Abstract
Metastasis is the primary cause of cancer-related death in oncology patients. A comprehensive understanding of the molecular mechanisms that cancer cells usurp to promote metastatic dissemination is critical for the development and implementation of novel diagnostic and treatment strategies. Here we show that the membrane protein RECK, controls breast cancer metastasis by modulating a novel, non-canonical and convergent STAT3-dependent angiogenic program. Neoangiogenesis and STAT3 hyperactivation are known to be fundamentally important for metastasis but the root molecular initiators of these phenotypes are poorly understood. Our study identifies loss of RECK as a critical and previously unknown trigger for these hallmarks of metastasis. Using multiple xenograft mouse models, we comprehensively show that RECK inhibits metastasis, concomitant with a suppression of neoangiogenesis at secondary sites, while leaving primary tumour growth unaffected. Further, with functional genomics and biochemical dissection we demonstrate that RECK controls this angiogenic rheostat through a novel complex with cell surface receptors to regulate STAT3 activation, cytokine signaling, and the induction of both VEGF and uPA. In accordance with these findings, inhibition of STAT3 can rescue this phenotype both in vitro and in vivo. Taken together, our study uncovers, for the first time, that RECK is a novel regulator of multiple well-established and robust mediators of metastasis; thus, RECK is a keystone protein that may be exploited in a clinical setting to target metastatic disease from multiple angles.
Introduction
Breast cancer is one of the most common cancers among women, causing over 400,000 deaths annually worldwide (1). Metastatic disease is a major cause of morbidity and mortality and is currently incurable, making it a primary obstacle to improving breast cancer outcomes (2). The molecular basis of metastasis remains incompletely understood. In recent years, a number of metastasis suppressor genes (MSGs) have been discovered. These genes are defined by their specific ability to inhibit metastasis without altering primary tumour growth. Both preclinical models and retrospective human studies suggest that MSGs play key roles in controlling the development of metastasis (3). Elucidating the molecular mechanisms by which these genes control the metastatic process offers valuable insight into tumour biology and may lead to new therapeutic options.
Reversion-inducing cysteine-rich protein with kazal motifs (RECK) is a putative MSG that is implicated in tumour progression (4). RECK is a membrane-anchored protein that is involved in several physiologic processes, including regulation of extracellular matrix integrity, vascular growth during development, and stabilization of tissue architecture (5, 6). In some cell systems, RECK can influence MMP function (7). RECK expression is frequently silenced in tumour cells (8, 9). Interestingly, a recent study focusing on genome-wide DNA methylation profiling of CpG islands in breast cancer identified the RECK gene as a common target of promoter hypermethylation (10). Despite documented effects of RECK on the behavior of tumour cells, the signaling pathways targeted by RECK and the specific mechanism by which RECK modulates metastasis remains elusive. Furthermore, while RECK-mediated invasion has been largely attributed to changes in MMP expression in vitro, most tumours and tumour cell lines express extremely low levels of MMPs, suggesting that additional mechanisms are likely at play (11, 12).
In this study, we apply a systematic, multifaceted strategy using human tumour samples, in vivo metastasis models, and multiple, unbiased high throughput analyses to provide a comprehensive analysis of the role of RECK during breast cancer metastasis. We generated new data sets which include large number of patient samples and used them to illustrate a relationship between RECK expression and disease-specific survival. Furthermore, analyses of matched pairs of primary human breast tumours and distant metastases reveal that RECK expression is further silenced during metastatic progression. We demonstrate using multiple mouse models that RECK reconstitution in vivo suppresses tumour metastases. Using several unbiased screens, we have identified novel pathways and binding partners that directly mediate the effects of RECK on metastasis. Finally, we show that RECK controls these phenotypes through STAT3 dependent regulation of an angiogenic program.
Results
Analysis of matched lymph node and distant metastases demonstrates that RECK expression is downregulated during metastatic breast cancer progression
We first sought to explore whether RECK expression was lost specifically during metastatic progression or whether the process of RECK silencing was already completed in the primary tumour. To answer this question, we first evaluated RECK gene expression in 36 primary tumours and lymph node metastasis pairs. Microarray analysis of these tumours revealed a significant decrease in RECK expression in metastatic lymph nodes compared to matched primary tumours in luminal A, luminal B and Her 2 subytpes (p=0.0009, Figure. 1A). These results were confirmed by qRT-PCR (p<0.01, Supplementary Figure. 1A). Interestingly, we did not see a significant difference in RECK expression in the basal subtype (Figure. 1A); however, the baseline expression of RECK in our basal primary tumours was already considerably lower than other subtypes (Supplementary Figure. 1B). This is not seen with other larger patient cohorts suggesting that there may be significant diversity in RECK expression between different basal tumours.
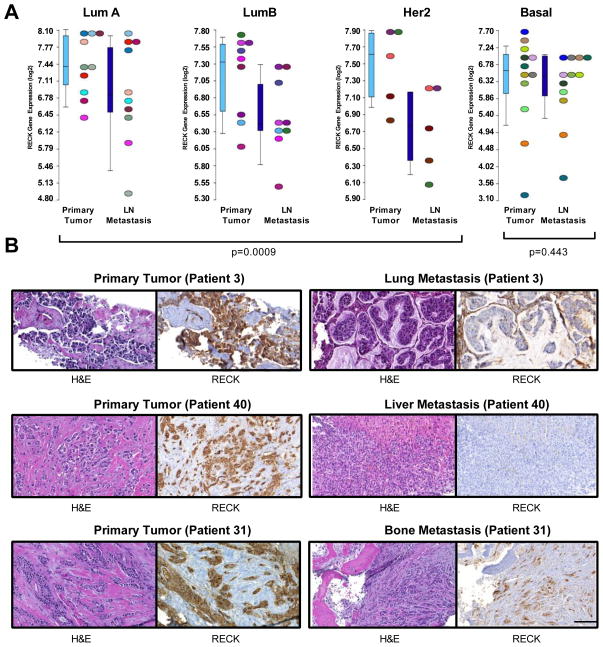
Figure 1. Analyses of matched pairs demonstrates that RECK is down-regulated during metastatic progression in both lymph nodes and distant sites.
(A) RECK is silenced during the establishment of nodal metastases. Expression levels of RECK in 36 matched pairs of primary breast tumours and lymph node metastases segregated based on PAM50 subtype (LumA, LumB, Her2 together (p=0.0009); LumA (p=0.4609), LumB (p=0.0078), Her2 (p=0.125), and Basal (p=0.443). Wilcoxon matched pairs signed rank test. Data is from microarray analysis.
(B) RECK is silenced in distant metastases. Representative images of H&E staining and RECK immunohistochemistry of matched primary breast tumours and distant metastases. RECK staining was scored and statistical quantification was based on Wilcoxon matched-pairs signed rank test (p=0.001, n=43).
We next wanted to determine if the RECK silencing was also evident in distant metastases. We performed immunohistochemistry for RECK on 43 matched primary breast tumours and distant metastatic lesions (including lung, liver and bone), and scored staining intensity. Distant metastases had significantly decreased RECK expression compared to matched primary tumours (p=0.001, Figure. 1B, Supplementary Figure. 1C–D). Together, these data show that RECK expression is lost in many tumors during progression from primary tumour to metastasis, consistent with a role for RECK in protecting against metastatic breast cancer.
RECK does not alter cell proliferation and loss of RECK alone is not sufficient to achieve cellular transformation
As RECK expression is low in most solid tumour cell lines, we next investigated the effects of reconstituting RECK expression (by constitutively expressing RECK via lentiviral transduction) in 3 highly invasive breast cancer cell lines (MDA-MB-231, LM2-4175 and BOM1-1833). RECK expression was confirmed by western blot (Figure. 2A) and proper membrane localization was visualized by immunocytochemistry and confocal microscopy (Figure. 2B). Altered RECK expression did not change the rate of cell growth in vitro (Figure. 2C–E) nor did it result in cell cycle changes (Figure. 2F–H). Given that RECK expression is lower in clinical tumour specimens compared to normal breast tissue, we next sought to examine if loss of RECK is sufficient for malignant transformation. Stable knockdown of RECK did not transform normal human mammary epithelial cells (HMECs) in xenograft assays (Figure. 2I–K). These results suggest that RECK loss is not sufficient to result in transformation or altered cell division but instead may act via other mechanisms.
Figure 2. RECK does not affect cell cycle or growth rate in invasive breast cancer cells and loss of RECK alone is insufficient to transform cells.
(A) Overexpression of RECK in breast cancer cell lines. Representative western blot analysis of MDA-MB-231, LM2-4175 and BOM1-1833 cells stably transduced with the lentiviral constructs indicated. β-actin was used as a loading control.
(B) Representative confocal immunofluorescent images of MDA-MB-231, LM2-4175 and BOM1-1833 cells stably transduced with lentiviral constructs indicated. IgG was used as a staining control. Scale bar represent 20μM.
Growth curve of (C) MDA-MB-231 (D) LM2-4175 and (E) BOM1-1833 cells stably transduced with lentiviral constructs. Data are presented as mean± SEM (n=3). Flow cytometric cell cycle analysis of (F) MDA-MB-231 (G) LM2-4175 and (H) BOM1-1833 cells stably transduced with lentiviral constructs indicated. Cell cycle curves and quantification of stages of cell cycle was based on the Watson (pragmatic) model (n>10000 events).
(I) Representative immunoblot of HMEC-hTERT stably infected with the lentivirus indicated.
HMEC-hTERT stably infected with the indicated lentiviral constructs were injected into the (J) lateral tail vein of of athymic nude mice (n=5) or (K) the mammary fat pad NSG mice (n=5). Representative BLI images at 15 weeks post-injection are shown.
RECK suppresses VEGF expression, endothelial recruitment in metastatic lesions, and metastatic burden in vivo
We next examined whether reconstituting RECK could reduce metastatic burden in an in vivo model of metastasis. Accordingly, we stably expressed RECK in LM2-4175 cells (an in vivo selected and highly lung metastatic cell line) (13, 14) and injected them into the lateral tail-vein of athymic nude mice. Bioluminescent imaging (BLI) of mice re-expressing RECK revealed a significant decrease in metastatic tumour burden compared to empty vector (EV) control (Figure. 3A,B). Given that a significant difference in metastatic burden was first evident via BLI at 4 weeks post-IV injection (Figure. 3B), we harvested lung tissue from each group at this time point for detailed analysis. Consistent with the BLI data at 4 weeks, H&E staining of lung tissue (Figure. 3C) revealed a significant decrease in tumour burden (Figure. 3D) and number of metastatic foci (Figure. 3E) in mice injected with cells re-expressing RECK.
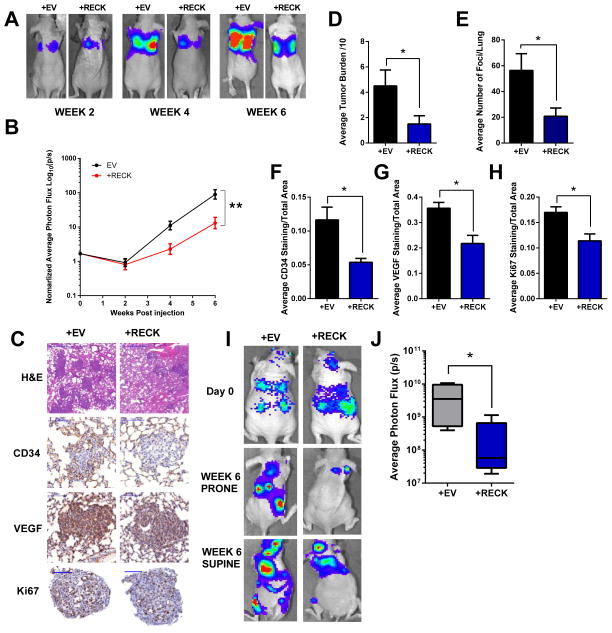
Figure 3. Reconstituting RECK in breast cancer cells results in decreased VEGF, vascular recruitment, and metastatic burden in vivo.
LM2-4175+EV or LM2-4175+RECK cells were injected into the lateral tail vein of athymic nude mice (n ≥7). BLI images were taken over a period of 6 weeks. (A) Representative images are depicted. (B) Graph of BLI quantification over time. Data is presented as mean± SEM (**p<0.01). (C) Mice were sacrificed at 4 weeks and lung tissue was stained with H&E, CD34, VEGF and Ki67. Scale bars represent 500μm, 100μm, 100μm, and 100μm, respectively.
(D) Scoring of metastatic tumour burden in H&E stained lungs. Data are presented as mean± SEM (*p<0.05, n≥3).
(E) Quantification of average number of metastatic foci per lung. Data are presented as mean± SEM (*p<0.05, n≥3).
Quantification of (F) CD34, (G) VEGF, (H) Ki67 staining. Data are presented as mean± SEM (*p<0.05, n≥3).
BOM1-1833+EV or BOM1-1833+RECK cells were intracardiac injected into the left ventricle of athymic nude mice (n=5). (I) Representative BLI of mice 6 weeks post injection. (J) Quantification of BLI at 6 weeks post injection. Data are presented as whisker box plots (*p<0.05, n=5).
RECK has been previously suggested to play a role in vascular organization (7). Therefore, we stained lung tissue for CD34 and VEGF. RECK-expressing metastatic foci had significantly less CD34 staining (Figure. 3C,F) suggesting RECK suppresses de-novo blood vessel formation or recruitment. This phenotype was concomitant with decreased VEGF (Figure. 3C,G) and Ki67 expression (Figure. 3C,H), demonstrating that RECK may interfere with metastatic outgrowth by reducing vascularization. A decrease in metastasis in vivo was also observed after intracardiac injection of BOM1-1833 cells (an in vivo selected and highly bone metastatic cell line) (13) re-expressing RECK (Figure. 3I,J).
RECK has been hypothesized to act in part through MMP-2, MMP-9 and MT1-MMP in some cell systems (15–17). Interestingly, in our breast metastatic model, immunostaining of metastatic nodules for MMP2- MMP-9 and MT1-MMP revealed no significant changes in expression (Supplementary Figure. 2A-C). However, as immunostaining does not discriminate between latent and active MMP-2 and MMP-9, we cannot discount the possibility that there may be differences in MMP activity. Nevertheless, MMP-2 and MMP-9 were undetectable in conditioned media from MDA-MB-231, LM2-4175 and BOM1-1833 by ELISA (data not shown), suggesting that any differences seen in vivo are likely from changes in secretion of these proteins from surrounding stroma and not from the tumour cells themselves. Together, these data suggest that the phenotypic effects of RECK expression in breast cancer metastases may involve novel mechanisms independent of MMPs.
Coordinate control of a cooperative neovascularization program by RECK via regulation of endothelial remodeling and multiple secreted regulators of angiogenesis
In order to gain further insight into the molecular changes associated with RECK expression, we analyzed the transcriptomes of MDA-MB-231 and LM2-4175 cells overexpressing RECK. Ingenuity pathway analysis (IPA) of differentially expressed genes between control and RECK overexpressing cells (Supplementary Table 1,2) revealed significant alterations in IL-6, IL-8 and IL-10 signaling pathways in both cell lines (Figure. 4A,B, Supplementary Table 5,6). In addition, VEGF and HIF-1α signaling pathways were also altered in MDA-MB-231 and LM2-4175 cells, respectively. All of these pathways are intimately involved in regulation of angiogenesis, consistent with our hypothesis that RECK can control transcriptional programs that influence tumour neovascularization (18–20).
Figure 4. RECK regulates angiogenic and cytokine transcriptional programs, endothelial reorganization, and a neovascular switch.
RECK expression resulted in significant changes in IL-6, IL-10, IL8, HIF1a and VEGF pathway genes. U133A 2.0 expression arrays were used to compare the transcriptomes of (A) MDA-MB-231+EV and MDA-MB-231+RECK cells or (B) LM2-4175+EV and LM2-4175+RECK cells. Differentially expressed genes were analyzed using IPA software.
(C) ELISA was used to determine VEGF concentrations in conditioned media from MDA-MB-231, LM2-4175 and BOM1-1833 cells stably transduced with lentiviral constructs indicated. Data is presented as mean± SEM (***p<0.001, n=3).
(D) RECK suppresses endothelial cell migration which is rescued by VEGF. Conditioned media from MDA-MB-231+EV, MDA-MB-231+RECK or MDA-MB-231+RECK+ 10 μM recombinant VEGF was used as a chemoattractant for HUVEC cells in a Boyden transwell chamber migration assay. Data is presented as mean± SEM (*p<0.05, **p<0.01, n=4).
(E) Same as in D but with LM2-4175 cells (*p<0.05, **p<0.01, n=4).
(F) RECK regulates endothelial cell reorganization. Conditioned media from MDA-MB-231, or LM2-4175 cells stably transduced with indicated lentiviral constructs was used to resuspend HUVEC cells in an in vitro tube formation assay. 5% BSA and 5% BSA+10 μM recombinant VEGF were used as negative and positive controls, respectively. Representative images are shown in (F).Scale bars represent 200μm. (G) Quantification of tube formation data is presented as mean± SEM (***p<0.001, n=3).
(H) RECK regulates a coordinated, convergent angiogenic switch. LM2-4175+EV and LM2-4175+RECK were placed at 0.5%O2 for 24h. Conditioned media from these cells was used to probe an angiogenesis array. Protein expression was quantified by densitometry and select candidates shown in heat map (I).
(J) ELISA was used to determine uPA concentrations in conditioned media from MDA-MB-231, LM2-4175 and BOM1-1833 cells stably transduced with indicated lentiviral constructs. Data are presented as mean± SEM (***p<0.001, n=3).
We next asked whether reconstituting RECK was sufficient to alter VEGF secretion or endothelial cell phenotypes in vitro. ELISAs were performed on conditioned media from MDA-MB-231, LM2-4175 and BOM1-1833 cells re-expressing RECK, and all were found to secrete significantly less VEGF compared to their respective controls (Figure. 4C). In addition, conditioned media from MDA-MB-231 and LM2-4175 cells re-expressing RECK inhibited chemotaxis of endothelial cells using Boyden transwell assays (Figure. 4D,E). Importantly, this phenotype was rescued with the addition of recombinant VEGF. Furthermore, conditioned media from MDA-MB-231 and LM2-4175 cells re-expressing RECK significantly reduced endothelial cell tube formation in vitro (Figure. 4F,G), showing that RECK can regulate endothelial organization. These data suggest that RECK may be an important modulator of tumour-mediated angiogenesis, consistent with the changes observed in vivo.
To determine if there are other potential angiogenic factors besides VEGF that are altered by RECK, we performed an antibody array screen of 55 known pro- or anti-angiogenic factors, including various ECM components, proteases, growth factors, and signaling molecules. LM2-4175 cells expressing either RECK or EV were subject to a low oxygen challenge (0.5% O2) to mimic the hypoxic tumour environment and stimulate release of angiogenic factors. After 24 hours, conditioned media was isolated from both cell lines and screened with the array. Strikingly, the array revealed that reconstituting RECK coordinately suppressed many pro-angiogenic factors and increased the secretion of anti-angiogenic factors (Figure. 4H,I). For example, reconstituting RECK potently suppressed the secretion of uPA, a major modulator of extracellular proteolysis and angiogenesis, and a known mediator of breast cancer metastasis (21). We confirmed RECK-mediated suppression of uPA secretion by ELISA in 3 breast cancer cell lines (Figure. 4J). Together these data demonstrate an important role for RECK in coordinately modulating multiple angiogenic factors, and for the first time reveal a link between RECK and regulation of uPA.
A spontaneous metastasis assay reveals that RECK suppresses metastasis in part through negative regulation of uPA
Given our finding that RECK modulates the secretion of uPA, a well-documented protein involved in extracellular matrix proteolysis and breast cancer metastasis, we next wanted to determine if reconstituting RECK in breast cancer cells could suppress spontaneous metastasis in vivo. Metastasis is a multistep process including initial invasion of surrounding tissue, intravasation/extravasation and survival and outgrowth at secondary sites (22). Although frequently modeled using tail vein injection, metastasis modeled via orthotopic injection may more accurately recapitulate the steps necessary for metastasis. MDA-MB-231 cells expressing either EV or RECK were orthotopically injected into the mammary fat pad of NOD/SCID/IL2Rγ-deficient (NSG) mice. Consistent with our in vitro proliferation results, primary tumour volume was unchanged following RECK re-expression (Figure. 5A). BLI of mice at day 40 revealed a significant decrease in spontaneous metastasis from tumours expressing high RECK versus control (Figure. 5B,C), which was confirmed by H&E staining and quantification of tumour burden in lung and liver (Figure. 5D,E). Immunohistochemistry (IHC) was used to confirm that RECK expression was high in primary tumours with reconstituted RECK at day 40 (Figure. 5F). Furthermore, IHC for uPA in primary tumours re-expressing RECK revealed a significant decrease in uPA staining intensity compared to controls (Figure. 5G), again suggesting a strong relationship between RECK, uPA and metastasis.
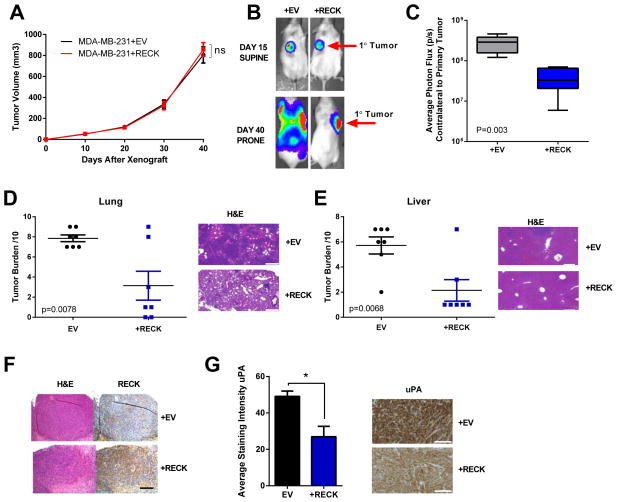
Figure 5. Orthotopic metastasis model demonstrates that RECK decreases lung and liver metastasis but does not significantly alter growth of the primary tumour.<.
br>MDA-MB-231+EV and MDA-MB-231+RECK cells were injected into the mammary fat pad of NSG mice.
(A) Quantification of primary tumour volume over time. Data are presented as mean ± SEM (ns=not significant, n=7).
(B) RECK suppresses metastatic dissemination in an orthotopic model. Representative BLI of mice at day 15 and day 40.
(C) Quantification of BLI on the contralateral side from primary tumour at day 40. Data are presented as whiskers min to max (p=0.003, n=7).
Lung and liver tissue was isolated from mice at day 40 and were sectioned and stained with H&E. Representative H&E sections and quantification of metastatic tumour burden are shown for both (D) lung and (E) liver. Scale bars represent 300μm. Data is presented as mean ± SEM (n=7).
(F) Representative image of primary tumours stained with H&E and immunohistochemistry for RECK. Data is presented as mean± SEM. Scale bars represent 200μm.
(G) RECK expression suppresses uPA. Representative immunohistochemistry of primary tumours for uPA and quantification of staining intensity. Scale bars represent 150μM. Quantification was performed using MetaMorph software. Data is presented as mean ± SEM (*p<0.05, n≥4).
Depletion of RECK promotes a uPA-dependent invasion and pro-angiogenic program
To evaluate the effect of RECK perturbation on VEGF and uPA, we knocked-down RECK in the breast cancer cell lines Hs343T and Hs606T. We chose these cell lines as they have significant endogenous levels of RECK expression (Cancer Cell Line Encyclopedia, Broad, Supplementary Figure. 3). Hs343t and Hs606t cells were transfected with scramble (control) or RECK siRNA. RECK knockdown was confirmed by western blot (Figure. 6A). Consistent with our RECK re-expression experiments, RECK knockdown had no significant effect on the growth of cells. Representative growth curves from Hs606t cells are shown in Figure. 6B. ELISA performed on conditioned media isolated from these cells demonstrated that RECK knockdown results in a significant increase in VEGF and uPA secretion (Figure. 6C,E). These changes were concurrent with changes in VEGF and PLAU gene expression (Figure. 6D,F). Furthermore, using a Matrigel™ transwell invasion assay, we revealed that RECK knockdown significantly increased invasion of both Hs343t and Hs606t lines, and that this was rescued by concomitant knockdown of uPA (siPLAU Figure. 6G,H). This suggests that the increased invasive potential associated with the loss of RECK could be in part due to increases in uPA secretion. Finally, to test the effects of RECK loss on angiogenic markers, we repeated the angiogenesis protein array using conditioned media from Hs606t cells transfected with either scramble (control) or RECK siRNA. Consistent with previous results, RECK knockdown resulted in high levels of pro-angiogenic factors in conditioned media, including VEGF and uPA, and reduced levels of anti-angiogenic factors (Figure. 6I,J).
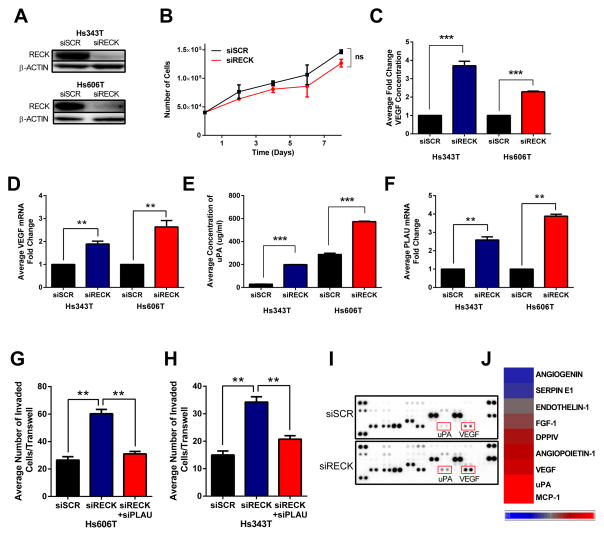
Figure 6. RECK depletion elevates VEGF and uPA secretion, activation of a coordinate pro-angiogenic program, and increases invasive potential but does not affect cell growth.
(A) Representative immunoblot of Hs343T and Hs606T cells 48h after transfection with SCR (scramble) or RECK siRNA.
(B) Growth curve of Hs606t cells transfected with indicated siRNA. Data is presented as mean ± SEM (ns=not significant, n=3).
(C) VEGF ELISA on conditioned media isolated from Hs343T and Hs606T transfected with indicated siRNA. Data is presented as mean ± SEM (***p<0.001, n=3).
(D) qRT-PCR of VEGF gene expression after transfection with indicated siRNA. Data is presented as mean ± SEM (**p<0.01, n=3).
(E) uPA ELISA on conditioned media isolated from Hs343T and Hs606T transfected with indicated siRNA. Data is presented as mean ± SEM (***p<0.001, n=3).
(F) qRT-PCR of PLAU gene expression after transfection with indicated siRNA. Data is presented as mean ± SEM (**p<0.01, n=3).
Quantification of Boyden transwell invasion of (G) Hs343T and (H) Hs606T cells transfected with indicated siRNAs. Data is presented as mean ± SEM (***p<0.001, n≥3).
Conditioned media from Hs606T cells transfected with indicated siRNA was used to probe an angiogenesis antibody array. (I) Image of array was used to quantify protein expression by densitometry, and altered candidates are shown in heat map (J).
RECK associates with multiple cell surface receptors, modulates cytokine signaling, and activates STAT3 signaling
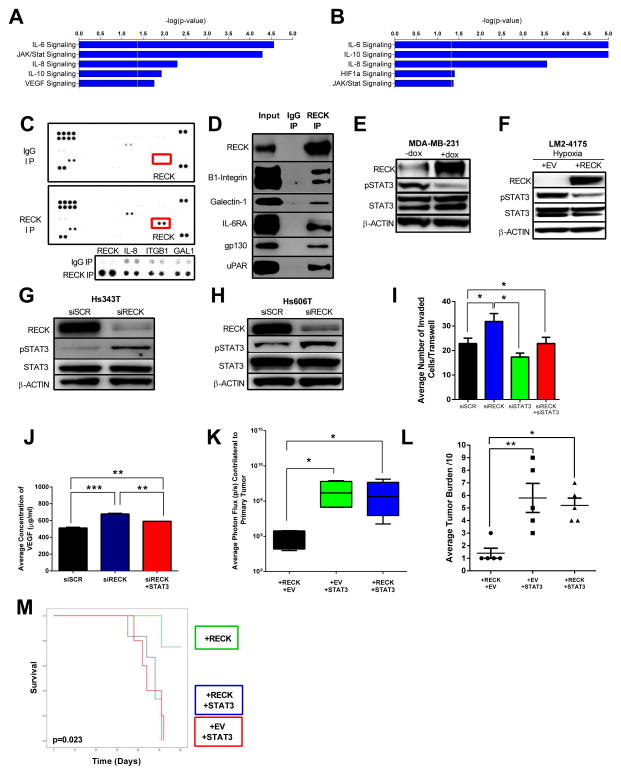
To further dissect the details of the signaling pathways which underlie the effects of RECK, we tested the results of RECK knockdown on global gene expression in Hs343t and Hs606t cells (Supplementary Table 3,4). Microarray and IPA analyses revealed that IL6, IL8, L10 and JAK/STAT signaling pathways were significantly altered in response to RECK knockdown in both cell lines tested (Figure. 7A,B, Supplementary Table 7,8). Importantly, these data are consistent with pathways altered by reconstituting RECK in previous experiments (Figure. 4A,B, Supplementary Table 1,2,9).
Figure 7. RECK binds to multiple cell surface receptors and regulates cytokine and STAT3 signaling to modulate metastasis and survival.
RECK knockdown resulted in significant changes in cytokine and angiogenesis pathways, including IL-6, IL-10, IL8, Jak/Stat and VEGF. Human Genome U133A 2.0 expression arrays were used to compare the transcriptomes of (A) scramble vs. siRECK in Hs343T cells and (B) scramble vs. siRECK in Hs606T cells. Differentially expressed genes were analyzed using IPA software. Immunoprecipitation using anti-RECK antibody in Hs606T cell lysate was used to probe a human soluble receptor antibody array. (C) Image of array highlighting enhanced RECK, IL-8, ITGB1, and GAL1 signal in IP-RECK compared to IgG (control). In the lower panel, data for each protein was taken at different exposures.
(D) RECK binds to novel receptors that signal through STAT3. RECK co-IP demonstrates that RECK binds to β1-integrin, Galectin-1, IL-6Ra, gp130, and uPAR.
(E) RECK decreases STAT3 activation. Representative immunoblot of lysates from MDA-MB-231 cells stably infected with a doxycyclin (dox) inducible RECK plasmid. Cells were treated with 1ug/ml dox for 18h prior to harvesting lysate. Whole cell lysate was used to probe for RECK, STAT3 and pSTAT3. β-actin was used as a loading control.
(F) RECK decreases STAT3 activation in hypoxic conditions. Representative immunoblot of LM2-4175 cells stably infected with lentiviral constructs indicated. Cells were placed in hypoxic conditions (0.5% O2) for 24h before harvesting lysates.
RECK depletion increases pSTAT3 levels. Representative immunoblot analysis of (G) Hs343t and (H) Hs606T cells transfected with siRNA indicated. (I) STAT3 knockdown reverses the increased invasion resulting from RECK depletion. Quantification of invasion of Hs606T cells transfected with indicated siRNAs. Data is presented as mean ± SEM (*p<0.05, n=3).
(J) STAT3 expression reverses increases in VEGF secretion post-RECK depletion. VEGF ELISA on conditioned media isolated from Hs606T cells transfected with indicated siRNA. Data is presented as mean ± SEM (**p<0.01, ***p<0.001, n=3).
(K) STAT3 reverses RECK-dependent suppression of metastasis. LM2-4175+RECK, LM2-4175+STAT3 and LM2-4175+RECK+STAT3 cells were injected into the mammary fat pad of NSG mice. Quantification of BLI on the contralateral side of the primary tumour was performed on day 30. Data are presented as whisker box plots with the line depicting the mean and the boxes showing 1 SD (n=5). (L) Lungs were isolated, sectioned and stained with H&E and the metastatic tumour burden was quantification out of 10.
(M) Kaplan-Meier of mice injected via tail vein with cells described in K. Increased survival conferred by RECK expression is reversed by concomitant overexpression of STAT3. p=0.023 (n=5, log rank).
Given that RECK is a GPI-anchored membrane protein, we hypothesized that RECK may be altering signaling pathways by directly binding other membrane proteins, extracellular cytokines or proteases. To test this, we modified a receptor antibody array to screen for potential RECK binding partners. RECK immunoprecipitated lysate from Hs606t cells was used to probe a panel of 119 soluble extracellular or membrane-bound proteins. Immunoprecipitation revealed that RECK bound to IL-8, β1-integrin, and galectin-1 (GAL-1) (Figure. 7C). As a positive control, we confirmed RECK binding in this modified system and by conventional western blot (Figure. 7C,D). We validated the interaction screen by immunoblot for β1-integrin and GAL-1 (Figure. 7D). Since one of the most significantly altered pathways common to all microarray analysis was IL-6 signaling, we next determined that RECK also directly complexes with IL-6RA and gp130, the principle mechanism of IL-6 signaling in breast cancer (Figure. 7D)(23). Finally, we identified that RECK also binds to the uPA receptor uPAR (Figure. 7D), providing a possible additional mechanism for RECK-mediated suppression of uPA activity.
To characterize RECK-induced signaling events in the cytoplasm, we used a phospho-protein array and compared lysate from MDA-MB-231+EV and MDA-MB-231+RECK cells. RECK expression results in decreased phosphorylation of a number or growth-related oncoproteins. Among the most significantly downregulated phospho-proteins was pSTAT3. RECK expression resulted in a loss of phosphorylation on the activating Y705 residue (Supplementary Figure. 4A). Interestingly, this is consistent with our protein and microarray results, as pSTAT3 is a well-described downstream effector of IL-6 and IL-8, and is capable of inducing both VEGF and uPA (24–27). We confirmed that pSTAT3 is regulated by RECK using a doxycycline-inducible construct, showing that RECK induction resulted in significantly decreased pSTAT3 levels (Figure. 7E). Furthermore, even under hypoxic conditions, which are known to potently activate STAT3 via phosphorylation (28, 29), pSTAT3 was suppressed upon RECK reconstitution in LM2-4175 cells (Figure. 7F). Consistent with this data, knockdown of RECK with siRNA in Hs343t and Hs606t cells resulted in increased pSTAT3 (Figure. 7G,H).
Modulation of metastasis and angiogenesis by RECK is dependent on STAT3 signaling
We next investigated whether the effects of RECK on angiogenesis, invasion and metastasis are STAT3 dependent. First, we determined that the increase in HS606t invasion caused by RECK knockdown was attenuated with the concurrent knockdown of STAT3, demonstrating that RECK-mediated invasiveness is in part mediated by STAT3 (Figure. 7I, Supplementary Figure. 4B). In addition, increased VEGF secretion conferred by RECK knockdown was also attenuated following concurrent knockdown of STAT3 (Figure. 7J). This shows that like the invasion phenotype, RECK-mediated VEGF secretion is also due at least in part to STAT3 activation. Lastly, we determined whether exogenously expressing STAT3 can reverse the ability of RECK to suppress metastasis. For this we again made use of our in vivo metastasis model. LM2-4175 cells which stably overexpressed RECK were transduced with STAT3 and injected into the mammary fat pad of NSG mice (spontaneous metastasis model). BLI and histological analysis of lung metastasis at day 30 revealed decreased metastasis in mice with cells expressing RECK alone, as expected (Figure. 7K,L). This phenotype was attenuated with the concomitant expression of STAT3 suggesting STAT3 is downstream of RECK and that metastatic suppressive activity of RECK is mediated by STAT3. Using our tail vein in vivo metastasis assay and these cells, we found that mice injected with cells expressing RECK has a significant survival advantage that was abrogated by co-expression of STAT3 and RECK (p=0.023, Figure. 7M). Together, these results demonstrate that the invasive and angiogenic phenotypes that result from RECK suppression are mediated through STAT3 signaling.
Suppression of RECK expression is associated with poor survival in human breast cancer
Finally, we sought to validate the association of RECK expression with breast cancer survival outcomes and to determine the relationship of RECK loss with pathologic covariates. Previous studies linking RECK with clinical outcomes used relatively small numbers of samples. We dichotomized 1586 breast cancer patients from the Curtis data set (30) into ‘RECK high’ and ‘RECK low” groups based on median RECK expression, and analyzed disease-specific survival (DSS). Here, high RECK expression conferred a significant survival advantage (p=0.025, Figure. 8A). This was confirmed in 2 independent, large breast cancer data sets using identical methodology (p=0.006, Figure. 8B, Supplementary Figure. 5A). Due to the unique size of the Curtis dataset, we were able to further categorized RECK expression as “normal” or “low,” based on values in normal breast tissue (Supplementary Figure. 5B). Here, “low” is defined as having an outlier level of RECK expression below the 10th percentile of RECK expression in normal breast tissue. Patients with primary tumours harboring normal RECK expression experienced significantly superior survival compared to those with tumours bearing low RECK (p=0.000083, Figure. 8C).
Figure 8. Clinical and pathologic correlates of RECK loss in breast cancer.
(A) Loss of RECK expression is associated with poor clinical outcome. Kaplan-Meier survival plot of 1586 breast cancer patients from the Curtis breast cancer dataset separated into ‘RECK High’ and ‘RECK Low’ groups based on the median RECK expression (p=0.025, log-rank).
(B) Kaplan-Meier survival plot of 295 breast cancer patients from the van de Vijver breast cancer dataset separated into ‘RECK High’ and ‘RECK Low’ groups based on the median RECK expression (p=0.006, log-rank).
(C) Kaplan-Meier survival plot of 1586 breast cancer patients from the Curtis breast cancer dataset separated into ‘RECK Normal’ and ‘RECK Low’ groups based on the RECK expression levels in normal breast tissue (p=0.000083, log-rank). See text for rationale.
RECK expression from the Curtis breast cancer dataset by (D) tumour grade in 1902 tumours (E) cellularity in 1927 tumours and (F) tumour size in 1972 tumours.
RECK expression in tumour sets segregated by (G) ER status in 1948 tumours (H) PR status and (I) HER2 status in 1992 tumours. All data are presented as mean± SEM (*p<0.05, **p<0.01, ****p<0.0001, ns=not significant).
(J) RECK expression based on histological subtype in 2037 breast cancer tumours. Data is presented as Tukey’s box plot.
(K) RECK expression based on PAM50 subtype in 1986 breast cancer tumours. Data is presented as Tukey’s box plot.
In addition to poorer survival, we found that higher primary tumour grade, cellularity, and size were also significantly associated with low RECK expression (Figure. 8D–F). RECK expression was not significantly associated with ER, PR, or HER2 status in the Curtis or TCGA breast cancer cohorts (Figure. 8G–I, Supplementary Figure. 5C–E). Notably, the expression of RECK was significantly decreased in all breast cancer subtypes compared to normal breast tissue (Figure. 8J,K, Supplementary Figure. 5F,G) suggesting that loss of RECK expression can occur across all breast cancer types. Together these results demonstrate that RECK expression is an important determinant of breast cancer survival, that decreased RECK expression is associated with aggressive tumour characteristics, and that RECK alteration is likely selected for in cancers.
Discussion
RECK is recognized as a frequently inactivated metastasis suppressor in a number of human malignancies. Although loss of RECK has been linked to aggressive tumour behavior and metastasis, the molecular mechanisms by which RECK affects these processes is incompletely understood. The effect of RECK on tumour cells has been thought to be attributable to changes in MMP activity. However, in most tumours, these proteases are not predominantly produced by the malignant cells themselves, but instead by adjacent stroma (31). Furthermore, MMP levels in our in vivo tumour models do not significantly associate with RECK expression. Accordingly, our data here suggest that, at least in metastatic progression, RECK functions by associating with a number of receptors that signal through STAT3, modulating STAT3 signaling, and promoting the coordinated expression of a multifaceted invasion and angiogenic program. As RECK is known to play key roles in normal development (32, 33), we reveal a molecular framework for how RECK regulates metastasis and highlight how metastatic cells can usurp RECK-dependent processes to promote tumour cell survival at distant sites.
We established a significant relationship between RECK expression and disease-specific survival in breast cancer. Through the use of matched pairs of primary human breast tumours and both distant and lymph node metastasis, we show that RECK expression is suppressed in metastatic lesions compared to primary tumours. This is critical as it demonstrates that loss of RECK in metastases is not simply due to sporadic differences between primary tumours but occurs during metastatic progression. Furthermore, since tumour heterogeneity often exists within breast tumours, it would be interesting to determine whether cells with the lowest levels of RECK expression within an individual primary tumour are in fact the founder cells in distant metastases.
Preclinical data and retrospective human studies suggest that MSGs have the potential to improve patient selection for therapy and may predict response to treatment (34). For example, mechanistic insights into the function of the MSG NM23 have led to the development of novel treatment strategies both exploiting aberrant NM23-associated cell signaling and reconstituting NM23 itself. Clinical trials using NM23 as an anti-metastasis therapy are currently underway (3). Notably, It has been shown in vitro that treating cell lines harboring epigenetically silenced RECK with HDAC or DNA methyltransferase inhibitors can lead to the re-expression of RECK (35, 36). In our study, RECK reconstitution in breast cancer cells significantly decreased metastatic burden in multiple in vivo models of metastasis. As such, it may be reasonable to explore new treatment strategies aimed at treating tumours with RECK silencing with epigenetically targeted therapies.
RECK not only affects invasive potential of cancer cells but its loss also supports the survival and expansion of tumour cells in distant organs and tissues. Angiogenesis is a hallmark of cancer, wound healing and various ischemic and inflammatory diseases (37, 38). VEGF is a potent inducer of angiogenesis and lymphangiogenesis, and is a highly specific mitogen for endothelial cells (39). Here, we reveal that RECK directly modulates VEGF secretion. Furthermore, RECK convergently regulates multiple pro-and anti-angiogenic factors in a coordinated fashion. These data therefore provide detailed mechanistic insights into the RECK-mediated angiogenic phenotype (40).
Our data reveal that RECK functions by regulating STAT3 activity. STAT3 activation has been linked to VEGF expression and oncogenesis but the mechanisms underlying this hyperactivity are not well understood. (41, 42). Two primary transcription factors that directly regulate VEGF expression are STAT3 and HIF1a (43). Induction of VEGF can be achieved by these transcription factors through diverse stimuli, including hypoxia and cytokines. Our data show that RECK binds to many receptors that are known activators of STAT3 signaling and angiogenesis, including IL-8, galectin-1, B1-integrin, uPAR and the primary activators of STAT3 signaling in breast cancer, IL-6RA and gp130 (23, 44–48). Interestingly, it has previously been reported that the inhibition of gp130 signaling in breast cancer blocks STAT3 activation, inhibiting malignancy in vivo (49). It seems that RECK plays a central role in coordinating and downregulating the extent of STAT3 activation downstream of these various receptors. This activity, in turn, regulates neovascular programs that are important for metastases. Of note, STAT3 can be activated by phosphorylation at Y705 and/or S727. pSTAT3-Y705, which was assessed in this study, is the STAT3 phosphorylation site that is associated with IL-6 and IL-8 signaling(50, 51). In contrast, STAT3-S727 phosphorylation is associated with cellular proliferation via ERK signaling and has been shown to inversely correlate with pSTAT3-Y705 levels(52–55). Here we show that RECK knockdown results in increased pSTAT3-Y705 and decreased pSTAT3-S727 (Supplementary Figure. 6A,B). This further supports our model of RECK mediated STAT3 regulation of metastatic processes via inhibition of IL-mediated STAT3 phosphorylation.
Interestingly, studies focusing on microRNA expression and metastasis have revealed that STAT3 can control microRNAs, which in turn regulates RECK expression (56). These data combined with the findings in our study point to a feedback loop between RECK, STAT3 and microRNAs. Of note, our study did not see changes in tumorigenecity or primary tumor growth by directly modulating RECK expression. In contrast, studies on microRNAs that modulate RECK do report changes in tumorigenecity, suggesting that RECK expression is just one of the many potential targets of microRNAs(57).
In our study, we showed that RECK mediated suppression of spontaneous metastasis in vivo, which suggests that RECK can affect multiple steps in the metastatic process. Interestingly, RECK suppression was concomitant with increased expression of uPA, a known inducer of breast cancer metastasis. In vitro, it has been shown that uPA itself has the capacity to increase the invasiveness of tumour cells (58), which we have confirmed here using breast cancer cell lines. The urokinase plasminogen activator system is a serine protease family that includes uPA, uPAR and plasminogen activator inhibitors (PAIs) (59). uPA catalyzes the activation of plasminogen to plasmin, which degrades the ECM and basement membranes either directly, or indirectly through activating pro-MMPs (as most MMPs are secreted as latent zymogens) (60, 61). This uPA-RECK relationship may partially explain how most tumours and tumour cell lines secrete very low levels of MMPs, yet retain strong invasive potential. Although MMPs have a clear role in metastasis, in vivo studies are complicated due to the presence of both inactive and active MMPs, whose conversion is catalyzed by other proteases. Further, the cellular origin of secreted MMPs in vivo is difficult to ascertain. As a result, we cannot exclude the importance of MMPs in our study, though our discovery of RECK-mediated uPA secretion provides both complimentary (uPA-induced MMP activation) and alternative (direct ECM proteolysis) mechanisms of RECK-associated suppression of metastasis.
In summary, RECK is a multifunctional and broadly important MSG that is frequently suppressed in cancer. Our observations address several ill-defined issues central to the development of metastasis: how a frequently inactivated MSG suppresses metastases, how metastases activate neovascular programs, what signaling mechanisms underlie RECK’s control of angiogenesis, and the molecular causes of STAT3 hyperactivity in metastatic lesions. Our work provides critical insights into our understanding of the metastatic process.
Methods
Expression Data Sets
The van de Vijver(62) and Stockholm (63) breast cancer datasets were downloaded from the Gene Expression Omnibus (GEO http://www.ncbi.nlm.nih.gov/geo). The Curtis dataset (30) and the TCGA breast cancer dataset (64) were downloaded from http://www.oncomine.org.
Primary Human Breast Cancer and Matched Lymph Node Metastases
Paired primary tumour and lymph node metastases were collected during surgical resection. All tissue was used with informed consent and banked at MSKCC in accordance with IRB approval. All samples were snap-frozen and stored at −80°C until use. All samples were H&E stained and independently reviewed by a breast cancer pathologist. Tumours were microdissected to obtain >70 % purity. RNA was isolated from fresh frozen samples using the RNEasy Plus™ mini prep kit (Qiagen). Nucleic acid quality was determined with the Agilent 2100 Bioanalyzer. Primary tumour and matched metastasis pairs, for which RNA of sufficient quantity and quality was available, were analyzed on Affymetrix GeneChip™ Human Genome U133 2.0 Array at the genomics core at Sloan-Kettering Institute according to the manufacturer’s protocol.
Primary Human Breast Cancer and Matched Distant Metastases
Paired primary tumour and distant metastases were collected with informed consent and banked at MSKCC in accordance with IRB approval. All samples were formalin-fixed and paraffin embedded until use. All samples were H&E stained and independently reviewed by a breast cancer pathologist. Samples were also sectioned (5μm) and immunostained with a RECK antibody and blindly scored.
Microarray Analysis of Cell Lines
Total RNA was isolated from cell lines using the RNEasy Plus™ mini prep kit (Qiagen). Nucleic acid quality was determined with the Agilent 2100 Bioanalyzer. Samples were analyzed on Affymetrix GeneChip™ Human Genome U133 2.0 Array at the genomics core at Sloan-Kettering Institute according to the manufacturer’s protocol.
Gene Expression Microarray Analysis
Gene expression analysis in cell lines was performed with the Human Genome U133A 2.0 microarray (Affymetrix). CEL files were imported into Partek Genomics Suite software and normalized using robust multiarray average (RMA) quartile normalization and log probe summarization. Differentially expressed genes were identified by analysis of variance (ANOVA). Genes that were differentially expressed at p<0.05 and had at least an absolute value of 1.2X fold change (1.5 Fold change for Hs343T) were considered significant. Enriched pathways were identified in the differentially expressed genes in Ingenuity Pathway Analysis (Ingenuity Systems) and determined significant at-log2(p-value)>1.3 (Supplementary Table 5-S8).
Animal Studies
All animal work was done in accordance with the Institutional Animal Care and Use Committee at MSKCC. Athymic Nu/Nu mice were obtained from Harlan Laboratories and NSG mice were obtained from Jackson Laboratories. All animals were injected between 5–7 weeks of age. Intracardiac injections of BOM1-1833 cells and intravenous injections of LM2-4175 cells were performed as previously described (13, 14). Single cell suspensions of indicated cell lines (1 × 106 cells) in 1:1 serum-free media/Matrigel™ (BD Biosciences) were injected orthotopically into the mammary fat pad of 6 week-old female NSG mice. Animals were imaged weekly via bioluminescence imaging (IVIS™, Xenogen).
Cell lines
Hs606.T and Hs343.T, HUVEC, HEK293T and HMEC-hTERT were purchased from The American Type Culture Collection (ATCC) and cultured according to ATCC recommended conditions. MDA-MB-231, LM2-4175 and BOM1-1833 were all generous gifts from Dr. Joan Massague (Memorial Sloan-Kettering Cancer Center, New York) and were cultured as described previously (13, 14).
Cell proliferation Assay
1 X 105 cells were plated in 6 cm plates in triplicate, and cell numbers were counted at indicated days using a ViCELL™ cell counter (Beckman).
Cell cycle Analysis
Cells were trypsinized, fixed with ice cold 70% ethanol and stained using 7-AAD. Cell cycle analysis was performed on a FACSCalibur™ (BD Biosciences). Analysis was performed using FlowJo software and cell cycle quantification was performed using the Watson (Pragmatic) model.
Immunoblotting
Cell lysates were prepared with CellLytic M™ (Sigma) containing protease and phosphotase inhibitor (Thermo Scientific). Debris was removed via centrifugation for 10 min at 14,000 r.p.m. in an Eppendorf Centrifuge 5424C at 4°C. Protein concentrations were determined by BCA protein assay kit (Thermo Scientific), and equal amounts of protein samples were loaded on NuPAGE™ Bis-Tris 4–12% SDS gels (Invitrogen) and blotted onto PVDF membrane (Millipore). Imaging was performed on a FujiFilm LAS4000. All blots were repeated 3 times and representative images were chosen for publication.
Chemotaxis Assay
Conditioned media from indicated cell types was placed in the bottom chamber of a 12-well plate. HUVEC cells were seeded at a density of 5 × 104/well in transwell inserts placed in the upper chamber (8 μm, BD Biosciences). After incubation for 12 hr, migrated cells were quantified by counting on a brightfield microscope.
ELISA
All ELISA kits were purchased from R&D Systems and performed according to manufacturer’s instructions. For isolation of conditioned media, equal number of cells was seeded and media was conditioned overnight. To remove cellular contaminant, conditioned media was collected and centrifuged at 300 x g for 10 min in an Eppendorf Centrifuge 5424C at 4°C.
Antibody Arrays
Proteome Profiler™ antibody arrays were purchased from R&D Systems and used according to manufacturer instructions. Conditioned media was generated as described above (ELISA). Imaging was performed on a FujiFilm LAS4000. Quantification of densitometry was achieved using ImageQuantTL software.
Virus Production and Generation of Stable Cell Lines
HEK 293T cells were transfected with lentiviral expression constructs using Fugene 6 (Promega) in combination with pCMV-dR8.2 (gag/pol)and VSV-G (env) expression vectors using Fugene 6 (Promega). Viral stocks were collected 48 hr post-transfection, filtered (0.45 μm) and placed on target cells for 8 h in the presence of 8 μg/ml polybrene. 48 hr post-infection, cells were selected in the presence of puromycin (2 μg/ml).
RNA Interference
Knock down using scramble and/or gene-specific siRNA (ON-TARGET SMARTpool™, Thermo Scientific) was performed using Lipofectamine RNAiMAX™ (Invitrogen). Stable knock down was achieved using scramble and RECK-specific shRNA constructs (Thermo Scientific) to generate virus as above (Virus Production).
Chemotaxis Assays
HUVEC cells were seeded on 12-well Boyden transwells in serum-free media (8 μm, BD Biosciences). Cells were allowed to migrate towards indicated conditioned media for 24 hours. Cells that did not migrate were removed using a cotton swab. The membranes were then fixed with methanol and stained with DAPI. The number of migrated cells was determined by manual counting using an inverted Nikon microscope.
Invasion Assays
Indicated cells were seeded in serum-free media on Matrigel™-coated transwell inserts (8 μm, BD Biosciences). Complete media was placed below transwell inserts to encourage cell invasion. After 24 hours, cells that did not invade were removed using a cotton swab. Membranes were then fixed with methanol and stained with DAPI. The number of invaded cells was determined by manual counting using an inverted Nikon fluorescent microscope.
Immunohistochemistry and Analysis
The immunohistochemical detection of indicated antibodies was performed at Molecular Cytology Core Facility of Memorial Sloan Kettering Cancer Center using Discovery XT processor (Ventana Medical Systems). The tissue sections were deparaffinized with EZPrep buffer (Ventana Medical Systems), antigen retrieval was performed with CC1 buffer (Ventana Medical Systems) and sections were blocked for 30 minutes with Background Buster solution (Innovex)(for rabbit, mouse and chicken Abs, for rat and goat Abs it should be 10% normal rabbit serum in PBS+2%BSA). Antibodies were applied and sections were incubated for 5 hours, followed by 60 minutes incubation with biotinylated goat anti-rabbit IgG (Vector labs, cat#PK6101) (biotinylated horse anti-mouse IgG (Vector Labs, cat# MKB-22258); biotinylated goat anti-chicken (Vector Labs, cat#BA-9010); biotinylated rabbit anti-goat IgG (Vector, cat #BA-5000)) at 1:200 dilution. The detection was performed with DAB detection kit (Ventana Medical Systems) according to manufacturer instruction. Slides were counterstained with hematoxylin and coverslipped with Permount (Fisher Scientific). Staining quantification was performed using MetaMorph Software.
Immunofluorescence
Cells were grown as monolayer cultures on D-polylysine-coated coverslips and were fixed with 4% PFA. After washing 3 times with PBS, cells were blocked for 30mins with 10% goat serum and incubated overnight at 4°C with primary antibodies against RECK (1:50, R&D). Cells were then stained with Alexa Fluor 563 secondary antibody (1:200, Invitrogen) for 1 hr at room temperature..Stained sections and cells were mounted in ProLong Gold™ antifade reagent (Invitrogen) with DAPI. Images were taken on a Leica TCS SP5 confocal microscope.
Quantitative Real-time PCR
Total RNA was isolated from cells using RNEasy Plus™ according to manufacturer instructions (Qiagen). cDNA was generated using 1 ug RNA and cDNA EcoDry Premix (Clontech). qRT-PCR was performed using a Realplex2 Mastercycler (Eppendorf) and analyzed by standard delta relative quantitation (ΔΔRQ). qRT-PCR parameters were as follows: initial denaturation at 95°C for 5 minutes, then 40 cycles of 95°C 15 seconds and 60°C 1 minute. Primers used: RECK – Forward 5′TGCAAGCAGGCATCTTCAAA-3′ Reverse 5′ACCGAGCCCATTTCATTTCTG-3′; GAPDH – Forward 5′-AATGAAGGGGTCATTGATGG-3′ Reverse5′-AAGGTGAAGGTCGGAGTCAA-3′
Endothelial Tube Formation Assay
Endothelial cells (HUVEC) were resuspended in indicated conditioned media and grown for 6 hr on Matrigel™ (BD biosciences). Tube formation was quantified by calculating the average number of branch points in 10 fields of view. Imaging was performed using a Nikon TE2000-E inverted microscope. Representative images were chosen for publication from 3 replicates.
Immunoprecipitation
For immunoprecipitation, 50 uL of protein G magnetic beads (PureProteome™, Millipore) were crosslinked with 10 ug anti-RECK goat polyclonal antibody (R&D) or goat polyclonal IgG antibody (R&D) using 5 mM BS3 according to manufacturer instructions (protocol guide, Millipore). Confluent Hs606.T whole cell lysates (500 ug total protein) were incubated with crosslinked beads at 4°C overnight with gentle agitation. The immunocomplexes were washed 3 times with TBS-T (0.1%), bound protein eluted in 60 uL 0.2 M glycine-HCl (pH 2.5), and neutralized using 5 uL of 1 M Tris (pH 8.5). Eluates were then used to probe an antibody array (Human Soluble Receptor Antibody Array, Non-Hematopoietic Panel, R&D). This array was modified by using IP eluate instead of conditioned media or cell lysate. Eluates were also subjected to SDS-PAGE and Western blotting analysis for RECK (goat antibody, R&D), uPAR (goat antibody, R&D), Galectin-1 (rabbit antibody, Cell Signaling), and Integrin β1 (rabbit antibody, Cell Signaling).
Statistical Analysis
For survival analysis, survival curves were analyzed using the Kaplan-Meier method and log rank test with IBM Statistica 20 software. All other statistical analysis was performed using Prism 6 Software (GraphPad Software).
Kinexus Antibody Microarray
Protein lysates were sent to Kinexus Bioinformatics Corporation and profiled using their Kinex™ 850 Antibody Microarray (KAM). All samples were normalized according to Kinexus normalization guidelines and a list of downregulated candidate proteins was generated.
Antibodies
| Antibody | Supplier | Cat. No |
|---|---|---|
| Actin | Sigma | A3853 |
| CD34 | abCAM | ab8158 |
| Galectin-1 | Cell Signaling | 5418S |
| Ki67 | abCAM | ab16667 |
| MMP-2 | R&D | AF902 |
| MMP-9 | R&D | AF911 |
| MT1-MMP | R&D | AF918 |
| pSTAT3 (Tyr705) | Cell Signaling | 9145S |
| pSTAT3 (Ser727) | Cell Signaling | 9136 |
| RECK | R&D | AF1734 |
| STAT3 | Cell Signaling | 9139S |
| uPA | R&D | AF1310 |
| uPAR | R&D | AF807 |
| VEGF | Santa Cruz | sc-507 |
| β1-Integrin | Cell Signaling | 4706P |
Microarray Accession Numbers
Gene Expression Omnibus (GEO) - accession numbers GSE56898.
Supplementary Material
Supplementary Table 1. Differentially expressed genes in MDA-MB-231+EV vs MDA-MB-231+RECK
Supplementary Table 2. Differentially expressed genes in LM2-4175+EV vs LM2-4175+RECK
Supplementary Table 3. Differentially expressed genes in Hs343T siSCR vs Hs343T siRECK
Supplementary Table 4. Differentially expressed genes in Hs606T siSCR vs Hs606T siRECK
Supplementary Table 5. Results of pathway analyses, based on genes differentially expressed after RECK overexpression in MDA-MB-231 cells
Supplementary Table 6. Results of pathway analyses, based on genes differentially expressed after RECK overexpression in LM2-4175 cells
Supplementary Table 7. Results of pathway analyses, based on genes differentially expressed after RECK knockdown in Hs343T cells
Supplementary Table 8. Results of pathway analyses, based on genes differentially expressed after RECK knockdown in Hs606T cells
Supplementary Table 9. IPA significantly altered pathways common to all 4 cell lines
Acknowledgments
We thank Agnes Viale and Russell Towers for technical assistance. We are indebted to Joan Massague (Memorial Sloan-Kettering Cancer Center) for generously providing cell lines and some constructs. We are grateful to the entire Tumour Procurement Service at MSKCC for facilitating tissue procurement and contributing valuable input. This work was supported by grants from the Department of Defense (DOD) (grant no. BC120568) (T.A.C), the Frederick Adler Fund (T.A.C.), the Avon Foundation (T.A.C.), the Elsa Pardee foundation (T.A.C.), the MSKCC Metastasis Center (T.A.C.), and the STARR Cancer Consortium (T.A.C.). L.A.W. was supported by The Canadian Institutes of Health Research PDF Award MFE-127325. D.M.R. was supported by the HHMI Research Fellows Program and NIH MSTP grant T32GM007739. A.S. was supported by the Ruth L. Kirschstein National Research Award (NRSA) Number T32CA009512 from the National Cancer Institute. S. T. was supported by an NIH T32 grant (5T32CA160001).
Footnotes
Author Contributions
L.A.W and D.M.R designed, performed and analyzed the experiments and aided in writing the manuscript. M.R. D.G. A.S. S.T. C.A.B. J.L. J.B. and T.A.K. contributed to the completion of various experiments. L.A.W and T.A.C. conceived the study, supervised the research and contributed to writing the manuscript.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Advances in experimental medicine and biology. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso F, Harbeck N, Fallowfield L, Kyriakides S, Senkus E, Group EGW. Locally recurrent or metastatic breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2012 Oct;23(Suppl 7):vii11–9. doi: 10.1093/annonc/mds232. [DOI] [PubMed] [Google Scholar]
- 3.Shoushtari AN, Szmulewitz RZ, Rinker-Schaeffer CW. Metastasis-suppressor genes in clinical practice: lost in translation? Nature reviews Clinical oncology. 2011 Jun;8(6):333–42. doi: 10.1038/nrclinonc.2011.65. [DOI] [PubMed] [Google Scholar]
- 4.Noda M, Takahashi C, Matsuzaki T, Kitayama H. What we learn from transformation suppressor genes: lessons from RECK. Future oncology. 2010 Jul;6(7):1105–16. doi: 10.2217/fon.10.80. [DOI] [PubMed] [Google Scholar]
- 5.Rabien A, Ergun B, Erbersdobler A, Jung K, Stephan C. RECK overexpression decreases invasive potential in prostate cancer cells. The Prostate. 2012 Jun 15;72(9):948–54. doi: 10.1002/pros.21498. [DOI] [PubMed] [Google Scholar]
- 6.Chang CK, Hung WC, Chang HC. The Kazal motifs of RECK protein inhibit MMP-9 secretion and activity and reduce metastasis of lung cancer cells in vitro and in vivo. Journal of cellular and molecular medicine. 2008 Dec;12(6B):2781–9. doi: 10.1111/j.1582-4934.2008.00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh J, Takahashi R, Kondo S, Mizoguchi A, Adachi E, Sasahara RM, et al. The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell. 2001 Dec 14;107(6):789–800. doi: 10.1016/s0092-8674(01)00597-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Ling Y, Zhang C, Xu Y, Gao L, Li R, et al. The silencing of RECK gene is associated with promoter hypermethylation and poor survival in hepatocellular carcinoma. International journal of biological sciences. 2012;8(4):451–8. doi: 10.7150/ijbs.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang HC, Cho CY, Hung WC. Silencing of the metastasis suppressor RECK by RAS oncogene is mediated by DNA methyltransferase 3b-induced promoter methylation. Cancer research. 2006 Sep 1;66(17):8413–20. doi: 10.1158/0008-5472.CAN-06-0685. [DOI] [PubMed] [Google Scholar]
- 10.Hill VK, Ricketts C, Bieche I, Vacher S, Gentle D, Lewis C, et al. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer research. 2011 Apr 15;71(8):2988–99. doi: 10.1158/0008-5472.CAN-10-4026. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa S, Takenaka K, Yanagihara K, Miyahara R, Kawano Y, Otake Y, et al. Matrix metalloproteinase-2 status in stromal fibroblasts, not in tumor cells, is a significant prognostic factor in non-small-cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004 Oct 1;10(19):6579–85. doi: 10.1158/1078-0432.CCR-04-0272. [DOI] [PubMed] [Google Scholar]
- 12.Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncology reports. 2009 May;21(5):1323–33. doi: 10.3892/or_00000358. [DOI] [PubMed] [Google Scholar]
- 13.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer cell. 2003 Jun;3(6):537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 14.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005 Jul 28;436(7050):518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, Zhang J, Xu ZY, Xie HL. Expression of RECK and matrix metalloproteinase-2 in ameloblastoma. BMC cancer. 2009;9:427. doi: 10.1186/1471-2407-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardeal LB, Boccardo E, Termini L, Rabachini T, Andreoli MA, di Loreto C, et al. HPV16 oncoproteins induce MMPs/RECK-TIMP-2 imbalance in primary keratinocytes: possible implications in cervical carcinogenesis. PloS one. 2012;7(3):e33585. doi: 10.1371/journal.pone.0033585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Q, Yu D, Yang CM, Jiang B, Zhang H. Expression of the reversion-inducing cysteine-rich protein with Kazal motifs and matrix metalloproteinase-14 in neuroblastoma and the role in tumour metastasis. International journal of experimental pathology. 2010 Aug;91(4):368–73. doi: 10.1111/j.1365-2613.2010.00724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zarogoulidis P, Yarmus L, Darwiche K, Walter R, Huang H, Li Z, et al. Interleukin-6 cytokine: a multifunctional glycoprotein for cancer. Immunome research. 2013 Aug 12;9(62):16535. doi: 10.1186/2090-5009-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenkilde MM, Schwartz TW. The chemokine system -- a major regulator of angiogenesis in health and disease. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2004 Jul-Aug;112(7–8):481–95. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- 20.Zeng L, O’Connor C, Zhang J, Kaplan AM, Cohen DA. IL-10 promotes resistance to apoptosis and metastatic potential in lung tumor cell lines. Cytokine. 2010 Mar;49(3):294–302. doi: 10.1016/j.cyto.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Tang L, Han X. The urokinase plasminogen activator system in breast cancer invasion and metastasis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2013 Mar;67(2):179–82. doi: 10.1016/j.biopha.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011 Mar 25;331(6024):1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 23.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast cancer research : BCR. 2007;9(3):R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bournazou E, Bromberg J. Targeting the tumor microenvironment: JAK-STAT3 signaling. Jak-Stat. 2013 Apr 1;2(2):e23828. doi: 10.4161/jkst.23828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Xie K. Crosstalk of Sp1 and Stat3 signaling in pancreatic cancer pathogenesis. Cytokine & growth factor reviews. 2012 Feb-Apr;23(1–2):25–35. doi: 10.1016/j.cytogfr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JH, Lee SC, Ro J, Kang HS, Kim HS, Yoon S. Jnk signaling pathway-mediated regulation of Stat3 activation is linked to the development of doxorubicin resistance in cancer cell lines. Biochemical pharmacology. 2010 Feb 1;79(3):373–80. doi: 10.1016/j.bcp.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Benasciutti E, Pages G, Kenzior O, Folk W, Blasi F, Crippa MP. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter element and endogenous gene transcription. Blood. 2004 Jul 1;104(1):256–62. doi: 10.1182/blood-2003-08-2661. [DOI] [PubMed] [Google Scholar]
- 28.Jung JE, Lee HG, Cho IH, Chung DH, Yoon SH, Yang YM, et al. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005 Aug;19(10):1296–8. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]
- 29.Kang SH, Yu MO, Park KJ, Chi SG, Park DH, Chung YG. Activated STAT3 regulates hypoxia-induced angiogenesis and cell migration in human glioblastoma. Neurosurgery. 2010 Nov;67(5):1386–95. doi: 10.1227/NEU.0b013e3181f1c0cd. discussion 95. [DOI] [PubMed] [Google Scholar]
- 30.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012 Jun 21;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zucker S, Cao J, Chen WT. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene. 2000 Dec 27;19(56):6642–50. doi: 10.1038/sj.onc.1204097. [DOI] [PubMed] [Google Scholar]
- 32.Prendergast A, Linbo TH, Swarts T, Ungos JM, McGraw HF, Krispin S, et al. The metalloproteinase inhibitor Reck is essential for zebrafish DRG development. Development. 2012 Mar;139(6):1141–52. doi: 10.1242/dev.072439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandana EP, Maeda Y, Ueda A, Kiyonari H, Oshima N, Yamamoto M, et al. Involvement of the Reck tumor suppressor protein in maternal and embryonic vascular remodeling in mice. BMC developmental biology. 2010;10:84. doi: 10.1186/1471-213X-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prabhu VV, Siddikuzzaman, Grace VM, Guruvayoorappan C. Targeting tumor metastasis by regulating Nm23 gene expression. Asian Pacific journal of cancer prevention : APJCP. 2012;13(8):3539–48. doi: 10.7314/apjcp.2012.13.8.3539. [DOI] [PubMed] [Google Scholar]
- 35.Jeon HW, Lee YM. Inhibition of histone deacetylase attenuates hypoxia-induced migration and invasion of cancer cells via the restoration of RECK expression. Molecular cancer therapeutics. 2010 May;9(5):1361–70. doi: 10.1158/1535-7163.MCT-09-0717. [DOI] [PubMed] [Google Scholar]
- 36.Cho CY, Wang JH, Chang HC, Chang CK, Hung WC. Epigenetic inactivation of the metastasis suppressor RECK enhances invasion of human colon cancer cells. Journal of cellular physiology. 2007 Oct;213(1):65–9. doi: 10.1002/jcp.21089. [DOI] [PubMed] [Google Scholar]
- 37.Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA. Vascular endothelial growth factor and angiogenesis. Pharmacological reviews. 2004 Dec;56(4):549–80. doi: 10.1124/pr.56.4.3. [DOI] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Byrne AM, Bouchier-Hayes DJ, Harmey JH. Angiogenic and cell survival functions of vascular endothelial growth factor (VEGF) Journal of cellular and molecular medicine. 2005 Oct-Dec;9(4):777–94. doi: 10.1111/j.1582-4934.2005.tb00379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasahara RM, Brochado SM, Takahashi C, Oh J, Maria-Engler SS, Granjeiro JM, et al. Transcriptional control of the RECK metastasis/angiogenesis suppressor gene. Cancer detection and prevention. 2002;26(6):435–43. doi: 10.1016/s0361-090x(02)00123-x. [DOI] [PubMed] [Google Scholar]
- 41.Carbajo-Pescador S, Ordonez R, Benet M, Jover R, Garcia-Palomo A, Mauriz JL, et al. Inhibition of VEGF expression through blockade of Hif1alpha and STAT3 signalling mediates the anti-angiogenic effect of melatonin in HepG2 liver cancer cells. British journal of cancer. 2013 Jul 9;109(1):83–91. doi: 10.1038/bjc.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bid HK, Oswald D, Li C, London CA, Lin J, Houghton PJ. Anti-angiogenic activity of a small molecule STAT3 inhibitor LLL12. PloS one. 2012;7(4):e35513. doi: 10.1371/journal.pone.0035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005 Aug 25;24(36):5552–60. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 44.Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nature reviews Cancer. 2008 Aug;8(8):604–17. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shain KH, Yarde DN, Meads MB, Huang M, Jove R, Hazlehurst LA, et al. Beta1 integrin adhesion enhances IL-6-mediated STAT3 signaling in myeloma cells: implications for microenvironment influence on tumor survival and proliferation. Cancer research. 2009 Feb 1;69(3):1009–15. doi: 10.1158/0008-5472.CAN-08-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Haene N, Sauvage S, Maris C, Adanja I, Le Mercier M, Decaestecker C, et al. VEGFR1 and VEGFR2 involvement in extracellular galectin-1- and galectin-3-induced angiogenesis. PloS one. 2013;8(6):e67029. doi: 10.1371/journal.pone.0067029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le QT, Shi G, Cao H, Nelson DW, Wang Y, Chen EY, et al. Galectin-1: a link between tumor hypoxia and tumor immune privilege. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005 Dec 10;23(35):8932–41. doi: 10.1200/JCO.2005.02.0206. [DOI] [PubMed] [Google Scholar]
- 48.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008 Nov 1;14(21):6735–41. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 49.Selander KS, Li L, Watson L, Merrell M, Dahmen H, Heinrich PC, et al. Inhibition of gp130 signaling in breast cancer blocks constitutive activation of Stat3 and inhibits in vivo malignancy. Cancer research. 2004 Oct 1;64(19):6924–33. doi: 10.1158/0008-5472.CAN-03-2516. [DOI] [PubMed] [Google Scholar]
- 50.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer research. 2003 Jun 1;63(11):2948–56. [PubMed] [Google Scholar]
- 51.Lee MM, Chui RK, Tam IY, Lau AH, Wong YH. CCR1-mediated STAT3 tyrosine phosphorylation and CXCL8 expression in THP-1 macrophage-like cells involve pertussis toxin-insensitive Galpha(14/16) signaling and IL-6 release. Journal of immunology. 2012 Dec 1;189(11):5266–76. doi: 10.4049/jimmunol.1103359. [DOI] [PubMed] [Google Scholar]
- 52.Chung J, Uchida E, Grammer TC, Blenis J. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Molecular and cellular biology. 1997 Nov;17(11):6508–16. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onishi A, Chen Q, Humtsoe JO, Kramer RH. STAT3 signaling is induced by intercellular adhesion in squamous cell carcinoma cells. Experimental cell research. 2008 Jan 15;314(2):377–86. doi: 10.1016/j.yexcr.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu FM, Li QL, Gao Q, Jiang JH, Huang XY, Pan JF, et al. Sorafenib inhibits growth and metastasis of hepatocellular carcinoma by blocking STAT3. World journal of gastroenterology : WJG. 2011 Sep 14;17(34):3922–32. doi: 10.3748/wjg.v17.i34.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang G, Yan H, Ye S, Tong C, Ying QL. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ES cell fates. Stem cells. 2013 Dec 3; doi: 10.1002/stem.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin HY, Chiang CH, Hung WC. STAT3 upregulates miR-92a to inhibit RECK expression and to promote invasiveness of lung cancer cells. British journal of cancer. 2013 Aug 6;109(3):731–8. doi: 10.1038/bjc.2013.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han L, Yue X, Zhou X, Lan FM, You G, Zhang W, et al. MicroRNA-21 expression is regulated by beta-catenin/STAT3 pathway and promotes glioma cell invasion by direct targeting RECK. CNS neuroscience & therapeutics. 2012 Jul;18(7):573–83. doi: 10.1111/j.1755-5949.2012.00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thummarati P, Wijitburaphat S, Prasopthum A, Menakongka A, Sripa B, Tohtong R, et al. High level of urokinase plasminogen activator contributes to cholangiocarcinoma invasion and metastasis. World journal of gastroenterology : WJG. 2012 Jan 21;18(3):244–50. doi: 10.3748/wjg.v18.i3.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Current pharmaceutical design. 2004;10(1):39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y, Lyons CE, Jr, Xiao A, Templeton DJ, Sang QA, Brew K, et al. Urokinase directly activates matrix metalloproteinases-9: a potential role in glioblastoma invasion. Biochemical and biophysical research communications. 2008 May 16;369(4):1215–20. doi: 10.1016/j.bbrc.2008.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choong PF, Nadesapillai AP. Urokinase plasminogen activator system: a multifunctional role in tumor progression and metastasis. Clinical orthopaedics and related research. 2003 Oct;(415 Suppl):S46–58. doi: 10.1097/01.blo.0000093845.72468.bd. [DOI] [PubMed] [Google Scholar]
- 62.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. The New England journal of medicine. 2002 Dec 19;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 63.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast cancer research : BCR. 2005;7(6):R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012 Oct 4;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Differentially expressed genes in MDA-MB-231+EV vs MDA-MB-231+RECK
Supplementary Table 2. Differentially expressed genes in LM2-4175+EV vs LM2-4175+RECK
Supplementary Table 3. Differentially expressed genes in Hs343T siSCR vs Hs343T siRECK
Supplementary Table 4. Differentially expressed genes in Hs606T siSCR vs Hs606T siRECK
Supplementary Table 5. Results of pathway analyses, based on genes differentially expressed after RECK overexpression in MDA-MB-231 cells
Supplementary Table 6. Results of pathway analyses, based on genes differentially expressed after RECK overexpression in LM2-4175 cells
Supplementary Table 7. Results of pathway analyses, based on genes differentially expressed after RECK knockdown in Hs343T cells
Supplementary Table 8. Results of pathway analyses, based on genes differentially expressed after RECK knockdown in Hs606T cells
Supplementary Table 9. IPA significantly altered pathways common to all 4 cell lines