Abstract
The extraordinary muscle growth potential of teleost fish, particular those of the Salmoninae clade, elicits questions about how the relatively highly conserved transcription factors of the myogenic program are regulated. In addition, the pseudotetraploid nature of the salmonid genome adds another layer of regulatory complexity, and this must be reconciled with epigenetic data to better understand how these fish achieve lifelong muscle growth. To this end, we identified three paralogous pax7 genes (pax7a1, pax7a2, and pax7b) in the rainbow trout genome. During in vitro myogenesis, pax7a1 transcripts remain stable, while pax7a2 and pax7b mRNAs increase in abundance, similarly to myogenin mRNAs and in contrast to the expression pattern of the mammalian ortholog. In addition, we profiled the distribution of repressive H3K27me3 and H3K9me3 and permissive H3K4me3 marks during in vitro myogenesis across these loci, finding that pax7a2 expression was associated with decreased H3K27 trimethylation, while pax7b expression was correlated with decreased H3K9me3 and −K27me3. Altogether, these data link the highly unique differential expression of pax7 paralogs with epigenetic histone modifications in a vertebrate species displaying growth divergent from that of mammals and highlight an important divergence in the regulatory mechanisms of pax7 expression among vertebrates. The system described here provides a more comprehensive picture of the combinatorial control mechanisms orchestrating skeletal muscle growth in a salmonid, leading to a better understanding of myogenesis in this species and across Vertebrata more generally.
Keywords: pax7, epigenetic regulation, histone methylation, ChIP, rainbow trout
INTRODUCTION
Many teleost fish possess the extraordinary ability to augment their skeletal musculature throughout life (termed indeterminate muscle growth) as a result of the continuous formation of new fibers (myofiber hyperplasia) and increased size by hypertrophy of existing myofibers (Mommsen 2001, Johnston et al. 2011). The muscle stem cells (called myosatellite cells, MSCs) are thought to be essential for these two processes (Lepper et al. 2011, Sambasivan et al. 2011). Upon commitment, these cells engage into a myogenic program, allowing them to proliferate, differentiate, and fuse with existing myofibers (hypertrophy) or with other myoblasts to form new myofibers (hyperplasia). It is assumed that MSCs are controlled by highly conserved molecular networks that regulate myogenesis across growth paradigm and taxa (determinate and indeterminate growth; mammalian, fish, and even invertebrate clades; Konstantinides and Averof 2014), but that these networks vary enough in expression patterns to allow MSCs from indeterminate growing organisms to continuously contribute to either hyperplasia and/or hypertrophy in the adult animal.
In this regard, we have shown that MSCs isolated from rainbow trout (an indeterminate growing fish) enter into the differentiation program by very quickly expressing the myogenic regulatory factor (MRF) transcription factors myoD1 and myogenin (Gabillard et al. 2010). Interestingly, an “unexpected” increase of pax7 expression (a well-accepted marker of quiescent MSCs in mammals) in culture of MSCs under differentiation into myotubes has been documented in a closely related species (Salmo salar), possibly reflecting the production of “quiescent reserve cells” (Bower and Johnston 2010). Likewise, we recently demonstrated that MSCs from the indeterminately growing giant danio (Devario aequipinnatus) in culture gradually increase pax3 expression (another marker of quiescent MSCs in mammals) during a similar progression to myotubes (Froehlich et al. 2013). These data support the hypothesis that the induced expression of these specific paired-box transcription factors (pax3/7) during the culture of MSCs is a common feature of indeterminate growing species and could indicate a high self-renewal of MSCs, a process needed for these cells to continuously contribute to hyperplasia and/or hypertrophy during the lifespan of these fishes (Froehlich et al. 2013). However, very few data exist detailing how the expression of these Pax and MRF transcription factors is regulated in indeterminate growing organisms.
In mammals, the regulation of several genes involved in MSC activity has been shown to be under the strict control of epigenetic mechanisms through histone methylation (Perdiguero et al. 2009, Saccone and Puri 2010). The consequences of histone methylation on transcriptional activation or repression depend on the site and degree of methylation. Histone lysine methylation can occur on histones H3 and H4 in monomethyl, dimethyl, or trimethyl states. Trimethylation of lysine 4, lysine 36, or lysine 79 of histone H3 (H3K4, H3K36, or H3K79) is generally associated with euchromatin and transcriptional activity (i.e., a ‘permissive’ state; Wang et al. 2001, Feng et al. 2002, Krogan et al. 2003, Krogan et al. 2003). Conversely, di- or trimethylation of lysine 9 or lysine 27 of histone H3 (H3K9 or H3K27) and trimethylation of lysine 20 on histone H4 (H4K20) are correlated with heterochromatin formation and transcriptional repression (Rea et al. 2000, Nakayama et al. 2001, Tachibana et al. 2001, Cao et al. 2002, Schultz et al. 2002, Schotta et al. 2004). Histone methylation, however, is not a permanent state. Thus, genomic regions are subjected to the opposing actions of methylation and demethylation in a dynamic manner, thereby ensuring a transcriptional response to altered organismal and/or environmental conditions. In this regard, the coordination of methylation events at H3K4, H3K9, and H3K27 in the promoters of genes coding for various Pax and MRF transcription factors (pax3/7, myf5, myoD1, and myogenin) have been demonstrated to regulate many aspects of MSCs activation, proliferation, and differentiation in mammals (Sebastian et al. 2009, Palacios et al. 2010, Stojic et al. 2011, Taberlay et al. 2011, Tao et al. 2011, Diao et al. 2012, Kawabe et al. 2012, Ling et al. 2012, Sdek et al. 2013, Wang et al. 2013). However, no information detailing the epigenetic regulation of myogenesis in indeterminate growing organisms is available in the literature. In the present study, we therefore sought to profile the distribution of H3K27me3, H3K9me3 and H3K4me3 marks during the in vitro myogenesis in rainbow trout across loci with very different roles in the control of myogenesis: pax7, in the commitment and survival of MSCs; and myogenin, in the terminal differentiation of myotubes.
MATERIALS AND METHODS
In silico analysis of pax7 genes
The presence of pax7 genes was queried against the rainbow trout genome (Berthelot et al. 2014) with BLAST search in SIGENAE databases (http://www.sigenae.org/). The new sequences of rainbow trout pax7a1, pax7a2 and pax7b genes are available in Genoscope database (www.genoscope.cns.fr/trout) under the numbers GSONMG00081386001, GSONMG00061433001 and GSONMG00027288001, respectively. The phylogenetic analysis was carried out with Pax7 amino acid sequences available on Ensembl Genome database (Homo sapiens, Ensembl ID: ENSP00000403389; Mus musculus, Ensembl ID: ENSMUSP00000030508; Danio rerio, Ensembl ID: ENSDARP00000123695 and ENSDARP00000108935; Astyanax mexicanus, Ensembl ID: ENSAMXP00000003905 and ENSAMXP00000025517) and GenBank (Branchiostoma floridae, GenBank ID: AAF89581.1). Phylogenetic tree was constructed by the Neighbor-Joining (NJ) method using the Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0 (Tamura et al. 2013). The reliability of the inferred tree was estimated using the bootstrap method with 500 replications. The amphioxus Pax7 sequence was used to root the tree. The Genomicus software program (http://www.genomicus.biologie.ens.fr/genomicus-trout-01.01/cgi-bin/search.pl) was used to identify syntenic genes that were located near the pax7 genes of different vertebrate species.
Isolation of trout myosatellite cells
For all studies, MSCs were isolated from juvenile rainbow trout (Oncorhynchus mykiss) (15-45 g, depending on culture and availability) essentially as previously described (Froehlich et al. 2014). Isolated MSCs were plated on poly-L-lysine and laminin-coated plates at 200,000 cells per cm2 in 6-well (chromatin immunoprecipitation, ChIP; or real-time quantitative PCR, RT-qPCR) or 24-well (immunocytochemistry) plates and cultured in 10% FBS-DMEM for 4 days (with cell fixation on days 2 and 4) and 2% FBS-DMEM for an additional 4 days (with cell fixation on day 8 post-isolation). Day 2 of culture represents MSCs, day 4 myoblasts, and day 8 nascent myotubes as shown previously (Gabillard et al. 2010).
Analysis of cell proliferation
On days 2, 4, and 8 of culture, 10 μM BrdU was added and cells were cultured for the following 24 h. The cells were then fixed with ethanol/glycine buffer (70% ethanol, 50 mM glycine, pH 2). The proliferation of the cells was measured by using a BrdU labeling and detection kit (Roche Diagnostics, no. 1 296 736). Briefly, the cells were incubated for 30 min at 37 °C with mouse anti-BrdU, washed, and then incubated with the secondary antibody anti-mouse FITC for 30 min. Cells were then mounted in Mowiol containing 0.5 μg/ml DAPI.
Immunofluorescence analysis
Cells on glass coverslips were briefly washed twice with phosphate-buffered saline (PBS) and fixed for 10 min with 4% paraformaldehyde in PBS. For permeabilization, cells were incubated 3 min in 0.1% Triton X-100/PBS. After three washes, cells were saturated for 1 h with 3% BSA, 0.1% Tween-20 in PBS (PBST). Cells were incubated for 3 h with the primary antibodies anti-myosin heavy chain (MyHC) and anti-myogenin diluted in blocking buffer (Gabillard et al. 2010). The secondary antibodies were diluted in PBST and applied for 1 h. Cells were mounted with Mowiol 4-88 (#475904, Calbiochem) containing Hoescht (0.5 μg/ml). Cells were photographed using a Canon digital camera coupled to a Canon 90i microscope.
Gene expression analysis
Total RNA samples were extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's recommendations. One microgram of the resulting total RNA was reverse transcribed into cDNA using the SuperScript III RNaseH- Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA) and random primers (Promega, Charbonnières, France), according to the manufacturer's instructions. Target gene expression levels were determined by RT-qPCR using specific primers (Table 1). The primer sequences were chosen based on the rainbow trout genes sequence available in the expressed sequence transcript database (http://www.sigenae.org/). To avoid amplification of genomic DNA, when possible, primer pairs included one intron-spanning oligonucleotide. Real-time RT-PCR was carried out on an iCycler iQ™ real time PCR detection system (BIO-RAD, Hercules, CA, USA) using iQ™ SYBR® Green Supermix. Relative quantification of the target gene transcripts was made using EF1α gene expression as reference and following the Pfaffl method with the Relative Expression Software tool (REST©) (Pfaffl 2001, Pfaffl et al. 2002). PCR was performed using 10 μl of the diluted cDNA mixed with five picomoles of each primer in a final volume of 25 μl. The PCR protocol was initiated at 95°C for 3 min for initial denaturation of the cDNA and hot-start iTaq TM DNA polymerase activation followed by a two-step amplification program (20 sec at 95°C followed by 30 sec at specific primer hybridization temperature) repeated 40 times. Melting curves were systematically monitored (temperature gradient at 0.5°C/10 sec from 55 to 94°C) at the end of the last amplification cycle to confirm the specificity of the amplification reaction. The different PCR products were initially sequenced to confirm the identities of the amplicons. Each PCR run included replicate samples (duplicate of reverse transcription and duplicate of PCR amplification) and negative controls (reverse transcriptase-free samples, NRT; RNA-free samples, NTC).
Table 1.
Sequences of primer pairs used for real-time quantitative RT-PCR
| Target | Forward primer (5′–3′) | Reverse primer (5′–3′) | Accession #a |
|---|---|---|---|
| pax7a1 | TGGGACTACGATTTTATTGTCTCC | TCGTGCAAAGTCCAGACAAG | GSONMG00081386001 |
| pax7a2 | TGGGACTACGATTTATAGTTCGATTT | TTCTTACTCGCGCAAAGTCC | GSONMG00061433001 |
| pax7b | CGTCAAAACAATTACCACAAACA | AAAGACGACTGCATTCTACAGC | GSONMG00027288001 |
| myoD1 | CTGGACCCGAGACTCGTTCA | TTCCTGCGATCAGCATTGGT | X75798 |
| PCNA | CCACGTCTCCCTAGTTCAGC | TCCTGGTTGAGTGTCTCGAA | BT046966 |
| myogenin | CACTACATTG AGAGGCTGCAGGCA | CTCACTCGACGACGACACGCTG | Z46912 |
| EF1α | TCCTCTTGGTCGTTTCGCTG | ACCCGAGGGACATCCTGTG | AF498320.1 |
Accession numbers are from www.genoscope.cns.fr/trout and http://www.ncbi.nlm.nih.gov/
Chromatin Immunoprecipitation (ChIP)
On days 2, 4, and 8 of culture, MSCs, myoblasts, or nascent myotubes were fixed in 1% methanol-free formaldehyde (16% diluted in serum-free DMEM immediately prior to fixation) for 10 min at room temperature. Formaldehyde was neutralized by the addition of 2.5 M glycine for 5 min. Fixed cells were then washed twice in ice-cold 0.01 M PBS. Next, 1 mL of ice-cold 0.01 M PBS containing protease inhibitors (HALT™; Pierce) was added, and three wells were pooled for each ChIP sample (i.e., negative/mock ChIP control, total H3, H3K4me3, H3K9me3, and H3K27me3) by scraping cells into a microcentrifuge tube and pelleting cells at 3000×g for 5 min. Samples were stored at −80°C until preparation of chromatin and subsequent immunoprecipitation.
Chromatin preparation and subsequent immunoprecipitation were completed using a commercial kit (Pierce™ Agarose ChIP Kit) according to the manufacturer's instructions (Pierce) with modifications. Nuclei were extracted using a membrane extraction buffer spiked with HALT™ cocktail (Pierce kit) and centrifuged at 6000×g for 3 min. Intact nuclei were then resuspended in 10 mM Tris/1 mM EDTA/1% SDS sonication buffer and sonicated 13-15 times on ice (15 sec pulses followed by 2 min rests) until chromatin was 100-800 bps in size, with the center being ~300 bp. Prior to incubation with primary antibodies, chromatin was cleared of any cross-reactivity to agarose by incubating the isolated chromatin with agarose beads (5x input ChIP volume; Pierce Agarose Negative Control Resin) for 6 hours at 4°C before passing chromatin through columns provided with the kit. Chromatin was diluted in 1x dilution buffer (provided with Pierce kit) 1:2 before the addition of antibodies. Cross-reactive antibodies, previously identified as appropriate for use in zebrafish (Wardle et al. 2006), were obtained from Abcam (Cambridge, MA) and used at dilutions recommended by the manufacturer (10 μg normal rabbit IgG; 10 μg anti-H3; 10 μg anti-H3K4me3; 10 μg anti-H3K9me3; 10 μg anti-H3K27me3 per ChIP) with overnight (~18 h) incubations. DNA-protein complexes were immunoprecipitated using protein A/G-agarose beads, blocked in sonicated DNA/native DNA isolated from CHO cells (~ 3 million per 0.6 ml of beads) and bovine serum albumin (10 mg/ml) prior to use, for 1 hour, followed by three 5 min washes with each buffer (3 buffers = 9 washes; Pierce kit). Elution from agarose beads was performed as recommended by the manufacturer. Formaldehyde crosslinks of protein to DNA were released by overnight (~18 hour) incubation at 65°C in 5M NaCl/elution buffer using proteinase K (20 mg/ml; Pierce). Following DNA isolation using the provided columns, upstream regions of interest, identified using previous mammalian data (Palacios et al. 2010, Seenundun et al. 2010), as well as the ChIP-seq data from the Encyclopedia of DNA Elements (ENCODE) database available at the UCSC Genome Browser (http://www. http://genome-euro.ucsc.edu/index.html), were analyzed by qPCR using primer sets designed in Primer3 (Rozen and Skaletsky 2000) (see Table 2) using the following cycling parameters: 1 cycle, 95°C for 3 min; 40 cycles, 95°C for 15 sec, 60°C for 30 sec, 72°C for 15 sec. Samples were run in duplicate, with the appropriate 10% input and DNA-free controls and dissociation curves following amplification. In advance of both types of PCR analysis, primer sets were validated using genomic DNA isolated from rainbow trout MSCs and found to amplify intended targets of interest. All primer sets were validated to recognize gene promoter regions enriched in H3 proteins using total H3 immunoprecipitation. Using Cq values, % input was calculated for each experimental and mock sample.
Table 2.
Sequences of primer pairs used for ChIP analysts
| Target | Forward primer (5′–3′) | Reverse primer (5′–3′) | Locationa |
|---|---|---|---|
| myogenin geneb | |||
| FAR | CTCCTGCTTGTCATTGAGGA | TGGTTGGACATTTTGACAGG | −2749/−2587c |
| DIS | CTTGAGGGATTAGCGACAGG | GGACACCCCTGAGTGACTGT | −1161/−989 |
| PRO | TCTCTGGGCTCGTCTAAAT | GAAGTTTAGGCGGCGAAGAC | −258/−13 |
| pax7 genesb | |||
| Pax7a1 | AGAGCAAAGAAGGAGTCGTG | TGTCGACCAATCAGACAACC | −652/−496 |
| Pax7a2 | TTTTTCCCCAGGTACAAGGA | TAGGACGTAAAGCACACATTTC | −633/−519 |
| Pax7b | TGAGTTTACTTCTACGTCCAAAACA | GCATGCGACGTAAAACACAT | −684/−532 |
The values indicate the location on the gene where each primer set localizes from the ATQ start codon.
Statistical analysis
Results for proliferation, differentiation and RT-qPCR are expressed as means ± SEM and analyzed by one-way ANOVA followed by Student-Newman-Keuls test. For ChIP data, enrichment is expressed as percent-input (% input) after correction for dilution (1:10; −3.32 Cq). Data were analyzed by one-way ANOVA followed by Tukey's post-hoc test. For all statistical analyses, the level of significance was set at P<0.05. All the experiments were performed twice. Within an experiment, results for each time point were obtained from three replicates (for the ChIP analysis), four replicates (for the proliferation and differentiation assays) and six replicates (for the RT-qPCR analysis).
RESULTS
Three orthologs of mammalian pax7 exist in rainbow trout genome
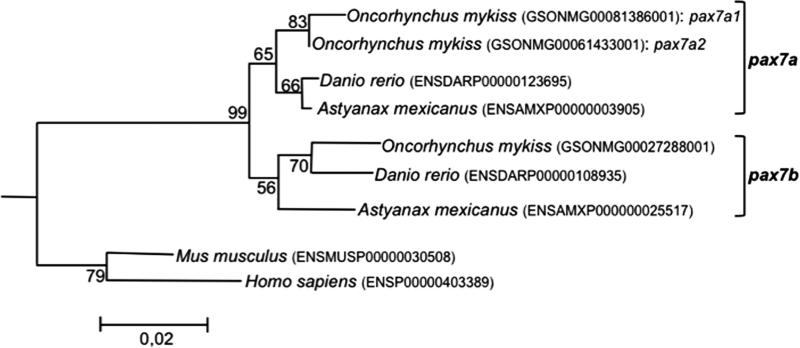
Using the recent availability of the rainbow trout genome assembly (Berthelot et al. 2014), we identified three genes (Genoscope accession number: GSONMG00081386001, GSONMG00061433001 and GSONMG00027288001) displaying high homology (E-value > 2e-40, Sigenae tblasn http://www.sigenae.org/) with the two trout sequences of pax7 previously available in GenBank (GenBank ID: NM_001258337 and NM_001258336). The phylogenetic analysis performed using the full-length Pax7 protein sequences of several vertebrate species and the amphioxus (Branchiostoma floridae) Pax3/7 sequence as an outgroup showed that the two trout sequences (GSONMG00081386001 and GSONMG00061433001), sharing the highest identity with the previously annotated NM_001258336 and NM_001258337 (99.7% and 100% identity, respectively), clustered with zebrafish (Danio rerio) and cave fish (Astyanax mexicanus) Pax7a, while the third sequence (GSONMG00027288001) was most similar to zebrafish and cave fish Pax7b (Fig. 1). These results suggested that the two former trout genes (GSONMG00081386001 and GSONMG00061433001) are paralogous genes and co-orthologous to the characid and cyprinid pax7a gene, while the last gene (GSONMG00027288001) was orthologous to the characid and cyprinid pax7b gene.
Figure 1. pax7 phylogenetic analysis.
The phylogenetic analysis was carried out using the Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0. Phylogenetic tree was constructed using the Neighbor-Joining (NJ) method. The reliability of the inferred tree was estimated using the bootstrap method with 500 replications. The amphioxus pax7 sequence (Branchiostoma floridae, GenBank ID: AAF89581.1) was used to root the tree. Proteins accession numbers are listed into the brackets.
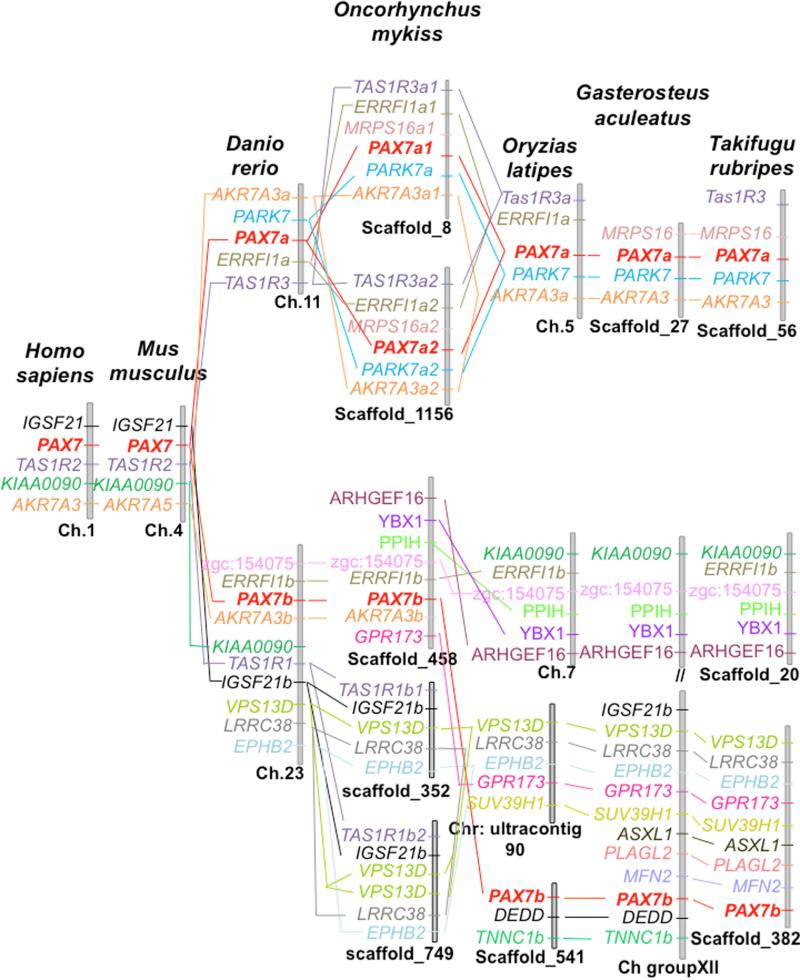
Our syntenic analysis showed that the tetrapod pax7 gene was localized in the igsf21-tas1r2-kiaa0090-akr7a3 syntenic group (Fig. 2). Interestingly, a syntenic conservation of this region was found with two distinct chromosomal regions of the zebrafish genome, which included the duplicated copies of pax7, tas1r3 and akr7a3 (Fig. 2). In addition, duplicated copies of errfl1 (a and b) were found to be syntenic with pax7a and pax7b. The syntenic region around the pax7a and pax7b loci in zebrafish was conserved in other teleost species but in a lesser extent. Considering the newly sequenced rainbow trout genome, our syntenic analysis showed that GSONMG00081386001 (pax7a1) and GSONMG00061433001 (pax7a2) genes were localized on two distinct scaffolds (scaffold_8 and scaffold _1156 carrying the genes GSONMG00081386001 and GSONMG00061433001, respectively). The synteny around pax7a locus in zebrafish and medaka (akr7a3a-park7-errfi1a-tas1r3) was well conserved around these two genes pointing out an additional duplication of pax7a in rainbow trout, thus giving birth to pax7a1 and pax7a2. As regard the third identified trout pax7 gene, the syntenic analysis showed that it was localized in the zgc:154075-errfi1b-pax7b-akr7a3b syntenic group conserved around the pax7b locus in zebrafish, confirming that this gene is related to pax7b of other teleosts. Interestingly, the synteny around the pax7b locus revealed a certain degree of chromosomal rearrangement in this genomic region between the zebrafish and the rainbow trout on one side and other teleost (e.g., medaka, stickleback and fugu) on the other, although the genetic environment is in overall conserved. Furthermore, we did not find any duplication of genes surrounding pax7b locus (errfi1b-pax7b-akr7a3b) in the trout genome, whereas our analysis revealed a duplication of the close locus tas1r1b1-igsf21b-vps13d. Taken together, these results revealed that the rainbow trout genome holds three pax7 paralogs, namely pax7a1 (GSONMG00081386001), pax7a2 (GSONMG00061433001) and pax7b (GSONMG00027288001).
Figure 2. pax7 syntenic analysis.
Conserved synteny around the pax7 loci in tetrapods and teleosts. The Genomicus software program (http://www.genomicus.biologie.ens.fr/genomicus-trout-01.01/cgi-bin/search.pl) was used to identify syntenic genes that were located near the pax7 genes of different vertebrate species.
pax7 paralogous genes are differently expressed during in vitro myogenesis
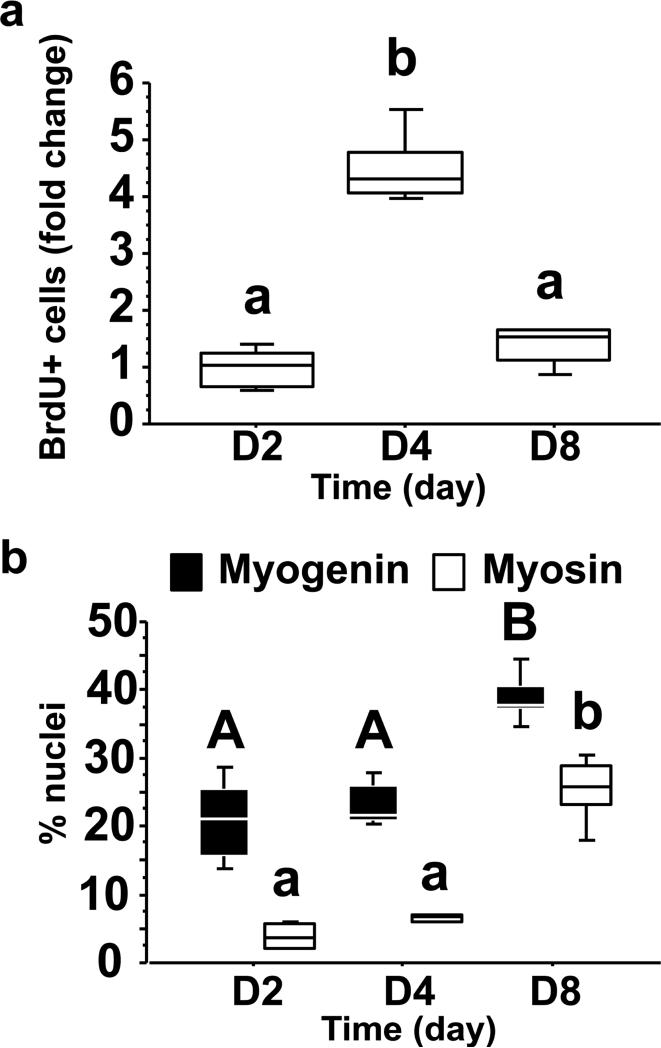
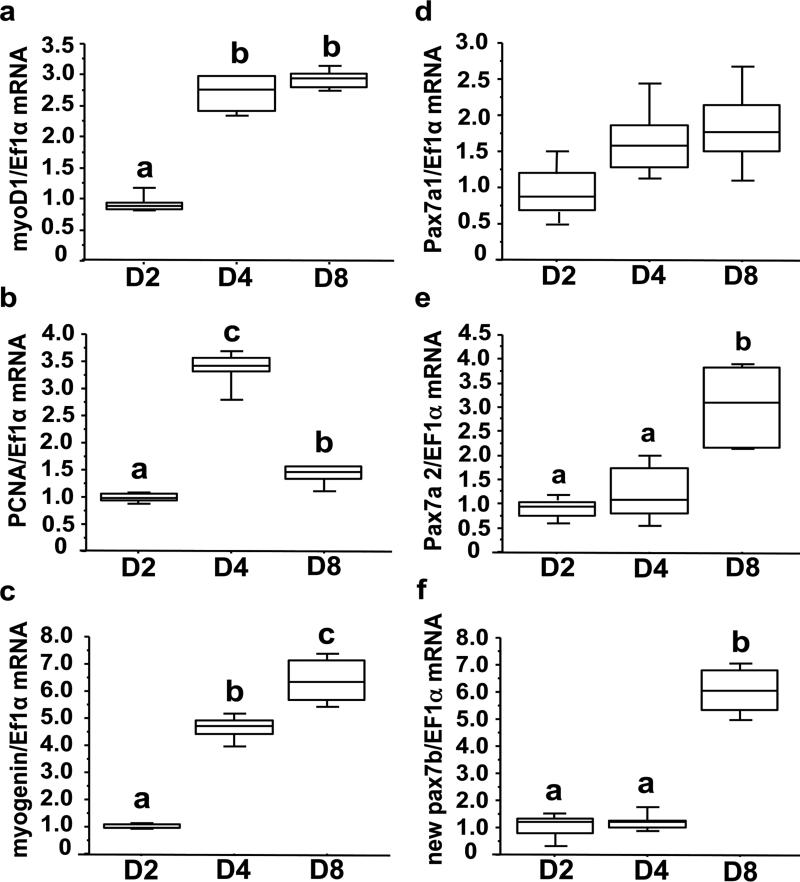
MSCs isolated from white muscle of juvenile rainbow trout were then used to monitor the expression of pax7a1, pax7a2 and pax7b during in vitro myogenesis. As shown in Fig. 3a, the cells (seeded at day 0) started to proliferate (as determined by measuring the incorporation of BrdU) at day 2 of culture and reached a peak at day 4. The proliferation rate subsequently decreased after 8 days of culture. Concurrently, the myogenic index of cultured cells (as determined by myogenin and myosin heavy-chain, MyHC, immunoreactivity) increases from day 2 to day 8 (Fig. 3b). These results demonstrated that isolated trout MSCs progress through the myogenic program to myotubes and validated their use as an in vitro model of myogenesis. In this regard, myoD1 and myogenin transcripts increased at day 8 of culture and those of the S phase marker PCNA paralleled the incorporation of BrdU (Figs 4a-c). Interestingly, pax7a1 transcripts remained stable during the culture period and those of pax7a2 and pax7b increased from 2 to 8 days (Figs 4d-f). Together these results suggest that the control of the expression of these pax7 paralogs diverged during evolution and make these genes relevant models for a better understanding of the evolutionary fate of duplicated genes.
Figure 3. Proliferation and differentiation of isolated trout MSCs cultured in DMEM with 10% fetal bovine serum (FBS) for 4 days and with 2% FBS for an additional 4 days.
(a), Proliferation of mononuclear myogenic cells at days 2, 4, and 8. Proliferation was determined by measuring the number of nuclei that incorporated BrdU during a period of 24 h. (b), Differentiation of myogenic cells at days 2, 4, and 8. Percentage of nuclei was determined by immunofluorescence analysis of myogenin (dark bars) and MyHC (clear bars). The different letters indicate significantly (P<0.05) different means. Capital letters represent pairwise comparisons between myogenin across days, while lowercase letters represent pairwise comparisons between myosin. Significant differences were determined by one-way ANOVA followed by Student-Newman-Keuls test for multiple comparisons. The presented experiment is representative of two experiments, and results for each time point were obtained from four replicates.
Figure 4. mRNA levels of myoD1 (a), PCNA (b), myogenin (c), pax7a1 (d), pax7a2 (e) and pax7b (f) in isolated trout MSCs cultured in DMEM with 10% fetal bovine serum (FBS) for 4 days and with 2% FBS for an additional 4 days.
mRNA levels of studied genes were estimated using real-time RT-qPCR at days 2, 4 and 8. Values are means ± SE, n = 6. They were analyzed by one-way ANOVA followed by Student-Newman-Keuls test for multiple comparison (P<0.05). Different letters represent significantly different values. The presented experiment is representative of two experiments.
H3K27me3, H3K9me3, and H3K4me3 profiling across the three pax7 loci reveals a complex regulatory schema
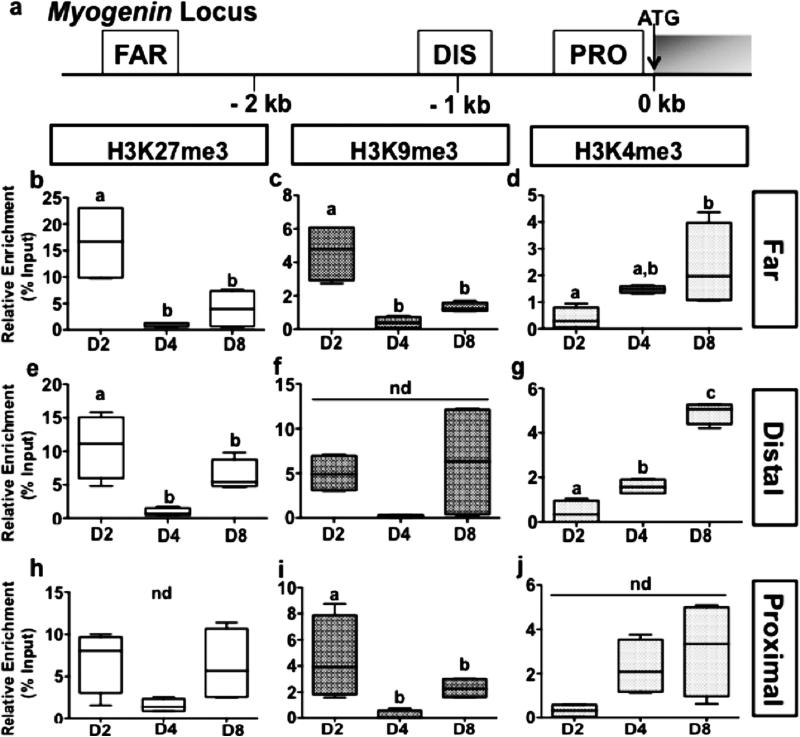
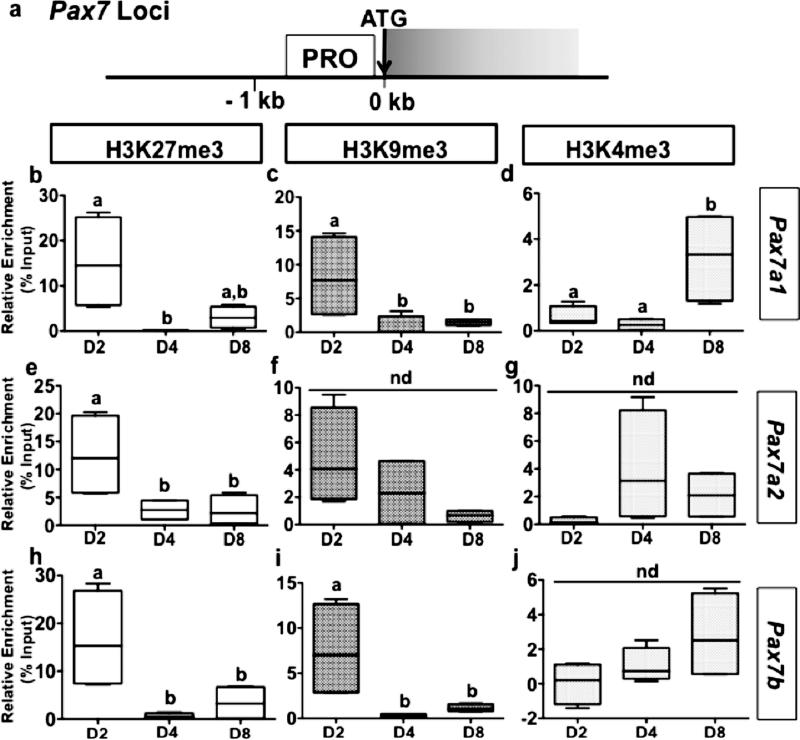
The regulation of the expression of several genes involved in myogenesis has been shown to be under the strict control of epigenetic imprinting through histone methylation. Therefore, ChIP was used to profile the distribution of H3K27me3, H3K9me3 and H3K4me3 across myogenin and pax7 loci (Figs 5a and 6a) during the in vitro myogenesis in rainbow trout. The results show an enrichment in repressive H3K27me3 and H3K9me3 marks throughout the upstream regions of myogenin in day 2, followed by an extensive loss of these histone modifications thereafter at day 4 and day 8 (Figs 5 b, c and c, i). In contrast, H3K4me3 levels remain relatively low at these loci in day 2 and display an increase at day 4 and 8 (Fig. 5d, g, j). Together these results fit with the increase of the mRNA levels of myogenin from day 2 to day 8 (Fig. 4c) and suggest that the expression of myogenin is regulated through the antagonistic action of repressive H3K9me3/H3K27me3 and permissive H3K4me3 marks. In contrast, pax7a1, which presented a stable expression over the culture period (Fig. 4d), switched at the targeted locus from a transcriptionally repressive state of H3K27me3 and H3K9me3 marked chromatin to the transcriptionally permissive state of H3K4me3 marked chromatin, during the progression of MSCs to myotubes (Fig. 6b-d), suggesting that regulation may occur at a different target locus or through different means. Conversely, the increase in pax7a2 and pax7b mRNA abundance (Fig. 4e and f) was associated to a statistical decrease in H3K27me3 levels for the former and in H3K27me3 and H3K9me3 levels for the last gene at the monitored locus (Figs 6e-j).
Figure 5. Methylation of H3 at the myogenin gene in isolated trout MSCs cultured in DMEM with 10% fetal bovine serum (FBS) for 4 days and with 2% FBS for an additional 4 days.
(a), Schematic representation of the myogenin gene. ATG represents the start codon and the gray box indicates the open reading frame. FAR (far upstream region), DIS (distal upstream region) and PRO (proximal upstream region) indicate the location on the gene where each of the different primer sets localizes for analysis of ChIP studies. (b - j), ChIP analyses of the temporal (day 2, 4 and 8) changes in H3K27me3 (b, e, h), H3K9me3 (c, f, i), and H3K4me3 (d, g, j) enrichment across the targeted myogenin regions FAR (b-d), DISTAL (e-g), and PROXIMAL (h-j). Chromatin isolated from trout myogenic cells at various stages of proliferation and differentiation (Day 2, 4 and 8) was subjected to immunoprecipitation using either an anti-H3K27me3, anti-H3K9me3, or anti-H3K4me3 antibody. Immunopurified DNA was then quantified by qPCR using primer sets that recognize the different regions indicated in the schematic in Figure 5a. Values are means ± SE, n = 3 (mean of 2 replications). They were analyzed by one-way ANOVA followed by Tukey's post-hoc test for multiple comparisons (P<0.05). Different letters represent significantly different values. nd = no difference detected.
Figure 6. Methylation of H3 at the pax7a1, pax7a2 and pax7b genes in isolated trout MSCs cultured in DMEM with 10% fetal bovine serum (FBS) for 4 days and with 2% FBS for an additional 4 days.
(a), Schematic representation of the pax7 paralogous genes. ATG represents the start codon and the gray box indicates the first exon. PRO (proximal upstream region) indicates the location on the pax7 genes where the used primer sets localize for analysis of ChIP studies. (b-j), ChIP analyses of the temporal (day 2, 4 and 8) changes in H3K27me3 (b, e, h), H3K9me3 (c, f, i), and H3K4me3 (d, g, j) enrichment across the pax7a1 (b-d), pax7a2 (e-g), and pax7b (h-j) genes. Chromatin isolated from trout myogenic cells at various stages of proliferation and differentiation (Day 2, 4 and 8) was subjected to immunoprecipitation using either an anti-H3K27me3, anti-H3K9me3, or anti-H3K4me3 antibody. Immunopurified DNA was then quantified by qPCR using the primer set that recognize specifically the targeted pax7 gene (a1, a2 or b) at the region indicated in the schematic in Figure 6a. Values are means ± SE, n = 3 (mean of 2 replications). They were analyzed by one-way ANOVA followed by Tukey's post-hoc test for multiple comparisons (P<0.05). Different letters represent significantly different values. nd = no difference detected.
DISCUSSION
Since Neil Stickland (Stickland 1983) first documented both hyperplastic and hypertrophic skeletal muscle growth in the postlarval rainbow trout, there has been much interest in understanding how these fish are able to form such a large number of myofibers outside of the embryonic and larval periods of development. Of particular importance have been in vitro studies, where MSCs can be isolated to assess their potential contribution to the muscle growth of these fish (Gabillard et al. 2010, Froehlich et al. 2014). Using such an in vitro model for experimentation, Neil Bower and Ian Johnston (Bower and Johnston 2010) documented that pax7 expression is up regulated during the differentiation of MSCs from Atlantic salmon (Salmo salar). Pax7, the canonical marker of MSCs in birds and mammals (Seale et al. 2000, Halevy et al. 2004), is rapidly down-regulated upon induction of MSC differentiation in rodents, birds, and Xenopus (Halevy et al. 2004, Zammit et al. 2004, Yamane and Nishikawa 2013), in agreement with its function in survival and quiescence of MSCs. Thus, the continued pax7 transcript expression in salmonid fishes is highly unusual in light of the current literature and warrants further investigation.
In the present study, we identified three pax7 paralogous genes in the recently available rainbow trout genome assembly (Berthelot et al. 2014). Based on phylogenetic analysis, we showed that two of these newly identified isoforms (pax7a1, −a2) belong to the pax7a clade while the third (pax7b) is orthologous to the characid and cyprinid pax7b gene. Previously, two pax7 cDNA sequences had been available in GenBank: NM_ 001258337.1, initially named pax7a and now renamed pax7a2; and NM_001258336.1 initially renamed pax7b and now renamed pax7a1. The single promoter sequence originally deposited in GenBank by P.Y. Rescan and C. Ralliere (FJ713022.1) is that of our newly coined pax7a2. Together, the phylogenetic analysis and synteny conservation data strongly suggest that a single copy of pax7 was present in the vertebrate ancestor and that a duplication of the pax7 gene occurred with the teleost-specific whole-genome duplication (WGD) 225 to 333 million years ago (Amores et al. 1998, Jaillon et al. 2004, Hurley et al. 2007, Santini et al. 2009, Near et al. 2012), giving rise to pax7a and pax7b paralogs in the common teleost ancestor. Our results also showed that the two identified trout pax7a1 and pax7a2 genes are co-orthologous to the zebrafish pax7a, suggesting that they resulted from the additional and relatively recent salmonid-specific WGD event (the 4th salmonid-specific WGD or Ss4R) around 100 million years ago (Berthelot et al. 2014, Macqueen and Johnston 2014). However, we failed to identify any duplicated gene for the syntenic group containing pax7b, possibly due to the recently described loss of half of the duplicated protein-coding genes in the rainbow trout genome (Berthelot et al. 2014). Taken together, these results demonstrate that the rainbow trout offers, with its three pax7 paralog genes, a unique opportunity to better understand the divergence in function of the best marker of MSCs currently known.
Real-time qPCR analysis revealed that these three pax7 paralogs are differentially expressed during the differentiation of isolated trout MSCs, in line with the recently described substantial number of Ss4R paralogs that has strongly diverged in terms of expression profiles and/or expression levels (Berthelot et al. 2014). While pax7a1 transcript levels remained stable over the culture period, pax7a2 and pax7b transcripts significantly increased on day 8 of culture, a time point in this cell culture model where myotubes have formed. Such an increase in pax7 expression during the myogenic progression to myotubes has been documented in the closely related species Salmo salar (Bower and Johnston 2010). Similarly, we recently demonstrated that expression of pax3 (another marker of MSCs in mammals) increases during a similar myogenic progression to myotubes in the indeterminately growing giant danio (Devario aequipinnatus) (Froehlich et al. 2013). In mammalian myogenic cell cultures, pax7 is not expressed in differentiated myotubes, but is maintained in a smaller accompanying population of undifferentiated cells that stops proliferating and returns to a non-proliferating state reminiscent of the quiescent reserve MSCs, which can then reenter the cell cycle, proliferate, and differentiate (Kitzmann et al. 1998, Yoshida et al. 1998). By analogy with these data, our results may indicate that the increase in pax3/7 expression during the myogenic progression to myotubes in indeterminate growing organisms may be involved with the continuous formation of new fibers through the enhanced production of reserve MSCs. In this regard, analyzing the mechanisms involved in the regulation of the expression of these genes is paramount to the understanding of how these fish are able to form such a large number of myofibers outside of the embryonic and larval stages of development and growth. An understanding of the mechanisms underlying differences in pax7 paralog expression is also of particular importance from an evolutionary point of view. Following WGD, it is well accepted that the remaining paralogs can acquire new expression patterns, potentially leading to neo- or subfunctionalization (Force et al. 1999). This suggests that evolution after WGD may also act on the regulatory regions of paralogous genes, leading to subsequent silencing, sub-, or neofunctionalization. Thus, the identification of the regulatory mechanisms affected by and/or modified between paralogous genes may lead to a better understanding of the evolutionary fate of genes within a duplicated vertebrate genome.
In mammals, the transcriptional state of several muscle-specific genes such as myogenin or pax7 has been shown to be associated with repressive H3K27me3 and H3K9me3 and permissive H3K4me3 chromatin marks (Mal 2006, Rampalli et al. 2007, Palacios et al. 2010, Mozzetta et al. 2011, Stojic et al. 2011). In the present study, we therefore sought to profile the distribution of H3K27me3, H3K9me3, and H3K4me3 marks across the three pax7 and myogenin loci during in vitro myogenesis in the rainbow trout. The obtained results show that, during the myogenic progression to myotubes, the myogenin locus appears to be modified from a transcriptionally repressive state (i.e., H3K27me3 and H3K9me3 enriched chromatin) to a transcriptionally permissive state (i.e., H3K4me3 enriched chromatin), in close agreement with previous findings obtained in C2C12 cell lines (Mal 2006, Rampalli et al. 2007, Seenundun et al. 2010). According to these studies, the proper temporal and spatial pattern of myogenin expression is established through the control of these repressive and permissive histone modifications mediated by the antagonistic functions of Polycomb group (PcG) and Trithorax group (TrxG) proteins (Mal 2006, Rampalli et al. 2007) as well as the action of histone lysine demethylases (Seenundun et al. 2010, Verrier et al. 2011). Although all of these factors remain to be characterized in fish, the parallel between the expression of myogenin and the monitored epigenetic marks in our cell culture model suggest a good conservation of these regulatory mechanisms among vertebrates. However, results obtained for the three trout pax7 genes appear to indicate a more complex regulatory schema. Indeed, pax7a1, with stable expression over the culture period, switched from a transcriptionally repressive state (i.e., H3K27me3 and H3K9me3 enriched chromatin) to a transcriptionally permissive state (i.e., H3K4me3 enriched chromatin) during in vitro myogenesis. Such a discrepancy between mRNA expression and histone methylation may be the result of timing, and an increase in the expression of pax7a1 may not become detectable until later time points. However, we cannot rule out the possibility that other post-translational modifications to exposed histone amino acid residues (acetylation, phosphorylation, mono-/di-/tri-methylation) may be at play in the control of the expression of this gene. Conversely, the increase in pax7a2 and pax7b mRNA abundance was clearly associated with a statistical decrease in H3K27me3 levels for the pax7a2 paralog and in H3K27me3 and H3K9me3 levels for the pax7b paralog. Interestingly, previous findings in other systems have demonstrated increasing levels of in the promoter region of pax7 during the myotube differentiation of murine MSCs (Palacios et al. 2010). These contrasting results highlight a possible divergence in the regulatory mechanisms of pax7 expression among vertebrates. Whether this difference in histone modifications and chromatin accessibility contributes to the increase in pax7a2 and pax7b expression during the in vitro myogenic program is worth investigating. Further studies are also warranted to describe a role for the “atypical” expression of pax7 paralogs in the extraordinary ability of trout to augment their skeletal musculature throughout life.
This report, linking the highly unique differential expression of pax7 paralogs with epigenetic histone modifications, provides a platform for future studies for a better understanding of myogenesis in this indeterminate growing species and other teleost fishes. Information from these studies will be critical not only to the mechanistic understanding of the indeterminate growth paradigm, but also to a more complete understanding of the evolutionary fate of duplicated genes (and genomes) in Vertebrata.
ACKNOWLEDGEMENTS
The authors would like to thank Ikram Belghit, Imad Aharchaou, and Alexandre Herman for their assistance in the setup of initial rainbow trout primary myoblast cultures.
FUNDING
This work was funded by the University of Alabama at Birmingham Nutrition and Obesity Research Center award #P30DK056336 and a University of Alabama at Birmingham Office of the Provost Faculty Development Grant to PRB.
REFERENCES
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282(5394):1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Berthelot C, Brunet F, Chalopin D, Juanchich A, Bernard M, Noel B, Bento P, Da Silva C, Labadie K, Alberti A, Aury JM, Louis A, Dehais P, Bardou P, Montfort J, Klopp C, Cabau C, Gaspin C, Thorgaard GH, Boussaha M, Quillet E, Guyomard R, Galiana D, Bobe J, Volff JN, Genet C, Wincker P, Jaillon O, Roest Crollius H, Guiguen Y. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower NI, Johnston IA. Paralogs of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am J Physiol Regul Integr Comp Physiol. 2010;298(6):R1615–1626. doi: 10.1152/ajpregu.00114.2010. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Diao Y, Guo X, Li Y, Sun K, Lu L, Jiang L, Fu X, Zhu H, Sun H, Wang H, Wu Z. Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11(2):231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JM, Fowler ZG, Galt NJ, Smith DL, Jr., Biga PR. Sarcopenia and piscines: the case for indeterminate-growing fish as unique genetic model organisms in aging and longevity research. Front Genet. 2013;4:159. doi: 10.3389/fgene.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JM, Galt NJ, Charging MJ, Meyer BM, Biga PR. In vitro indeterminate teleost myogenesis appears to be dependent on Pax3. In Vitro Cell Dev Biol Anim. 2013;49(5):371–385. doi: 10.1007/s11626-013-9616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JM, Seiliez I, Gabillard JC, Biga PR. Preparation of primary myogenic precursor cell/myoblast cultures from basal vertebrate lineages. J Vis Exp. 2014;(86) doi: 10.3791/51354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabillard JC, Sabin N, Paboeuf G. In vitro characterization of proliferation and differentiation of trout satellite cells. Cell & Tissue Research. 2010 doi: 10.1007/s00441-010-1071-8. [DOI] [PubMed] [Google Scholar]
- Halevy O, Piestun Y, Allouh MZ, Rosser BW, Rinkevich Y, Reshef R, Rozenboim I, Wleklinski-Lee M, Yablonka-Reuveni Z. Pattern of Pax7 expression during myogenesis in the posthatch chicken establishes a model for satellite cell differentiation and renewal. Dev Dyn. 2004;231(3):489–502. doi: 10.1002/dvdy.20151. [DOI] [PubMed] [Google Scholar]
- Hurley IA, Mueller RL, Dunn KA, Schmidt EJ, Friedman M, Ho RK, Prince VE, Yang Z, Thomas MG, Coates MI. A new time-scale for ray-finned fish evolution. Proc Biol Sci. 2007;274(1609):489–498. doi: 10.1098/rspb.2006.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431(7011):946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Johnston IA, Bower NI, Macqueen DJ. Growth and the regulation of myotomal muscle mass in teleost fish. J Exp Biol. 2011;214(Pt 10):1617–1628. doi: 10.1242/jeb.038620. [DOI] [PubMed] [Google Scholar]
- Kawabe Y, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11(3):333–345. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M, Carnac G, Vandromme M, Primig M, Lamb NJ, Fernandez A. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle-specific expression in muscle cells. J Cell Biol. 1998;142(6):1447–1459. doi: 10.1083/jcb.142.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinides N, Averof M. A common cellular basis for muscle regeneration in arthropods and vertebrates. Science. 2014;343(6172):788–791. doi: 10.1126/science.1243529. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23(12):4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138(17):3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling BM, Gopinadhan S, Kok WK, Shankar SR, Gopal P, Bharathy N, Wang Y, Taneja R. G9a mediates Sharp-1-dependent inhibition of skeletal muscle differentiation. Mol Biol Cell. 2012;23(24):4778–4785. doi: 10.1091/mbc.E12-04-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen DJ, Johnston IA. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc Biol Sci. 2014;281(1778):20132881. doi: 10.1098/rspb.2013.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25(14):3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommsen TP. Paradigms of growth in fish. Comp Biochem Physiol B Biochem Mol Biol. 2001;129(2-3):207–219. doi: 10.1016/s1096-4959(01)00312-8. [DOI] [PubMed] [Google Scholar]
- Mozzetta C, Consalvi S, Saccone V, Forcales SV, Puri PL, Palacios D. Selective control of Pax7 expression by TNF-activated p38alpha/polycomb repressive complex 2 (PRC2) signaling during muscle satellite cell differentiation. Cell Cycle. 2011;10(2):191–198. doi: 10.4161/cc.10.2.14441. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292(5514):110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci U S A. 2012;109(34):13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, Sartorelli V, Puri PL. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7(4):455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Sousa-Victor P, Ballestar E, Munoz-Canoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4(8):541–550. doi: 10.4161/epi.4.8.10258. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14(12):1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Saccone V, Puri PL. Epigenetic regulation of skeletal myogenesis. Organogenesis. 2010;6(1):48–53. doi: 10.4161/org.6.1.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138(17):3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol. 2009;9:194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18(11):1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdek P, Oyama K, Angelis E, Chan SS, Schenke-Layland K, MacLellan WR. Epigenetic regulation of myogenic gene expression by heterochromatin protein 1 alpha. PLoS One. 2013;8(3):e58319. doi: 10.1371/journal.pone.0058319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci U S A. 2009;106(12):4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29(8):1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickland NC. Growth and development of muscle fibres in the rainbow trout (Salmo gairdneri). J Anat. 1983;137(Pt 2):323–333. [PMC free article] [PubMed] [Google Scholar]
- Stojic L, Jasencakova Z, Prezioso C, Stutzer A, Bodega B, Pasini D, Klingberg R, Mozzetta C, Margueron R, Puri PL, Schwarzer D, Helin K, Fischle W, Orlando V. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin. 2011;4:16. doi: 10.1186/1756-8935-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Kelly TK, Liu CC, You JS, De Carvalho DD, Miranda TB, Zhou XJ, Liang G, Jones PA. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147(6):1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276(27):25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, Zhang Y, Jin SW, Wang DZ. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol. 2011;194(4):551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier L, Escaffit F, Chailleux C, Trouche D, Vandromme M. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 2011;7(6):e1001390. doi: 10.1371/journal.pgen.1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH, Zare H, Mousavi K, Wang C, Moravec CE, Sirotkin HI, Ge K, Gutierrez-Cruz G, Sartorelli V. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J. 2013;32(8):1075–1086. doi: 10.1038/emboj.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8(6):1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol. 2006;7(8):R71. doi: 10.1186/gb-2006-7-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Nishikawa A. Differential muscle regulatory factor gene expression between larval and adult myogenesis in the frog Xenopus laevis: adult myogenic cell-specific myf5 upregulation and its relation to the notochord suppression of adult muscle differentiation. In Vitro Cell Dev Biol Anim. 2013;49(7):524–536. doi: 10.1007/s11626-013-9635-z. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111(Pt 6):769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166(3):347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]