat searches were performed with the Java interface (J)EMBOSS package (26) at http://www.hgmp.mrc.ac.uk/Software/EMBOSS/Jemboss/.
Table 2. PCR primers for mtDNA amplification.
| Sequence 5′→3′ | 5′ start | Location | |
|---|---|---|---|
| (a) Short oligonucleotide primers for fragment amplification of the mtDNA control region and cyt b used for sequencing | |||
| 35L | 5′-GGAGCTCTCCATGCATTTGG-3′ | 35 | Control region D-loop |
| 611H | 5′-CAGTGTATTGCTTTGAGGAGG-3′ | 611 | Control region D-loop |
| 14682L | 5′-CACGGACTACAACCACGACC-3′ | 14 682 | Cyt b |
| 15516H | 5′-GTATAATTGTCTGGGTCGCCTAGG-3′ | 15 516 | Cyt b |
| (b) Primers used for amplification of multiple mtDNA deletions | |||
| FR31 | 5′-CTTCCCACAACACTTTCTCGGCCTA-3′ | 7177 | COX I |
| 611H/l | 5′-CAGTGTATTGCTTTGAGGAGGTAAGCTACATAA-3′ | 611 | Control region D-loop |
| 15996H | 5′-GCTTTGGGTGCTAATGGTGG-3′ | 15 996 | tRNAPro |
| 15516H | 5′-GTATAATTGTCTGGGTCGCCTAGG-3′ | 15 516 | Cyt b |
| 14338H | 5′-GGTGGTTGTGGTAAACTTTAA-3′ | 14 338 | ND 6 |
Numbers refer to the Cambridge reference sequence (25). L and H refer to the strand of the sequence, not to the complementary strand.
RESULTS
High mutation levels in the control region of muscle mtDNA from PEO patients
To determine the point mutation levels of mtDNA from ad/arPEO patients with multiple deletions, we sequenced mtDNA from PEO patients’ tissues, healthy controls and patients with single sporadic deletions, followed by cloning and sequencing of individual plasmids, and comparison with the Cambridge reference sequence to score mutations. It is important to note here that all of the healthy controls except control 6 were close relatives of the patients 3–5 and 7–11 (see Fig. 1 and Table 1 for family pedigrees and additional patient information). Therefore, their maternally inherited mtDNA was similar, and the possible differences in mtDNA seen between controls and patients are due to the inherited nuclear factors.
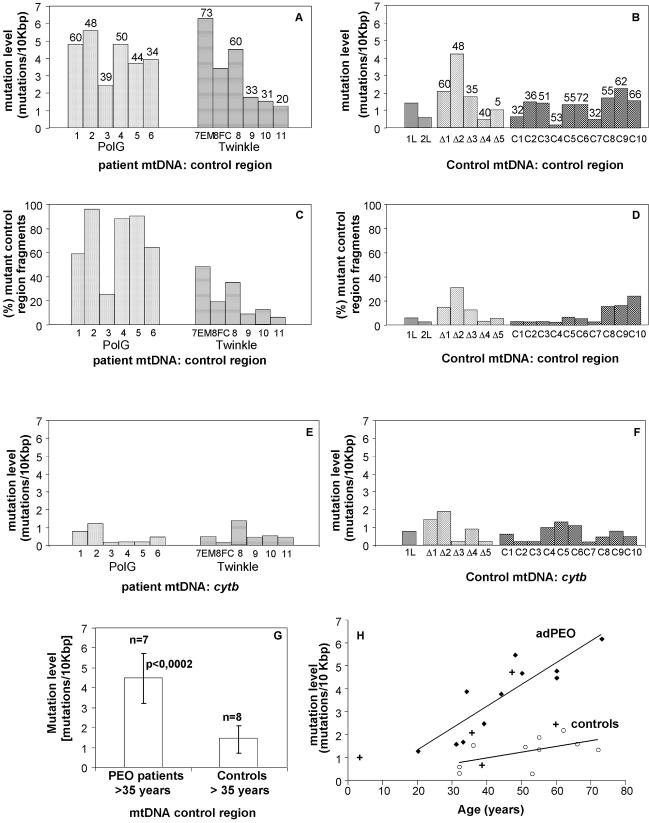
We analyzed two regions of mtDNA, a fragment of the control region that includes part of the D-loop [nucleotides 35–611 of the Cambridge reference sequence (25)] and a fragment of the cyt b gene (nucleotide 14 682–15 516). The mutation level was defined as the number of different mutations per 10 kb of sequenced cloned fragments of the control region. This is a measure of sequence variability, but not necessarily a strict measure of mutation burden since repeated clones with an identical mutation are not counted separately (see also below). The Twinkle/PEO patient samples included several relatively young individuals, one of whom, patient 11 (Fig. 1), had not yet presented disease symptoms at the age of biopsy. The biopsy DNA sample had, however, shown weak multiple mtDNA deletions by Southern blot and clear deletions by long-range PCR (23). The youngest patients had the lowest levels of control region point mutation of the patient samples, with levels being similar to those of elderly controls (Fig. 1). With increasing age (≥35 years), control region mtDNA of PEO patients showed a significantly higher mutation level (P < 0.0002) than that of the control samples (>35 years) (Fig. 2A, B and G). The mutation level in patients ranged from 1.6 (patient 11) to 6.3 (patient 7) mutations/10 kb, whereas in the 10 healthy control individuals of varying ages, the level was lower than 2.2 mutations/10 kb (controls 1–10), closer to the PCR-induced mutation background for Pfu polymerase (27). A weak tendency to higher mutation levels with increasing age was observed in the controls (Fig. 2H), while the ad/arPEO patients showed a strong increase of control region mutations during aging (Fig. 2H). Three additional controls, from patients with a sporadic single mtDNA deletion, showed mutation levels of 4.3, 2.1 and 1.7 mutations/10 kb, which is higher than the levels in controls of a similar age (Fig. 2H). Point mutation levels in leukocyte DNA of two PEO patients (1L and 2L) were much lower than in muscle DNA, comparable with controls.
Figure 2.
Point mutation levels in the control and cyt b region of PEO patient muscle mtDNA. (A and B) Number of different mutations detected per 10 kb of control region mtDNA in PEO patients (A) and in different types of controls (B). Numbers above the bars indicate the age at the time of biopsy. (C and D) Number of cloned mtDNA control region fragments carrying at least one point mutation in PEO patients (C) and in controls (D), expressed as a percentage of the total number of clones that were sequenced. (E and F) Number of different mutations detected per 10 kb of cyt b gene sequence in PEO patients (E) and in controls (F). Patients and abbreviations: 1–6, POLG/PEO patients, skeletal muscle; 7EM, Twinkle/PEO patient, extraocular muscle; 8FC, Twinkle/PEO patient, frontal cortex; 1L, 2L, POLG/PEO patients 1 and 2, leukocytes; Δ1–Δ5, patients with sporadic single mtDNA deletions, skeletal muscle; C1–C10, healthy control samples, skeletal muscle. (G) Number of different control region mutations in muscle mtDNAs of PEO patients >35 years old grouped together, and in the >35-year-old controls. The probability (P) of both groups being identical was calculated using the unpaired Student’s t-test. The indicated results show that mutation level differences between both groups are highly significant. (H) Mitochondrial control region mutation levels in PEO patients (filled diamonds), healthy controls (open circles) and single deletion controls (+), related to the age at the time of biopsy, based on the values also indicated in (A) and (B). Lines for the PEO patients and healthy controls indicate trend lines.
Analysis of the sequence data revealed that mutations found in the control region were not randomly distributed and recurred in several clones from the same patient, suggesting that a considerable proportion of mtDNAs carried these mutations (Fig. 3). The amount of mutant A189G was verified by Genescan analysis and found to be in good agreement with the levels based on the frequency of the mutation in the cloned fragments (data not shown). The total mutation load was calculated by counting the total number of all clones of a sample with one or more mutations. This is a direct measure of the percentage of the total mtDNA population with one or more point mutations (Fig. 2C and D). This analysis of total mutant load showed that in the tissues of PEO patients, 5% (patient 11) to 96% (patient 2) of mtDNA carried control region mutations (Fig. 2C), whereas in the controls this load was <7% for most samples (Fig. 2D), except for three of the oldest individuals who showed a mutant load of between 18 and 25%. Similar to the mutation level analysis, total mutant load in PEO patients was always higher than that of controls of a similar age. Two of five single deletion patients also showed a higher percentage of mutant fragments than did controls of a similar age. mtDNA from leukocytes of POLG-PEO patients 1 and 2 showed similar values to control muscle mtDNA (2L, 3%; 1L, 6%).
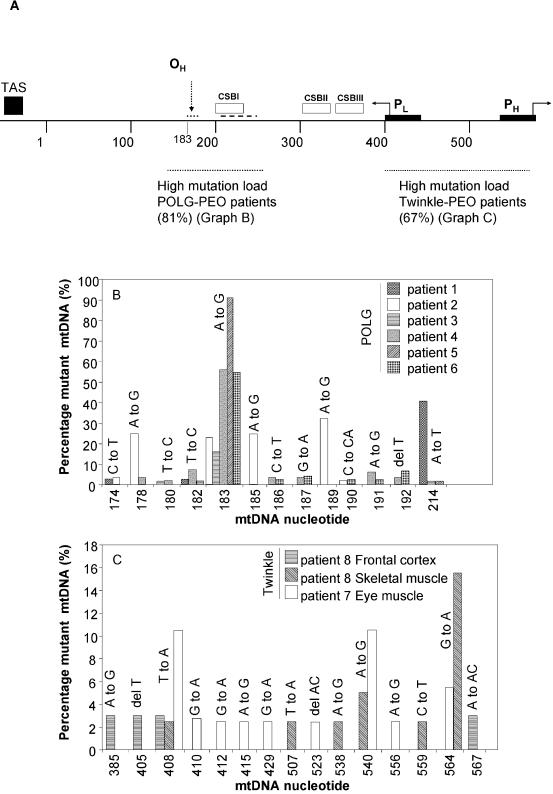
Figure 3.
Non-random distribution and heteroplasmy levels of control region mutations. (A) Diagram of the mtDNA control region and mutational hotspots. Abbreviations and symbols: OH, heavy strand replication origin; PH and PL, H and L strand promoters; TAS, termination-associated sequence; CSB I, II and III, conserved sequence blocks I, II and III; bold dotted lines, the regions where most mutations were found in POLG/PEO and Twinkle/PEO patients’ muscle mtDNA; percentages in parentheses, the percentage of mutated fragments with mutations in the indicated region; small dotted line adjacent to OH, A to G conversion mutational hotspot region of patient 2; dashed line, position of the duplication hotspot in patient 1. (B) Pattern of mutations in the mtDNA replication control region of POLG/PEO patients between nucleotides 174 and 214. (C) Mutation pattern in the mtDNA transcription control region of Twinkle patients 7 and 8. The percentage of mutant mtDNA in (B) and (C) indicates the number of clones with the indicated mutation in the indicated patients relative to the total number of clones sequenced for each patient.
Comparable low mutation levels in the cyt b region in muscle mtDNA from PEO patients and controls
Contrary to the mtDNA control region, no statistically significant differences were observed in mutation levels between patients and controls in the cyt b region (Fig. 2E and F). The mutation levels in patients ranged from 0.2 (patient 3) to 1.4 (patient 8) mutation/10 kb. In skeletal muscle from the 10 healthy individuals and lymphocyte mtDNA from patient 1, the mutation levels were also within this range. Total mutation loads revealed no difference between PEO patients and controls (data not shown). Partial analyses of other mtDNA regions, COX I (7203–7445) and HVR1 (16 000– 16 569), showed levels in PEO patients comparable with those observed for the cyt b region (not shown). Similar results to those in skeletal muscle were obtained in mtDNA control and cyt b regions in brain and extraocular muscle of one of the Twinkle/PEO patients (see also Fig. 2). However, no appropriate controls were available for these two tissues.
Distribution of heteroplasmic control region mutations
The heteroplasmic control region mutations found in the patients were non-randomly distributed (Fig. 3). In POLG/PEO patients, the majority of mtDNA mutations (86%) accumulated in the region from nucleotide 150 to 250, particularly close to the presumptive heavy strand replication origin OH and in conserved sequence block I (CSB I). Specific mutation hotspots at the area could be identified (Fig. 3B). Most striking were the mutations found in PEO patients 1 and 2, carrying the recessive POLG R3P and A467T mutations in the compound heterozygous state, and the dominant Y955C POLG mutation, respectively. Patient 1 had several duplications (2–32 bp, Supplementary fig. 1B available at NAR Online), partly overlapping with CSB I. Patients 3 and 4 each also showed one clone with a small duplication (Supplementary fig. 1B). Patient 2 presented an A to G transition ‘hotspot’ close to OH at nucleotides 178, 183 and 189 (∼20% heteroplasmy). In addition, based on the leukocyte mtDNA sequence, patient 2 had the less common 185A polymorphism. This polymorphic nucleotide 185 also appeared as an A to G transition hotspot in this patient. Although several clones showed two of these transitions, most clones contained only one, sometimes in combination with an additional point mutation. The unrelated patient 3, with the same Y955C mutation in POLG, only carried the mtDNA A183G mutation at a high percentage. This change was also detected at levels >15% of total mtDNA in POLG/PEO patients 3, 4, 5 and 6, but was not observed in Twinkle/PEO patients’ tissues or controls, nor is it a recognized polymorphism or disease-associated mutation (http://www.mitomap.org/). An aging-associated nucleotide change, A189G (28), was detected at high levels in POLG patient 2 and in one of about 50 clones from each of the two elderly Twinkle patients’ samples, but not in any other POLG/PEO patients or the younger Twinkle/PEO patients. This mutation was also frequently found in the control samples, where it represented 53% of all mutated clones of these samples. The T408A mutation represented 16% of all mutated clones in the controls.
The distribution of control region point mutations in the Twinkle/PEO patients (patients 7–11) differed from POLG/PEO patients, since the majority of the Twinkle-associated mutations (67%) were located between nucleotides 385 and 570. In the two elderly Twinkle patients 7 and 8, who showed the highest point mutation levels, this value was 87%. For samples 7EM and 8, of the total number of clones sequenced, 42 and 31% had mutations between 385 and 570. For the POLG patients, these values were typically around 5%, with the exception of patient 1 who had 24% of all clones with a mutation between 385 and 570. Individual point mutations were generally present at low levels (note the difference in scale between Fig. 3B and C), but the number of different mutations, at least for the oldest Twinkle patients, was nevertheless comparable with POLG/PEO patient mutation levels (compare Fig. 2A and C). The 564 G to A transition mutation, which was seen in the highest percentage of clones in patients 7 and 8, was detected in all Twinkle/PEO patients, including patients 9–11, but only in the POLG patient 1 and one of the elderly controls. The above data are not skewed by large differences in the number of sequenced clones or differences in total number of bases that were sequenced: from each sample for each region, approximately 50 clones were sequenced resulting in ∼30 kb of sequence, except for a young Twinkle/PEO patient 9, of whom we sequenced ∼150 kb. Because point mutations in the samples from the young Twinkle patients are rare and mutation levels are only marginally higher than those of controls of a similar age, the analysis of regional variation could not be done with confidence. Nevertheless, the analysis of patient 9 showed that she had already had several of the same mutations observed in her older relatives with PEO. Since her maternally inherited mtDNA came from her unaffected mother, this suggests that the occurrence of these mutations at young age are associated with the nuclear disease allele. The sporadic patients with a single mtDNA deletion (Δ1–5) showed a distribution of control region point mutations similar to the Twinkle/PEO patients.
Comparison of mutation load in deleted and non-deleted mtDNA molecules
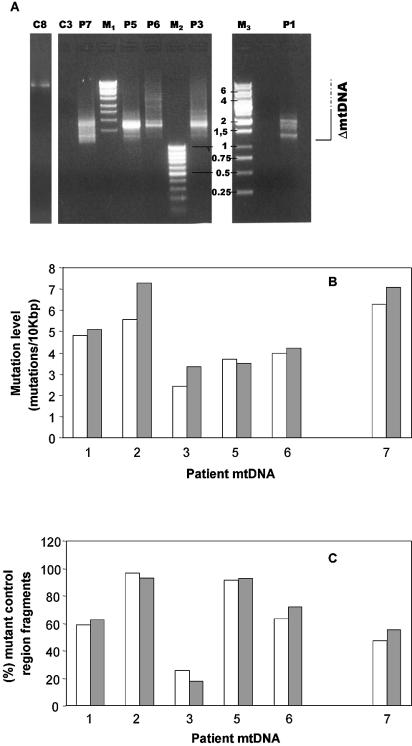
The low mutation load in the cyt b gene suggested close to normal mutant POLG fidelity (see Discussion). To study whether control region point mutations predispose the molecules to deletion formation, we specifically amplified deleted mtDNA molecules including the control region. We compared control region mutation levels in these deleted molecules with the levels in the total population of mtDNA molecules obtained by the amplification and sequencing of just the control region, as already shown in Figure 2. The outermost PCR primers used (Table 2b) create an ∼10 kb fragment using wild-type mtDNA and the appropriate PCR conditions (not shown). However, a short extension time was used in the PCR to promote preferential amplification of deleted mtDNA molecules. In all samples, a set of products generally shorter than 4 kb was amplified (Fig. 4A), cloned and the control region sequenced. Figure 4B and C shows the mutation levels and total mutational loads in deleted molecules compared with the values for the total population of mtDNA molecules (as shown in Fig. 2) for patients 1–3, 5, 6 (POLG/PEO) and 7 (Twinkle/PEO). No significant POLG- or Twinkle-specific differences were observed.
Figure 4.
Detection of multiple mtDNA deletions in PEO patients by long-range Pfu PCR and comparison of control region mutation levels between the deleted and the total mtDNA population. (A) DNA fragments amplified from muscle mtDNA of patients 1, 3, 5, 6 and 7 and control muscle mtDNA were separated on a 1% agarose gel and stained with ethidium bromide. Abbreviations: M1, M2 and M3, DNA size markers indicated in kb; C3 and C8, controls 3 and 8. Deletion PCR was performed as described in Materials and Methods using primers FR31 and 611H/l (Table 2b), amplifying the mtDNA region between nt 7177 and 611. Note that the PCR from C8 did show a faint band corresponding to the full-length mtDNA fragment. (B) The number of point mutations in the control region detected in deleted mtDNA molecules (gray bars) and in the total population of mtDNA molecules as determined in Figure 1 (white bars) of patients 1–3 and 5–7. (C) The number of mtDNA fragments carrying a mutation in skeletal muscle mtDNA of patients 1–3 and 5–7.
Mapping of multiple mtDNA deletions
Another prediction of strand slippage, resulting from decreased replication fidelity, is the occurrence of direct repeats flanking the deletions. In addition, with the current knowledge of the patients’ nuclear gene mutations, we asked whether different mutations in one gene such as POLG or mutations in different genes, such as POLG and C10orf2 (Twinkle), result in systematic differences in deletion breakpoints, possibly pointing to different mechanism(s) of deletion formation. Finally, we questioned the hypothetical relationship between particular control region point mutations and specific mtDNA deletions. For these purposes, we mapped the breakpoints of mtDNA deletions from the cloned fragments of deleted molecules that were also used to determine the mutation levels in the control region of the deleted molecules (see Table 3 and Supplementary fig. 1). Many breakpoints were located in the mitochondrial genes from cyt b to COX I. Using the outermost primer pair (611H/l-FR31), which also amplified the control region, a major 3′ (L-strand orientation) deletion breakpoint was identified around bp 16 070 at or near the ‘termination-associated sequence’ (TAS). This breakpoint was identified in the first patients identified with adPEO (6), most of whom carried the POLG Y955C mutation (14). Our results indicate that bp 16 070 is a persistent breakpoint in both POLG and Twinkle/PEO patients. The breakpoint is rarely observed in sporadic cases with single mtDNA deletions, but is frequently encountered in so-called ‘sublimons’ from control tissues (29). However, with the PCR primers used here, these molecules are probably over-represented since they are short and preferentially amplified in PCR. In addition, the ligation–cloning procedure used here strongly favors the shortest PCR products, thus enriching for the largest mtDNA deletions in the derived bacterial clones. In fact, the shortest fragments visible in our deletion PCR (Fig. 4A) from ∼1200–1300 bp very probably represent those molecules with breakpoints at around 16 070 at the 3′ end and close to 7177 at the 5′ end. To avoid the strong prevalence of the 16 070 breakpoint, we used several primer pairs for more extensive mapping of deletions from patient 1. In this patient, breakpoints of deletions in different regions of the mitochondrial genome showed the same general features. We detected short or imperfect direct repeats at most breakpoints (Table 3 and Supplementary fig. 1A) and occasional imperfect palindromic sequences (data not shown). Most notably, frequent short homopolymeric runs of four or more nucleotides were seen close to the breakpoint. Several breakpoints showed microsatellite-like sequences (Supplementary fig. 1A). Allowing one different nucleotide in a run of six nucleotides, 111 out of a total of 142 deletions had runs within 10 nucleotides of the breakpoint at one or both ends. We analyzed the occurrence of homopolymeric runs at the ‘deletion’ region (bp 7177– 16 100), as defined by the region we amplified with the outermost PCR primer pair and assuming no deletion breakpoints occur beyond the 16 070 region. Over the entire length of the 7177–16 100 region, 816 bp are in randomly distributed perfect homopolymeric runs of four or more nucleotides. Based on this, the predicted frequency of randomly occurring deletions within or at the precise boundary of a homopolymeric run is 1:9.0, assuming that a run of four nucleotides has five positions at which the break can occur. Analysis of the deletion boundaries of the patient samples showed that 60 of 284 3′ or 5′ breakpoints are at the exact end or within a perfect homopolymeric run of four or more nucleotides, indicating a frequency of 1:4.8. When excluding all the boundaries located at the PEO deletion hotspot area around mtDNA bp 16 070 from this analysis, 60 out of 186 breakpoints are within or precisely flanked by a homopolymeric run, giving a frequency of 1:3.1, which is 3-fold higher than predicted by random deletion occurrence. The bias towards deletions in the vicinity of homopolymeric runs would have been even more pronounced had we included imperfect but long homopolymeric runs, such as the cgccctccc at nt 7815 (28 of 142 5′ breakpoints). The most frequently encountered 5′ breakpoints around nt 7400 (30 of 142 5′ breakpoints) included in the calculation occurred within a sequence stretch ccccccaccc. This is a perfect 6 nt homopolymeric run and a 9 out of 10 nt imperfect run. No association of particular control region point mutations with any specific mtDNA deletions were detected (data not shown). No obvious differences were observed in types of deletion breakpoints between Twinkle/PEO and POLG/PEO patients (Supplementary fig. 1). Finally, no point mutations were observed in the deletion boundaries of any of the 142 deletions we sequenced (data not shown).
Table 3. Statistics of deletion mapping.
| Deletion PCR | Mapped deletions | Flanking repeat length | Frequent breakpoint elements | ||||
|---|---|---|---|---|---|---|---|
| >5 bp | >6 bp | >7 bp | (3′) 16 070 | (5′) 7400 ccccccaccc | (5′) 7815 cgccctccc | ||
| FR31–611H/l (7177–611) | 105 | 29 | 0 | 0 | 98 | 20 | 28 |
| FR31–15 996H (7177–15 996) | 10 | 5 | 0 | 0 | NA | 0 | 0 |
| FR31–15 516H (7177–15 516) | 14 | 2 | 2 | 0 | NA | 4 | 0 |
| FR31–14 338H (7177–14 338) | 13 | 1 | 1 | 0 | NA | 6 | 0 |
| Total | 142 | 37 | 3 | 0 | 30 | 28 | |
Multiple mtDNA deletions were amplified using the indicated primer pairs, yielding PCR products of varying length, as shown for example in Figure 4A. The PCRs were subsequently used for direct bacterial cloning, with random clones picked for sequence analysis (see Materials and Methods for details). Primer pair FR31–611H/l was used on DNA from patients 1–3 and 5–7, while the other primer pairs were only used on DNA from patient 1. Detailed mapping of the deletions is shown in Supplementary figure 1. The table shows the absence of long flanking direct repeats in this series of deletions, while the most frequent 5′ or 3′ breakpoint elements are indicated (see Results and Discussion for more details).
DISCUSSION
Here, we catalog mtDNA point mutations accumulated in PEO patients’ post-mitotic tissues because of POLG and Twinkle defects. Our results strongly suggest that the mutation mechanism in the two pathological conditions is closely related but is not caused by generalized replication infidelity as previously suggested. Instead, we present evidence for the hypothesis that multiple deletion formation in PEO is initiated by frequent replication stalling.
A recent study (17) showed that in vitro, the Y955C POLG mutation affected the accuracy of mtDNA synthesis, resulting in enhanced point mutation levels caused by misinsertion. By analogy with mutations in the synonymous amino acid in the Klenow fragment of Escherichia coli DNA polymerase I, POLG Y955 is an important residue for base discrimination and binding [(18) and references therein]. Misinsertions following direct repeats were suggested to promote slipped mispairing and hence deletion formation. On the other hand, the same study showed that the enhanced misinsertion rate was counteracted by the proofreading capabilities of the POLG exonuclease domain. In order to assess the fidelity of POLG in PEO in vivo, we established the point mutation levels in the tissues of the patients carrying either POLG or Twinkle mutations. In addition, we mapped the breakpoints of 142 ad/arPEO-associated mtDNA deletions in order to analyze: (i) the occurrence and types of direct repeats; (ii) a possible correlation between point mutation and deletion occurrence; and (iii) a possible correlation between disease genotype and mtDNA genotypes. The results lead to the following conclusions: (i) POLG and Twinkle PEO patients, as well as three out of five patients with single sporadic mtDNA deletions, show increased levels of point mutations in the mtDNA control region compared with controls of a similar age, but not in the cyt b-encoding gene region, strongly suggesting that the PEO patients do not have a generalized decrease in replication/repair accuracy. (ii) The number and types of point mutations in deleted molecules do not differ from the number and type in the total population of mtDNA molecules, arguing against the idea that point mutations predispose to deletions. (iii) Deletion breakpoint mapping in skeletal muscle showed no obvious differences between POLG/PEO and Twinkle/PEO patients, suggesting a similar mechanism of deletion formation. (iv) Analysis of deletion-flanking sequences does not show prominent features suggestive of a strand slippage mechanism, such as precisely flanked long (8–13 nt) direct repeats. Instead, the occurrence of frequent deletion breakpoints at known problem sites for DNA polymerases, most notably homopolymeric runs, suggests that deletions in ad/arPEO might arise following replication stalling.
Increased mutational load in the mtDNA control region but not in the cyt b region of ad/arPEO skeletal muscle
Dominant mutations in POLG cluster in and around the polymerase motif B, while the recessive mutations are mostly not in recognized functional domains, although they are generally closer to the exonuclease domains (13,14,30,31). In vitro studies on the consequences of the polymerase B domain Y955C mutation suggest that POLG mutations in PEO generally cause decreased accuracy of the POLG protein during replication and/or repair, resulting in an increased nucleotide misinsertion rate leading to point mutations (17). Furthermore, our previous cell culture model, which over-expressed a mutant POLG lacking proofreading activity [(27); S. Horttanainen, S. Wanrooij, J. Kurkela, M. Jokela, H. T. Jacobs and J. N. Spelbrink, unpublished data), predicts that reduced POLG fidelity causes misinsertion mutations randomly throughout the mitochondrial genome. This is now also confirmed for a physiologically more relevant model system, the (mtDNA mutator) mouse (32). However, our present data show that in the muscle of the POLG/PEO patients, this is not the case: point mutations accumulate in the control region, but not in the coding region. No evidence on other pathological mutations of mtDNA support selection against the mutant mtDNA in post-mitotic tissues, such as muscle, whereas in leukocytes that may be the case (33). Post-mitotic tissues are usually quite resistant to mutant mtDNA, and become affected only when high proportions of mutant mtDNA are reached (3). mtDNA mutator mice indeed do not show any evidence of rapid selection against coding region mutations in the post-mitotic tissues brain, heart and liver (32). On the contrary, point mutation levels at 2 or 6 months of age are ∼2-fold higher in the cytochrome b gene region compared with the control region. Nucleotide pool imbalance has been implied to predispose to replication errors in the MNGIE syndrome. In this case, point mutations in the coding regions were shown to occur in a wide variety of tissues (21). Thus, the present findings of enhanced point mutation levels in the mtDNA control region, but not in the cyt b gene or other mtDNA regions, refute the idea of increased replication infidelity in ad/arPEO.
The increased level of control region mutations begs the question, cause or effect? If control region point mutations did predispose deletion formation, then they should occur more frequently in deleted molecules. This was not the case; all mtDNA, deleted and full length, showed an increased control region mutation burden. That enhanced levels of control region mutations were also detected in patients with a sporadic single deletion of mtDNA suggests that the mere presence of deleted mtDNA in a cell can induce mutagenesis, without the need for a nuclear gene mutation. The observed low point mutation levels in the control region of young Twinkle/PEO patients, who already had multiple mtDNA deletions and showed disease symptoms, further supports this hypothesis.
A further argument against replication infidelity as a cause of increased point mutation levels is the non-random distribution of mutations within the control region, and the observation that several of the mutations we have detected have also been described to accumulate with increasing age [see, for example, Wang et al. (28), Michikawa et al. (34) and Chomyn and Attardi (35)]. The POLG/PEO patients accumulate mutations around the heavy strand replication origin (OH) and CSB I (bp 150–250), whereas Twinkle/PEO patients showed more mutation accumulation in the region containing the L-strand and major H-strand promoter, which are important for transcription initiation (36). However, since the Twinkle/PEO patients studied here were related, other Twinkle/PEO families should be studied to confirm this difference in the mutation distribution pattern from POLG/PEO patients.
OH and CSB I are important replication initiation sites in the strand-asynchronous model of mtDNA replication. Although this model is now hotly debated (37,38), both OH and CSB I are conserved features of mammalian mtDNA, suggesting that they do play a regulatory role. OH and CSB I are also sites of DNA–protein interactions (39). The occurrence and frequency of point mutations in this region suggest that some of these mutated molecules could have a replicative advantage, as previously proposed for the C150T mutation and possibly other mutations that accumulate during aging (40), or are mutational hotspots (41). This might explain the high proportions of some of these mutations.
Two POLG/PEO patients showed either an A to G transition hotspot (patient 2, Y955C) or a duplication mutation hotspot (patient 1, R3P/A467T). The absence of such clear mutation hotspots in the other samples could suggest that the hotspots are related to the specific mutations found in POLG. This was, however, not supported by the fact that two unrelated patients with the Y955C mutation did not share a hotspot. A lower number of individual mutations in patient 3, as well as a considerably lower point mutation load could reflect a difference in lifestyle, genetic background or age.
The A189G mutation was previously shown to accumulate with increasing age (28). In our material, it was usually present at low levels, with the exception of patient 2, who showed multiple A to G transitions near OH including A189G. In all our other cases, the percentage of mtDNA with A189G was not particularly high, comparable with age-related reported percentages (28). This finding is somewhat different from a report in which patients with multiple mtDNA deletions showed a higher risk for the A189G transition at ages <53 years (42). The novel A183G mutation was recently also identified in three out of eight POLG patients, two of them with a high percentage of mtDNA showing this change (43). In our samples, it was present at 15% or more of total mtDNA in all but one POLG/PEO patient. The absence of this mutation in all controls and in Twinkle/PEO patients suggests that this nucleotide is normally not mutation prone. Although close to OH, the function of the A183 nucleotide has, to our knowledge, not been established.
In a recent paper, Del Bo and co-workers also used a sequencing strategy for mutation analysis in PEO patients (43). In sharp contrast to our results, they failed to detect significant differences between PEO patients carrying mutations in the POLG polymerase motifs or in Twinkle, and control samples. However, in their study, the mutation load in controls with an average age of 45 years was 30%. In our study, it was just 8.2% in controls with an average age of 51 years, three elderly controls contributing significantly to this percentage. The difference probably reflects their use of an error-prone Taq polymerase and high PCR cycle number, which probably has masked this finding [see also Chinnery (44)]. Although our data oppose the idea of replication infidelity as a major problem in the PEO patients studied here, we do not exclude that some PEO patients would show such a defect as a result of a clearly deficient POLG proofreading activity caused by one of more mutations in the conserved exonuclease motifs. This has indeed been observed by Del Bo et al. (43), but their generalization that this shows the predisposing effect of high mtDNA point mutation levels to multiple deletion formation is supported neither by their own nor by our data.
Enhanced control region point mutation levels in some of the sporadic single deletion patients shows that the presence of deleted mtDNA alone is enough to provoke this effect. The most obvious explanation for this is an enhanced production of reactive oxygen species (ROS), caused by a progressive OXPHOS deficiency. Perhaps the best evidence to suggest that mitochondrial dysfunction can result in enhanced ROS generation in mitochondria, which subsequently may result in mtDNA damage, comes from mouse genetic studies (45,46). Why particularly the mtDNA control region would be prone to ROS remains an open question, but perhaps has something to do with the mtDNA topology in that region. For example, single-stranded DNA is possibly as much as a 1000-fold more prone to DNA damage by ROS than double-stranded DNA (47). The POLG and Twinkle PEO mutations could perhaps have an effect on mtDNA topology by stalling at inappropriate sites, in the replication or the transcription control region, respectively, increasing the sensitivity of the region to DNA damage.
Deletion breakpoint analysis suggests replication stalling as a major event leading to deletions
Single, large mtDNA rearrangements are usually associated with strictly sporadic mitochondrial disorders such as KSS and isolated PEO, and often flanked by relatively long, direct or inverted repeats. Deletions in these sporadic cases previously have been put into two classes. Class I represents deletions with perfect direct repeats that flank the deletion boundaries precisely; class II contains deletions without flanking repeats, with imperfect repeats, with perfect repeats that do not flank the deletions precisely and also with some palindromic sequences (48,49). Notably, the majority of sporadic deletions (∼70%) belong to the first category. Patients with single deletions usually also show evidence of partial duplications (50) whereas, to our knowledge, in PEO cases with multiple deletions, duplications have not been observed. Some studies have excluded duplications in multiple deletion cases (23,51), which was taken to suggest that single and multiple deletions might arise by different mechanisms. Our analysis of deletion breakpoints in ad/arPEO and published deletion breakpoints of adPEO patients [e.g. Zeviani et al. (6)] suggest that the majority of multiple deletions do not correspond to class I of single sporadic deletions. Most direct repeats we observed were short, often imperfect, and hardly ever flanked the deletions precisely.
By which mechanism could POLG and Twinkle dysfunction cause the late-onset multiple deletion pattern of ad/arPEO? Our data and that from published studies suggest that deletion formation might be induced by replication stalling, which in turn has been shown to induce double-strand breaks (DSBs) (52,53). Replication stalling is supported, first of all, by the 2- to 3-fold over-representation of homopolymeric runs at deletion breakpoints. This can be understood for the POLG Y955C mutation by the reported 45-fold reduction in affinity for the incoming nucleotide (17). Stalling at homopolymeric runs could be explained by frequent reiteration of a single nucleotide, which might result in local depletion of that nucleotide and make it difficult for a deficient polymerase to incorporate the subsequent nucleotide. It was shown that a mutation of the corresponding Y844 in adenovirus 5 DNA polymerase also results in a dramatic decrease in polymerase activity and DNA-binding capacity (54), supporting a propensity to stalling. Several other dominant POLG mutations affect the polymerase B domain and probably result in similar defects. Such expectations are supported by extensive mutation studies of the orthologous Klenow and T7 polymerase, as well as their crystal structures [for a recent review, see Copeland et al. (18)]. In addition to homopolymeric runs, several deletions were flanked by microsatellite types of repeats, which are known sites of replication stalling and genome instability [see, for example, Gordenin and Resnick (55)]. A further finding in favor of the idea of increased DSBs in the case of POLG dysfunction is the higher proportion of restriction enzyme-undigested but ‘spontaneously’ linearized full-length muscle mtDNA molecules in POLG/PEO patients (including patient 2 of this study) when compared with control muscle mtDNA (56). Finally, the nt 16 070 breakpoint boundary is compatible with a replication stalling model since it is conceived as a fork barrier as it the defines the boundary of the TAS element and could thus be a frequent replication stalling site, as previously suggested (6). This is consistent with it being a conserved protein-binding site, as shown by footprinting analysis (57,58). It is also in line with the recent suggestion that the mtDNA D-loop is a fork arrest region (59). In that model, the TAS region could be the 5′ boundary of this region.
Our data on Twinkle patients’ mtDNA suggest that the pathogenetic pathways leading to deletion formation are closely linked. The role of Twinkle as a 5′–3′ helicase was recently suggested (12) and this, together with its high homology to replication helicases, supports its role in mtDNA replication. Since the precise role(s) for Twinkle in mtDNA replication and/or repair is still to be established, the mechanism of deletion formation by defective Twinkle remains speculative. Apart from a direct role in replication fork movement, it could be directly involved in DSB repair, by analogy with, for example, the Werner syndrome helicase (60), or it could play a role in re-initiation of stalled replication forks.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
J.N.S. wishes to thank Howy Jacobs for continuous interest and discussion. We are grateful to the patients and family members who volunteered for this study. Muscle DNA from the two sporadic PEO patients used in this study was kindly provided by Jean-Jacques Martin (Department of Neurology and Neuropathology, University Hospital and University of Antwerp). Ann Löfgren is gratefully acknowledged for POLG gene sequencing of Belgium PEO samples. This research has been funded by the Academy of Finland, Centre of Excellence program to J.N.S. and A.S., and the Sigrid Juselius Foundation and Biocentrum-Helsinki to A.S.
REFERENCES
- 1.Satoh M. and Kuroiwa,T. (1991) Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res., 196, 137–140. [DOI] [PubMed] [Google Scholar]
- 2.Garrido N., Griparic,L., Jokitalo,E., Wartiovaara,J., Van Der Bliek,A.M. and Spelbrink,J.N. (2003) Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell., 14, 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grossman L.I. and Shoubridge,E.A. (1996) Mitochondrial genetics and human disease. Bioessays, 18, 983–991. [DOI] [PubMed] [Google Scholar]
- 4.Mandel H., Szargel,R., Labay,V., Elpeleg,O., Saada,A., Shalata,A., Anbinder,Y., Berkowitz,D., Hartman,C., Barak,M. et al. (2001) The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nature Genet., 29, 337–341. [DOI] [PubMed] [Google Scholar]
- 5.Saada A., Shaag,A., Mandel,H., Nevo,Y., Eriksson,S. and Elpeleg,O. (2001) Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nature Genet., 29, 342–344. [DOI] [PubMed] [Google Scholar]
- 6.Zeviani M., Servidei,S., Gellera,C., Bertini,E., DiMauro,S. and DiDonato,S. (1989) An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature, 339, 309–311. [DOI] [PubMed] [Google Scholar]
- 7.Zeviani M., Bresolin,N., Gellera,C., Bordoni,A., Pannacci,M., Amati,P., Moggio,M., Servidei,S., Scarlato,G. and DiDonato,S. (1990) Nucleus-driven multiple large-scale deletions of the human mitochondrial genome: a new autosomal dominant disease. Am. J. Hum. Genet., 47, 904–914. [PMC free article] [PubMed] [Google Scholar]
- 8.Bardosi A., Creutzfeldt,W., DiMauro,S., Felgenhauer,K., Friede,R.L., Goebel,H.H., Kohlschutter,A., Mayer,G., Rahlf,G., Servidei,S. et al. (1987) Myo-, neuro-, gastrointestinal encephalopathy (MNGIE syndrome) due to partial deficiency of cytochrome-c-oxidase. A new mitochondrial multisystem disorder. Acta Neuropathol. (Berl.), 74, 248–258. [DOI] [PubMed] [Google Scholar]
- 9.Hirano M., Silvestri,G., Blake,D.M., Lombes,A., Minetti,C., Bonilla,E., Hays,A.P., Lovelace,R.E., Butler,I., Bertorini,T.E. et al. (1994) Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): clinical, biochemical and genetic features of an autosomal recessive mitochondrial disorder. Neurology, 44, 721–727. [DOI] [PubMed] [Google Scholar]
- 10.Kaukonen J., Juselius,J.K., Tiranti,V., Kyttala,A., Zeviani,M., Comi,G.P., Keranen,S., Peltonen,L. and Suomalainen,A. (2000) Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science, 289, 782–785. [DOI] [PubMed] [Google Scholar]
- 11.Spelbrink J.N., Li,F.Y., Tiranti,V., Nikali,K., Yuan,Q.P., Tariq,M., Wanrooij,S., Garrido,N., Comi,G., Morandi,L. et al. (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nature Genet., 28, 223–231. [DOI] [PubMed] [Google Scholar]
- 12.Korhonen J.A., Gaspari,M. and Falkenberg,M. (2003) Twinkle has 5′ to 3′ DNA helicase activity and is specifically stimulated by mtSSB. J. Biol. Chem., 278, 48627–48632. [DOI] [PubMed] [Google Scholar]
- 13.Van Goethem G., Dermaut,B., Lofgren,A., Martin,J.J. and Van Broeckhoven,C. (2001) Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nature Genet., 28, 211–212. [DOI] [PubMed] [Google Scholar]
- 14.Lamantea E., Tiranti,V., Bordoni,A., Toscano,A., Bono,F., Servidei,S., Papadimitriou,A., Spelbrink,H., Silvestri,L., Casari,G. et al. (2002) Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann. Neurol., 52, 211–219. [DOI] [PubMed] [Google Scholar]
- 15.VanGoethem G., Schwartz,M., Lofgren,A., Dermaut,B., Van Broeckhoven,C. and Vissing,J. (2003) Novel POLG mutations in progressive external ophthalmoplegia mimicking mitochondrial neurogastrointestinal encephalomyopathy. Eur. J. Hum. Genet., 11, 547–549. [DOI] [PubMed] [Google Scholar]
- 16.VanGoethem G., Martin,J.J., Dermaut,B., Lofgren,A., Wibail,A., Ververken,D., Tack,P., Dehaene,I., Van Zandijcke,M., Moonen,M. et al. (2003) Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromusc. Disord., 13, 133–142. [DOI] [PubMed] [Google Scholar]
- 17.Ponamarev M.V., Longley,M.J., Nguyen,D., Kunkel,T.A. and Copeland,W.C. (2002) Active site mutation in DNA polymerase gamma associated with progressive external ophthalmoplegia causes error-prone DNA synthesis. J. Biol. Chem., 277, 15225–15228. [DOI] [PubMed] [Google Scholar]
- 18.Copeland W.C., Ponamarev,M.V., Nguyen,D., Kunkel,T.A. and Longley,M.J. (2003) Mutations in DNA polymerase gamma cause error prone DNA synthesis in human mitochondrial disorders. Acta Biochim. Pol., 50, 155–167. [PubMed] [Google Scholar]
- 19.Nishino I., Spinazzola,A. and Hirano,M. (1999) Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science, 283, 689–692. [DOI] [PubMed] [Google Scholar]
- 20.Spinazzola A., Marti,R., Nishino,I., Andreu,A.L., Naini,A., Tadesse,S., Pela,I., Zammarchi,E., Donati,M.A., Oliver,J.A. et al. (2002) Altered thymidine metabolism due to defects of thymidine phosphorylase. J. Biol. Chem., 277, 4128–4133. [DOI] [PubMed] [Google Scholar]
- 21.Nishigaki Y., Marti,R., Copeland,W.C. and Hirano,M. (2003) Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J. Clin. Invest., 111, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishigaki Y., Marti,R. and Hirano,M. (2004) ND5 is a hot-spot for multiple atypical mitochondrial DNA deletions in mitochondrial neurogastrointestinal encephalomyopathy. Hum. Mol. Genet., 13, 91–101. [DOI] [PubMed] [Google Scholar]
- 23.Suomalainen A., Majander,A., Wallin,M., Setala,K., Kontula,K., Leinonen,H., Salmi,T., Paetau,A., Haltia,M., Valanne,L. et al. (1997) Autosomal dominant progressive external ophthalmoplegia with multiple deletions of mtDNA: clinical, biochemical and molecular genetic features of the 10q-linked disease. Neurology, 48, 1244–1253. [DOI] [PubMed] [Google Scholar]
- 24.Luoma P., Melberg,M., Rinne,J.O., Kaukonen,J.A., Nupponen,N.N., Chalmers,R.M., Oldfors,A., Rautakorpi,I., Peltonen,L., Majamaa,K. et al. (2004) Parkinsonism, premature menopause and mitochondrial DNA polymerase gamma mutations. Lancet, in press. [DOI] [PubMed] [Google Scholar]
- 25.Anderson S., Bankier,A.T., De Bruijn,M.H.L., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H. et al. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 26.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 27.Spelbrink J.N., Toivonen,J.M., Hakkaart,G.A., Kurkela,J.M., Cooper,H.M., Lehtinen,S.K., Lecrenier,N., Back,J.W., Speijer,D., Foury,F. et al. (2000) In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem., 275, 24818–24828. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Michikawa,Y., Mallidis,C., Bai,Y., Woodhouse,L., Yarasheski,K.E., Miller,C.A., Askanas,V., Engel,W.K., Bhasin,S. et al. (2001) Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc. Natl Acad. Sci. USA, 98, 4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajander O.A., Rovio,A.T., Majamaa,K., Poulton,J., Spelbrink,J.N., Holt,I.J., Karhunen,P.J. and Jacobs,H.T. (2000) Human mtDNA sublimons resemble rearranged mitochondrial genomes found in pathological states. Hum. Mol. Genet., 9, 2821–2835. [DOI] [PubMed] [Google Scholar]
- 30.VanGoethem G., Martin,J.J. and Van Broeckhoven,C. (2003) Progressive external ophthalmoplegia characterized by multiple deletions of mitochondrial DNA: unraveling the pathogenesis of human mitochondrial DNA instability and the initiation of a genetic classification. Neuromol. Med., 3, 129–146. [DOI] [PubMed] [Google Scholar]
- 31.Agostino A., Valletta,L., Chinnery,P.F., Ferrari,G., Carrara,F., Taylor,R.W., Schaefer,A.M., Turnbull,D.M., Tiranti,V. and Zeviani,M. (2003) Mutations of ANT1, Twinkle and POLG1 in sporadic progressive external ophthalmoplegia (PEO). Neurology, 60, 1354–1356. [DOI] [PubMed] [Google Scholar]
- 32.Trifunovic A., Wredenberg,A., Falkenberg,M., Spelbrink,J.N., Rovio,A.T., Bruder,C.E., Bohlooly,Y,.M., Gidlöf,S., Oldfors,A., Wibom,R. et al. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature, 429, 417–423. [DOI] [PubMed] [Google Scholar]
- 33.Rahman S., Poulton,J., Marchington,D. and Suomalainen,A. (2001) Decrease of 3243 A→G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am. J. Hum. Genet., 68, 238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michikawa Y., Mazzucchelli,F., Bresolin,N., Scarlato,G. and Attardi,G. (1999) Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science, 286, 774–779. [DOI] [PubMed] [Google Scholar]
- 35.Chomyn A. and Attardi,G. (2003) MtDNA mutations in aging and apoptosis. Biochem. Biophys. Res. Commun., 304, 519–529. [DOI] [PubMed] [Google Scholar]
- 36.Clayton D.A. (1992) Transcription and replication of animal mitochondrial DNAs. Int. Rev. Cytol., 141, 217–232. [DOI] [PubMed] [Google Scholar]
- 37.Bogenhagen D.F. and Clayton,D.A. (2003) The mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci., 28, 357–360. [DOI] [PubMed] [Google Scholar]
- 38.Holt I.J. and Jacobs,H.T. (2003) Response: the mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci., 28, 355–356. [DOI] [PubMed] [Google Scholar]
- 39.Ghivizzani S.C., Madsen,C.S., Nelen,M.R., Ammini,C.V. and Hauswirth,W.W. (1994) In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol. Cell. Biol., 14, 7717–7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J., Asin-Cayuela,J., Fish,J., Michikawa,Y., Bonafe,M., Olivieri,F., Passarino,G., De Benedictis,G., Franceschi,C. and Attardi,G. (2003) Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes. Proc. Natl Acad. Sci. USA, 100, 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoneking M. (2000) Hypervariable sites in the mtDNA control region are mutational hotspots. Am. J. Hum. Genet., 67, 1029–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Bo R., Bordoni,A., Martinelli Boneschi,F., Crimi,M., Sciacco,M., Bresolin,N., Scarlato,G. and Comi,G.P. (2002) Evidence and age-related distribution of mtDNA D-loop point mutations in skeletal muscle from healthy subjects and mitochondrial patients. J. Neurol. Sci., 202, 85–91. [DOI] [PubMed] [Google Scholar]
- 43.Del Bo R., Bordoni,A., Sciacco,M., Di Fonzo,A., Galbiati,S., Crimi,M., Bresolin,N. and Comi,G.P. (2003) Remarkable infidelity of polymerase gammaA associated with mutations in POLG1 exonuclease domain. Neurology, 61, 903–908. [DOI] [PubMed] [Google Scholar]
- 44.Chinnery P.F. (2003) Mitochondrial disorders come full circle. Neurology, 61, 878–880. [DOI] [PubMed] [Google Scholar]
- 45.Esposito L.A., Melov,S., Panov,A., Cottrell,B.A. and Wallace,D.C. (1999) Mitochondrial disease in mouse results in increased oxidative stress. Proc. Natl Acad. Sci. USA, 96, 4820–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melov S., Coskun,P., Patel,M., Tuinstra,R., Cottrell,B., Jun,A.S., Zastawny,T.H., Dizdaroglu,M., Goodman,S.I., Huang,T.T. et al. (1999) Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc. Natl Acad. Sci. USA, 96, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frederico L.A., Kunkel,T.A. and Shaw,B.R. (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry, 29, 2532–2537. [DOI] [PubMed] [Google Scholar]
- 48.Mita S., Spadari,S., Moraes,C.T., Shanske,S., Arnaudo,E., Fabrizi,G.M., Koga,Y., DiMauro,S. and Schon,E.A. (1990) Recombination via flanking direct repeats is a major cause of large scale deletions of human mitochondrial DNA. Nucleic Acids Res., 18, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solano A., Gamez,J., Carod,F.J., Pineda,M., Playan,A., Lopez-Gallardo,E., Andreu,A.L. and Montoya,J. (2003) Characterisation of repeat and palindrome elements in patients harbouring single deletions of mitochondrial DNA. J. Med. Genet., 40, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schon E.A. (2000) Mitochondrial genetics and disease. Trends Biochem. Sci., 25, 555–560. [DOI] [PubMed] [Google Scholar]
- 51.Carrozzo R., Hirano,M., Fromenty,B., Casali,C., Santorelli,F.M., Bonilla,E., DiMauro,S., Schon,E.A. and Miranda,A.F. (1998) Multiple mtDNA deletion features in autosomal dominant and recessive diseases suggest distinct pathogeneses. Neurology, 50, 99–106. [DOI] [PubMed] [Google Scholar]
- 52.Michel B., Ehrlich,S.D. and Uzest,M. (1997) DNA double-strand breaks caused by replication arrest. EMBO J., 16, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis L.K. and Resnick,M.A. (2000) Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res., 451, 71–89. [DOI] [PubMed] [Google Scholar]
- 54.Liu H., Naismith,J.H. and Hay,R.T. (2000) Identification of conserved residues contributing to the activities of adenovirus DNA polymerase. J. Virol., 74, 11681–11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gordenin D.A. and Resnick,M.A. (1998) Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat. Res., 400, 45–58. [DOI] [PubMed] [Google Scholar]
- 56.Van Goethem G., Martin,J.-J., Löfgren,A., Dehaene,I., Tack,P., Van Zandijcke,M., Ververken,D., Ceuterick,C. and Van Broeckhoven,C. (1997) Unusual presentation and clinical variability in Belgian pedigrees with progressive external ophthalmoplegia and multiple deletions of mitochondrial DNA. Eur. J. Neurol., 4, 476–484. [Google Scholar]
- 57.Saccone C., Attimonelli,M. and Sbisa,E. (1987) Structural elements highly preserved during the evolution of the D-loop-containing region in vertebrate mitochondrial DNA. J. Mol. Evol., 26, 205–211. [DOI] [PubMed] [Google Scholar]
- 58.Madsen C.S., Ghivizzani,S.C. and Hauswirth,W.W. (1993) Protein binding to a single termination-associated sequence in the mitochondrial DNA D-loop region. Mol. Cell. Biol., 13, 2162–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowmaker M., Yang,M.Y., Yasukawa,T., Reyes,A., Jacobs,H.T., Huberman,J.A. and Holt,I.J. (2003) Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem., 278, 50961–50969. [DOI] [PubMed] [Google Scholar]
- 60.Oshima J., Huang,S., Pae,C., Campisi,J. and Schiestl,R.H. (2002) Lack of WRN results in extensive deletion at nonhomologous joining ends. Cancer Res., 62, 547–551. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.