Abstract
Marine cyanobacteria are an ancient group of organisms and prolific producers of bioactive secondary metabolites. These compounds are presumably optimized by evolution over billions of years to exert high affinity for their intended biological target in the ecologically relevant organism but likely also possess activity in different biological contexts such as human cells. Screening of marine cyanobacterial extracts for bioactive natural products has largely focused on cancer cell viability; however, diversification of the screening platform led to the characterization of many new bioactive compounds. Targets of compounds have oftentimes been elusive if the compounds were discovered through phenotypic assays. Over the past few years, technology has advanced to determine mechanism of action (MOA) and targets through reverse chemical genetic and proteomic approaches, which has been applied to certain cyanobacterial compounds and will be discussed in this review. Some cyanobacterial molecules are the most-potent-in-class inhibitors and therefore may become valuable tools for chemical biology to probe protein function but also be templates for novel drugs, assuming in vitro potency translates into cellular and in vivo activity. Our review will focus on compounds for which the direct targets have been deciphered or which were found to target a novel pathway, and link them to disease states where target modulation may be beneficial.
1 Introduction
Natural products have historically been utilized to develop new drugs, and it remains to be one of the most successful approaches to find small molecules for the drug discovery pipeline. Approximately 50% of new drugs introduced from 1981–2010 were derived from Nature, the majority being antiproliferative agents and antibiotics.1 The discovery of small molecule therapeutics can be undertaken using either a target-based (reverse genetics) or a phenotypic-based (forward genetics) approach.2 In the target-based approach, purified proteins with disease-relevance are utilized to screen for modulators of activity.3 This screening approach has the advantage of knowing the direct target of the small molecule; however, it poses the possibility of the in vitro activity not translating to in vivo potency and the cellular effects being defined later in the drug discovery process.2,3 Since target-based screening is usually done in cell-free systems, it has limited utility for discovery of prodrugs and in providing preliminary insights on the pharmacokinetic properties of small molecule hits. Several of the limitations in target-based screening are addressed by employing phenotypic-based assays. Relying on phenotypic assays, however, complicates the search for the cellular targets of small molecules and continues to be a bottleneck for this approach.2,4 Equally important to defining the potency of natural products, discovered using either method, is determining unintended off-targets, which is critical to predicting possible side effects. Since natural products are regarded as privileged structures, capable of binding to multiple proteins with unrelated structures, these small molecules may have multiple targets.5 Rigorous characterization of cellular targets and mechanism of bioactivity is then necessary to achieve a comprehensive assessment of the potency, efficacy and pharmacology of bioactive small molecules. Natural products have been central to the discovery of novel drug targets and represent a unique source of chemical probes to investigate proteins and signaling networks.6 For example, the natural products trapoxin7 and trichostatin A8,9 were pivotal to elucidating the structure and functional role of histone deacetylases (HDACs). An affinity matrix based on trapoxin B, K-trap, allowed for the purification of HDACs from bovine thymus and permitted the molecular characterization of HDACs.7 Trichostatin A, on the other hand, was instrumental to the structural analysis of HDACs, providing the first X-ray cocrystal structures of histone deacetylase-like proteins and HDAC8, crucial in defining the critical structural elements of HDACs for pharmacological interventions.8,9 These discoveries were instrumental to revolutionizing epigenetics and in defining the role of HDACs in cancer. Today, HDACs represent a novel molecular target and mechanism to modulate malignancies and are also being pursued for non-cancer diseases where gene expression changes may be beneficial.10
Proteins represent the majority of molecular targets of marketed drugs, with enzymes and G-protein coupled receptors accounting for almost 75% of these molecular targets.11 In contrast, non-protein targets such as DNA, RNA, ribosomes, metabolites and physicochemical mechanisms represent only close to 5% of the molecular targets of marketed drugs.11 The rest of the molecular targets of marketed drugs include ion channels, transport proteins and non-GPCR receptors. The overrepresentation of proteins as druggable targets and challenges in exploiting non-protein targets have initiated protein-centric experimental methods for target identification that are geared towards interrogating the proteome as well as the genome and transcriptome, in relation to protein expression and the observed phenotypic effects and MOA.2,4,11,12 In recent years, significant improvements in the omics technologies, specifically genomics, transcriptomics and proteomics, have allowed for the systematic and unbiased assessment of the molecular targets of small molecules. These techniques have been demonstrated to be amenable for mammalian cellular systems as well as for model organisms such as Saccharomyces cerevisiae (yeast), Caenorhabditis elegans (nematode worm) and Danio rerio (zebrafish).2,4,11,12
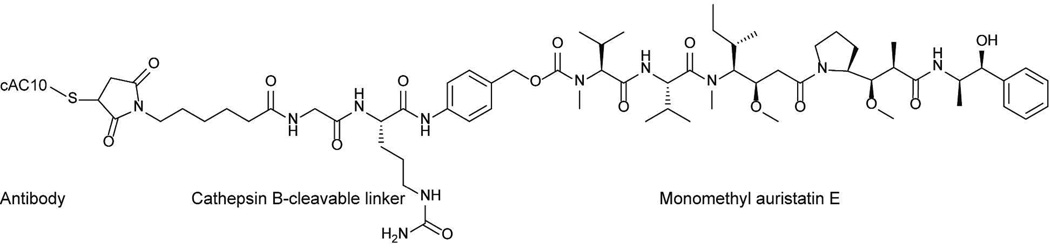
Cyanobacteria, or blue-green algae, represent a unique source of small molecules since these organisms are believed to be the most ancient organisms on Earth and may thus represent chemical factories with highly evolved machineries for secondary metabolite production. In addition, cyanobacteria are also known to affect the biosynthesis of compounds from marine invertebrates such as sponges, ascidians and shell-less mollusks through either endosymbiosis or diet-derived enrichment.13–15 A survey of natural products with FDA approval and those in clinical trials indicated that ~20% of these small molecules are likely to have cyanobacteria as predicted biosynthetic sources.16 Cyanobacteria are also a validated source of new drugs, with the FDA-approved antibody-drug conjugate brentuximab vedotin (Fig. 1), inspired from the cyanobacteria compound dolastatin 10.17
Fig. 1.
The structure of brentuximab vedotin (Adcetris). The small molecule portion, monomethyl auristatin E, is designed based on the cyanobacterial molecule dolastatin 10.
Cyanobacteria as source organisms may then represent a treasure chest of new small molecules that can potentially be tapped for biomedical and pharmacological applications, either as drug leads or chemical probes.18–20 The majority of these small molecules was derived using bioactivity-guided purification, showing diversity of chemistry and biological activity.18–20 The true value of this resource may be fully realized with rigorous studies on the MOA to define the intended therapeutically-relevant and possible cellular off-target effects. In this review, we examine cyanobacterial small molecules with established molecular targets and MOA and discuss future prospects for several of these potent pharmacological agents.
2 Methods in Target Identification and Elucidation of Mechanism of Action
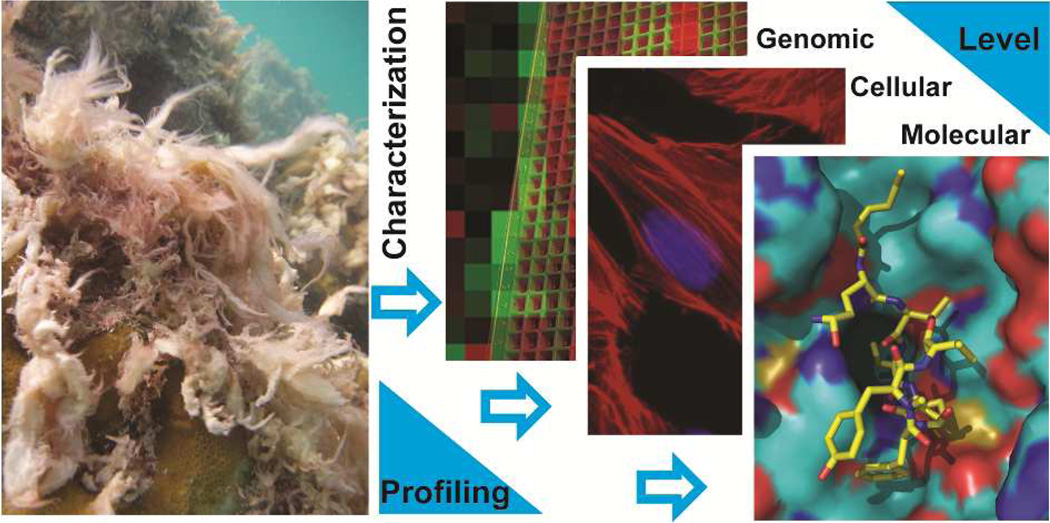
Insights into the direct cellular target of small molecules have typically been derived from target-based assays, chemical structure and phenotypic response similarity to other small molecules with defined MOA. These methods, however, have limited utility for small molecules with novel chemical structures and MOA. The last twenty years have seen a revolution in the development of global and unbiased methods to elucidate the direct target and MOA of small molecules. These methods are complementary to one another and should be used hand-in-hand to provide a comprehensive picture of the cellular consequences of small molecule treatment. Variants of several of the techniques discussed in this section have also been applied to determine targets and MOA of natural products from marine cyanobacteria (Fig. 2). In addition, hits derived from both indirect and direct techniques for target identification should be rigorously validated using biochemical evaluation and/or X-ray cocrystallography studies.
Fig. 2.
Screening platforms for bioactivity assessment, mechanism of action and target identification of small molecules from marine cyanobacteria.
2.1 Indirect Approach
One of the earliest comprehensive approaches to elucidating the MOA of natural products is through the National Cancer Institute (NCI) 60 cell line screen (NCI-60) and COMPARE algorithm.21 This is based on the similarity in phenotypic response for structurally unrelated small molecules, with similar MOA.21 The NCI-60 screen employs measurement of the antiproliferative effects of small molecules against a panel of 60 tumor cell lines consisting of leukemia, small cell lung, non-small cell lung, colon, central nervous system, melanoma, ovarian, and renal. The differential cytotoxicity of the small molecule towards each cell line is evaluated and plotted to obtain a “fingerprint”. This “fingerprint” differential cytotoxicity profile is compared to the extensive database of previously screened small molecules of NCI using the COMPARE algorithm.21 Compounds with unique differential cytotoxicity profile are likely to possess a novel MOA, while compounds with similar MOA can be clustered together.21 The utility of this method was first demonstrated for halichondrin B.22 The differential NCI-60 profile of halichondrin B showed striking similarity with the known antimitotic agent maytansine, indicating analogous MOA.22 Drug susceptibility can be correlated with the unique gene expression profiles of the cell lines and compounds thus linked to molecular target and modes of resistance.23–25 While this approach has been successfully utilized for several natural products, this is obviously limited for small molecules with significant antiproliferative effects towards cancer cells. Also, the NCI-60 screen provides information on the MOA, rather than the direct target. This method provides little information on the MOA of small molecules with potentially unique MOA.
The principle of the NCI-60 screen has been expanded, using high content screening, to provide insights into the MOA of prefractionated extract libraries of natural products and for identification of compounds with possible novel bioactivity or chemistry. The high-content cell morphology database Morphobase utilizes similarity in chemical-genetic phenotype of cancer cell lines treated with small molecules for rapid target identification.26
The MOA of small molecules has also been interrogated via the transcriptome. Microarray analysis has been used to monitor global changes in transcript levels of mammalian cells, in response to small molecule treatment. Changes to the transcriptome, together with a network analysis of significantly modulated transcripts provide insights into the MOA of the small molecule.27 Recent progress in next-generation sequencing technology has also allowed for the comprehensive and unbiased profiling of the transcriptome of resistant colonies of mammalian tumor cells derived from prolonged exposure to small molecule treatment.28 The observed resistance was demonstrated to arise from mutations in the small molecule target and/or overexpression of drug efflux pumps.28 These mutations were limited to single nucleotide variations and short insertions and/or deletions present in the coding sequences.28 The utility of transcriptome profiling for target identification was successfully demonstrated as proof-of-concept for bortezomib and BI-2356.28 Five bortezomib-resistant clones were isolated and transcriptome analysis indicated 15–28 single nucleotide variations in five genes. The highest frequency in mutation was observed for PSMB5, which encodes for the proteasome, thus validating this new method for target identification.28 Other changes in the transcriptome of resistant cells, independent of the drug target or the drug efflux transporters were, however, not completely elucidated in this seminal paper. Modulating transcript levels either via cDNA overexpression or siRNA-mediated mRNA knockdown has also been utilized to elucidate the target of small molecules.29 This method relies on the correlation between transcript and protein levels such that overexpression would lead to an increase of the target protein, while silencing of the target gene will cause a corresponding decrease or depletion of target protein.29 Changes in the level of the target protein will lead to either resistance or susceptibility to the small molecule treatment, arising from overexpression or silencing, respectively.29
2.2 Direct Approach
Interrogation of the direct target of small molecules relies on chemical proteomics – combination of affinity chromatography and mass spectrometry – for purification and identification of direct binding proteins.2 The success of affinity chromatography is dependent on the binding affinity of the small molecule and the abundance of the target protein.2,30 Small molecules with high affinity for their target are generally favored for affinity-based purification. While the binding affinity of small molecules may not always be directly correlated to the IC50 or EC50, compounds with nanomolar IC50s are ideal for affinity chromatography.30
Affinity chromatography requires the immobilization of the small molecule on a solid support such as sepharose beads and, hence, necessitates chemical derivatization of the small molecule.30 Extensive structure-activity relationship (SAR) studies are required to determine the appropriate site for modification, which does not significantly affect the biological activity of the small molecule. Affinity chromatography is then prohibitive for very small molecules with no appropriate sites for chemical modification. For natural products with limited amounts and not readily prepared through chemical synthesis, the choice of derivatization site may be limited to functional groups that can easily be modified such as terminal alkynes which rapidly and selectively react in a CuSO4-catalyzed azide-alkyne cycloaddition click chemistry. A linker moiety bridges the small molecule to the solid support and is required not solely for immobilization but also to avoid steric interference of matrix and target protein.4 Common linkers include polymethylene, polyethylene and polyproline chains with varying lengths.4
The pulldown affinity probe is incubated with either prefractionated or whole cell lysates, the former providing preliminary enrichment of proteins localized in specific cellular compartments.4 After incubation, the probe is washed extensively to remove non-specific binding proteins. This is the crucial step in affinity purification and dependent on the balance of the affinity of the small molecule for its target and the abundance of the protein target.30 Captured proteins are monitored using gel electrophoresis and/or mass spectrometric analysis. In order to differentiate nonspecific binding proteins from the bona fide small molecule target, competition experiments and a control probe may be utilized. For competition experiments, excess amounts of the unmodified small molecule are added to the cell lysates.4 The unmodified small molecule will then compete with the probe for the target protein; hence, a lower amount of a real hit will be captured.4 The use of a control probe, on the other hand, entails design of a pulldown probe based on an inactive analog of the small molecule.4 The direct target is unable to bind to the control probe and thus significant enrichment of the real protein target will be observed in incubations using the bioactive probe. A combination of affinity chromatography with mass spectrometry-compatible labeling techniques such as stable isotope labeling by amino acids in cell culture (SILAC) and isobaric tag for relative and absolute quantitation (iTRAQ) has also been utilized to differentiate nonspecific binding proteins from the real direct target of small molecules.31 SILAC utilizes either natural (light isotope) or 13C6-labeled (heavy isotope) lysine during cell culture and, in the process of translation, will be incorporated into proteins. The proteins labeled with the light isotope are incubated with the affinity probe, while proteins with the heavy isotope are incubated with the affinity probe and free small molecule. The relative abundance of the captured protein is assessed based on the difference in signal intensity between the light- and heavy-isotope labeled proteins. In iTRAQ, proteins are labeled after trypsin digestion using isobaric tags to differentiate proteins from the active and inactive probe treatments.32,33 The relative abundance of proteins is determined based on the ratio of the isobaric tags between treatments. Protein identification has been facilitated by significant technological advancements in mass spectrometry and also in sample preparation prior to MS/MS analysis, which allows sensitive detection and enrichment of low abundance proteins.
Drug affinity responsive target stability (DARTS) is another mass spectrometry-based target protein identification method. DARTS represents an alternative method to chemical derivatization and probe design to elucidate the direct target of small molecules, while still interrogating the proteome.34 DARTS utilizes the unmodified small molecule as the probe and is based on the decreased susceptibility of proteins to protease degradation upon binding of the small molecule. Resistance to protease degradation may be due to either stabilization of the target protein via ligand binding or the small molecule masking the protease recognition site.34 DARTS has been utilized for several natural products of marine and terrestrial origins such as didemnin B, FK506, rapamycin and resveratrol.34 The marine tunicate-derived didemnin B was utilized as a model compound to validate the utility of DARTS for target identification.34 Jurkat cells were treated with 1 µg/mL didemnin B or solvent control. Whole cell lysates were subjected to thermolysin- or mock-digestion and subsequently profiled by SDS-PAGE and visualized by Coomasie Blue staining.34 Significant enrichment of the protein band with molecular weight ~50 kDa was observed in didemnin B-treated cells and identification using MS indicated that the primary protein present at high abundance in didemnin B-treated cells was EF-1α, in accord with the observed direct target of this small molecule using chemical proteomics.34 Despite the validation of DARTS for target identification of several small molecules, its full potential for target identification is yet to be realized.34 The potential drawbacks of DARTS will be related to the binding affinity of the small molecule for its target and also the inherent variability in the susceptibility of proteins to proteolysis because of the conformational energy landscape.34
3 Mechanisms of Action and Direct Cellular Targets of Biologically Active Cyanobacterial Metabolites in Mammalian Cells
3.1 Classical Anticancer Drug Targets
Inhibition of microtubule and microfilament assembly is one of the well-represented mechanisms of action of antiproliferative natural products.35 Apart from antimetabolites and topoisomerase poisons, tubulin-targeting agents from Nature were among the first small molecules to achieve approval for clinical use. In contrast, small molecules that act on actin have failed to gain clinical approval but have largely been used as pharmacological probes to study actin function. Actin- and tubulin-targeting agents are among the first compounds to have defined MOA. Tubulin-targeting agents were utilized as first-line drugs for cancer because of their ability to halt uncontrolled cell division by causing mitotic arrest that culminates in programmed cell death (Fig. 4).36,37 This method of curbing the growth of malignancies, while effective, may pose detrimental effects to normal cells since tubulin is also present in these cells. Both actin and tubulin are critical in maintaining cell shape, motility and cell division, and these proteins are largely abundant in cells, making the observation of their pharmacological modulation relatively easy by monitoring the tubulin or actin network and cellular morphology by microscopy.38 The abundance and ease in monitoring of tubulin and actin may have posed as biases in the discovery of modulators of these proteins during the dawn of the development of anticancer agents.
Fig. 4.
Tubulin-targeting agents from marine cyanobacteria.
3.1.1 Tubulin-Targeting Agents
The discovery of tubulin-targeting agents continues to be an active research field because of the chemodiversity of active small molecules. Small molecules that disrupt microtubule proteins do not significantly change the microtubule mass, and rather, suppress the microtubule dynamics.35 Tubulin poisons from cyanobacteria either bind to the Vinca domain or colchicine binding site, thereby acting in a comparable manner to the vinca alkaloids or colchicine, respectively.35 Despite several tubulin-targeting agents gaining clinical approval, there are concerns on the use of these small molecules for chemotherapeutic intervention related to tumor specificity, undesired effects, most notably peripheral neuropathy and drug resistance evoked by increased expression of P-glycoprotein.37 In addition to pursuing their tumor growth inhibitory activity, tubulin inhibitors are also being explored for their antivascular effects leading to inhibition of angiogenesis.36
Vinca domain binding agents
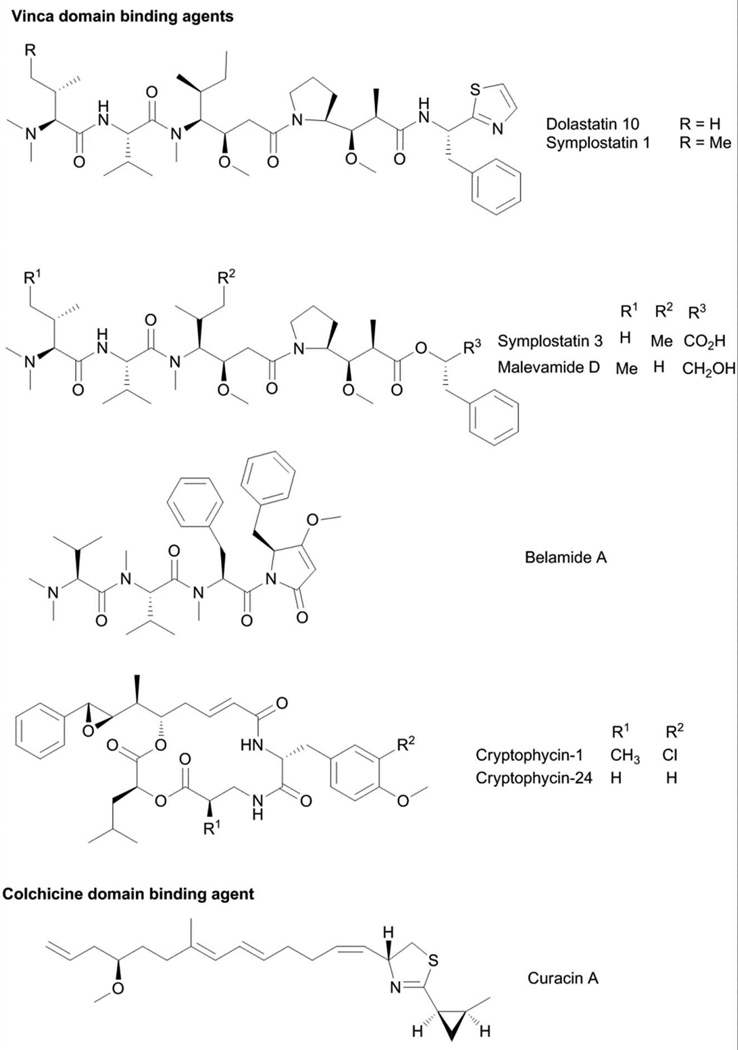
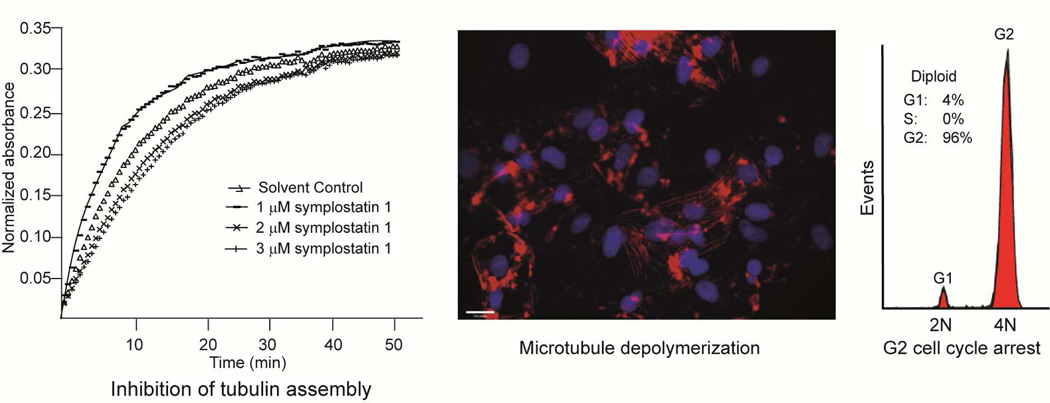
The modified linear peptides belonging to the dolastatin 10-type of compounds are among the predominant class of tubulin-targeting agents from marine cyanobacteria (Fig. 4).39 Dolastatin 10 is characterized by a terminal dimethylated amino acid residue and several nonproteinogenic amino acids (Fig. 4).40 Dolastatin 10 was initially isolated from the sea hare Dolabella auricularia, but the low yield and subsequent purification of the related compound symplostatin 1 from the cyanobacteria Symploca hydnoides indicated the dietary origins of dolastatin 10.15,40–42 Symplostatin 1 and dolastatin 10 only differed in the N-terminal amino acid residue, with N,N-dimethyl Ile in the former and N,N-dimethyl Val in the latter (Fig. 4).41 Dolastatin 10 was isolated as the antiproliferative component in D. auricularia, following large-scale purification, and was demonstrated to arrest the cells in metaphase.39 Dolastatin 10 and symplostatin 1 both exhibited potent antiproliferative activity, with pico- to nanomolar IC50s against a wide array of cancer cells.39,43 Dolastatin 10 was shown to affect the polymerization of purified tubulin. Structural similarities between dolastatin 10 and the tubulin inhibitor phomopsin A suggested that dolastatin 10 may bind to the Vinca domain of tubulin, analogous to phomopsin A.39 To probe this hypothesis, the binding of [3H]-labeled vincristine and [3H]-labeled colchicine to tubulin was monitored in the presence of dolastatin 10.39 Dolastatin 10 inhibited the binding of radiolabeled vincristine without any effect on radiolabeled colchicine. In addition to tubulin binding, dolastatin 10 was also shown to inhibit critical events during tubulin assembly such as tubulin-dependent GTP hydrolysis and nucleotide exchange on β-tubulin. To determine possible binding sites of dolastatin 10 on α- and β-tubulin, a combination of molecular dynamics simulation and molecular modeling studies was undertaken.44 Despite the high homology between α- and β-tubulin, dolastatin 10 only demonstrated extensive molecular interactions with β-tubulin and occupied a binding pocket adjacent to the exchangeable GTP site that is composed of the amino acid residues: Ser172, Lys174, Val175, Asp177, Asn204, Glu205, Tyr208, Asp209, Phe212, Pro220 and Tyr222.44 The binding pocket occupied by dolastatin 10 is the same as that for cryptophycins 1 and 52, hemiasterlin and phomopsin A.44 This was corroborated by experimental observations that these structurally unrelated modified peptides competitively inhibit each other from binding to tubulin. The dolastatin 10 analog, symplostatin 1, also caused disruption of microtubule proteins. The effects of both compounds on cellular microtubules in A10 rat aortic smooth muscle cells were probed by indirect immunofluorescence using monoclonal β-tubulin antibody.43 Both compounds caused almost complete loss of cellular microtubules at low nanomolar concentrations and the formation of abnormal mitotic spindles.43
Due to its potent activity, dolastatin 10 was pursued for clinical trials for hormone refractory adenocarcinoma and recurring platinum-sensitive ovarian carcinoma. Further development of dolastatin 10 was discontinued, however, due to marginal efficacy and also peripheral neuropathy.45 The preclinical studies on symplostatin 1 in two murine solid tumor models indicated a delay in tumor growth in response to treatment.43 However, symplostatin 1 was poorly tolerated by test animals and also caused gastrointestinal and liver toxicity.43 Dolastatin 10 was not pursued for clinical development, but it served as the template for the design of synthetic analogs that have reduced toxicity and were intended for targeted delivery. The dolastatin 10 analog, monomethyl auristatin E, served as the drug portion of the antibody-drug conjugate brentuximab vedotin, which was approved in 2011 for clinical use in Hodgkin’s and systemic anaplastic large cell lymphoma (Fig. 1).17,46 Brentuximab vedotin consists of the drug monomethyl auristatin E linked to a cathepsin B-cleavable linker and a CD30-targeting antibody.17,46 This targeted approach allows for selective delivery of monomethyl auristatin E to tumor cells, which express higher levels of CD30 antigen and have low cross-reactivity in normal cells. Four molecules of monomethyl auristatin E are attached to cAC10 interchain Cys residues.17 Through receptor-mediated endocytosis, the antibody-drug conjugate is taken up by the cell and by the proteolytic action of lysosomal enzymes such as cathepsins, the free drug is released.17
Several other modified linear peptides (Fig. 4) belonging to the dolastatin 10-type of compound class were purified from marine cyanobacteria, such as symplostatins 3 and 447,48, malevamide D49 and belamide A.50 These compounds showed varying antiproliferative activity and, consequently, tubulin disrupting effects. The dolastatin 10/15 hybrid, symplostatin 4, exhibited moderate antiproliferative effects with IC50 values of 12 and 53 µM against HeLa and HT29 cells.48 The low potency of symplostatin 4 can be attributed to several structural modifications in both N-terminal and C-terminal regions of symplostatin 4, relative to dolastatins 10 and 15.48 Cryptophycins (Fig. 4) are a class of cyanobacterial depsipeptides distinguished by a modified octenoic acid moiety with a distinctive 7,8-epoxy-8-phenyl terminus.51 To date, more than 25 members of this compound class have been purified from Nostoc spp.52 The major metabolite, cryptophycin-1, showed potent antifungal activity against Cryptococcus but not Candida.51,53 An analog of cryptophycin-1, cryptophycin-24 or arenastatin A (Fig. 4) was isolated as the bioactive component of the marine sponge Dysidea arenaria. This highlighted the close association between the source sponge and a cyanobacterial symbiont in the production of secondary metabolites.54
Initial toxicity assessment in mice indicated that cryptophycin-1 is a potent, toxic compound with TD50 of 6.25 mg/kg.53 Subsequent studies on cryptophycin-1 indicated that it is indeed a potent cytotoxic agent, equipotent in drug sensitive and resistant cell lines, with subnanomolar IC50s.55 Indirect immunofluorescence studies using A10 vascular smooth muscle cells indicated significant depletion of cellular microtubules and reorganization in vimentin filaments, without any effect on cytoskeletal components.55 The effect of cryptophycin-1 on in vitro tubulin polymerization was extensively studied by several groups. Among the known tubulin poisons, the MOA of cryptophycin-1 closely resembles that of vinblastine, acting as a microtubule disrupting agent. Cryptophycin-1 is a noncompetitive inhibitor of [3H]vinblastine with Ki of 3.9 µM, while it is a competitive inhibitor of [3H]dolastatin 10 with Ki of 2.1 µM, indicating that cryptophycin-1 occupies the peptide binding site of the vinca domain of tubulin. The interaction of arenastatin A with porcine brain tubulin was likewise probed using [3H]arenastatin A.56 Binding experiments indicated that there is one binding site for arenastatin A per tubulin heterodimer, and the tubulin poisons rhizoxin was a competitive inhibitor while vinblastine partially competed with arenastatin A.56
The potent activity of the natural cryptophycins served as the basis for the design of more potent analogs to pursue for further development. Among these, cryptophycin-52 is regarded as the most potent inhibitor of microtubule dynamics (requiring only 5–6 molecules to sufficiently decrease microtubule dynamicity by 50%) which has a high affinity with tubulin and dissociates slowly, with Kd of 47 nM.57 Cryptophycin-52 (LY355703) reached Phase II clinical trials for non-small cell lung carcinoma (NSCLC) and ovarian cancer previously treated with platinum-based drugs.58,59 Poor response and unacceptable toxicity was seen in NSCLC patients, while better prognosis was observed in ovarian cancer. This indicated that the dose and schedule of cryptophycin-52 treatment is crucial for activity and in minimizing undesired side effects.58,59
Colchicine domain binding agents
From a bioassay-guided purification, the antiproliferative compound curacin A (Fig. 4) was obtained as the active principle.60 Curacin A is a unique fatty acid that is modified by a terminal thiazoline-methyl-cyclopropane moiety.60 Evaluation of its differential cytotoxicity profile in the NCI-60 screen followed by COMPARE analysis indicated similarity with anti-tubulin agents and, hence, suggested microtubule proteins as the molecular target of curacin A.60 Like other tubulin targeting agents, curacin A also caused G2/M cell cycle arrest. Assessment of the effects of curacin A on purified tubulin corroborated its inhibitory activity on tubulin polymerization. Curacin A competed with radiolabeled colchicine for binding to tubulin, but not with vinblastine, thus suggesting that curacin A occupies the colchicine binding site, distinct from tubulin-targeting cyanobacterial peptides.61 Radiolabeled curacin A was prepared through biosynthesis using Na[14C]acetate as precursor and used to probe the interactions of curacin A with tubulin.61 Binding studies using [14C]curacin A indicated rapid association with tubulin, with kf 4.4 × 103 M−1s−1.61 Dissociation from tubulin is very slow with a half-life of 50–70 h at 32°C. However, this was not due to formation of a curacin-tubulin covalent adduct, as [14C]curacin A is released from tubulin following urea treatment. Scatchard analysis indicated significant superstoichiometric amounts of [14C]curacin A bound to tubulin and suggested the presence of two binding sites for curacin A in tubulin.61 The binding of curacin A to tubulin resembles that of combrestatin A-4 and podophyllotoxin, while its dissociation from tubulin is similar to the behavior of colchicine.
3.1.2 Actin-Targeting Agents
The ability of actin to control cytokinesis (the separation of two daughter cells) made it a logical target in cancer chemotheraphy.62 While actin targeting agents have yet to gain approval for clinical use, these small molecules are perhaps among the most prevalent molecular probes to study actin function. The fungi-derived compound cytochalasin was initially used to define the functions of actin in maintaining cell shape and cellular motion.63 In addition, the visualization of microfilament assembly is commonly accomplished using a fluorescent derivative of the natural product phalloidin.63
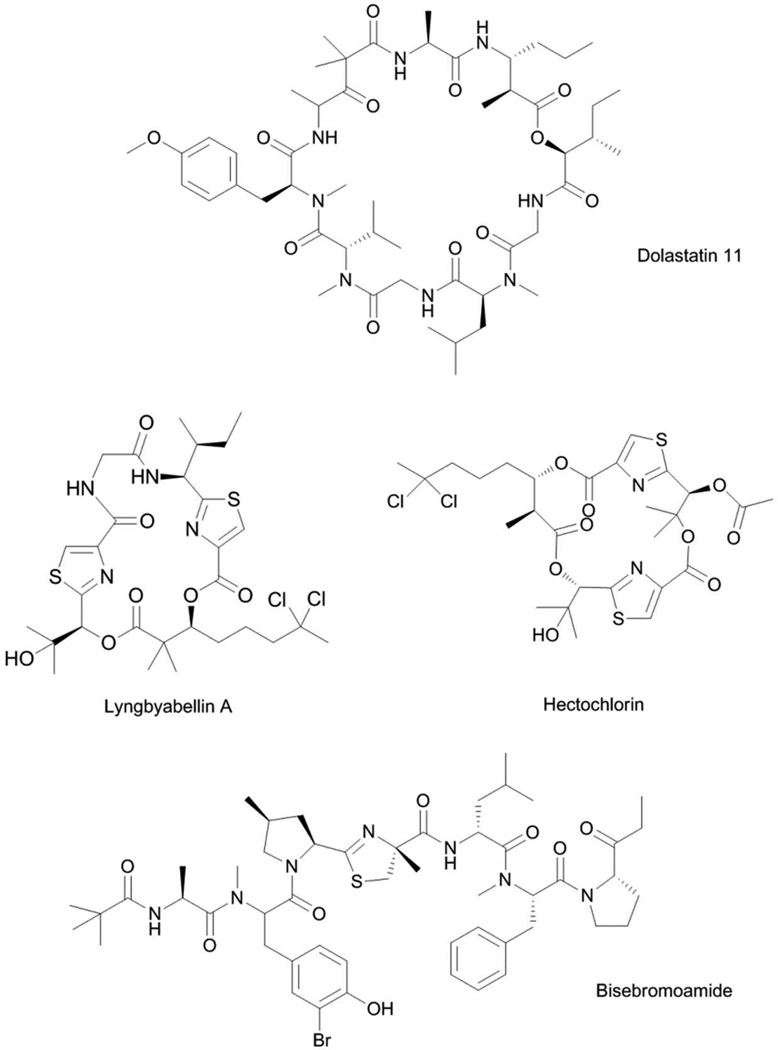
The family of antiproliferative cyclodepsipeptides belonging to the dolastatin 11-class (Fig. 5) was purified from both cyanobacteria and D. auricularia.64 The compounds are different from dolastatins 10 and 15 since this family of compounds did not induce mitotic arrest or any interaction with tubulin or microtubule proteins. Instead, dolastatin 11 caused a rapid change in cell shape, characterized by extensive retraction of the cytoplasm, leading to the formation of binucleated cells.65 These morphological changes indicated that dolastatin 11 potentially interacted with the cytoskeletal protein actin. The antiproliferative activity of dolastatin 11 against PtK1 cells was compared with the known actin-targeting agents jasplakinolide and latrunculin B. Dolastatin 11 showed better biological activity and was close to 3- and 12-fold more potent than jasplakinolide and latrunculin B, respectively.65 PtK1 cells were incubated with jasplakinolide, dolastatin 11 or solvent control at varying durations, using IC50 and 10× IC50 concentrations, and the effects on microfilaments of PtK1 cells were evaluated by immunofluorescence detection using FITC-labeled anti-actin antibody. Cytoplasmic retraction was not observed at lower concentrations for both compounds for all time points, and little difference was observed in the cellular effects of the two compounds. At higher concentrations, dolastatin 11 and jasplakinolide showed significant differences in the rate of inducing their cellular effects. Dolastatin 11 caused few stress fibers to remain after 30 min, and microfilaments were no longer observable at later time points. The effects of dolastatin 11 on the assembly of purified actin were evaluated using pyrenyl-labeled actin.65 Incorporation of pyrenyl-labeled actin on growing unmodified actin leads to an enhancement in fluorescence that can be readily monitored. In the presence of inducing salts (50 mM KCl, 2 mM MgCl2, 1 mM ATP) and 10 µM actin/pyrenyl-labeled actin, dolastatin 11 stimulated actin polymerization, with no significant difference to the effects of jasplakinolide or phalloidin.65 Further comparison of the effects of these three compounds on actin polymerization was done in the absence of inducing salts in the polymerization reaction. Dramatic differences in the activities of dolastatin 11, jasplakinolide and phalloidin were observed; the actin stimulatory activity paralleled the observed cytotoxicity for these compounds, with dolastatin 11 being the most potent.65 Further differences between dolastatin 11 and jasplakinolide were observed for the inhibition of binding of FITC-labeled phalloidin to actin polymer. Dolastatin 11, unlike jasplakinolide, was not able to displace FITC-labeled phalloidin from actin polymer, and suggested that dolastatin 11 occupies a distinct binding site.65 The binding of dolastatin 11 to actin was examined using X-ray diffraction diagrams that were derived from oriented filament sols.66 The results corroborated the biochemical analysis, wherein dolastatin 11 did not compete with phalloidin, evident from the different binding sites occupied by these two natural products in the same region of the F-actin strand. Both dolastatin 11 and phalloidin occupy opposite sides of the gap between two long-pitch F-actin strands.66 This indicated that modulation of the interaction between F-actin strands is a key aspect in controlling microfilament assembly. Several analogs of dolastatin 11 were also purified from marine cyanobacteria such as majusculamide C67 and lyngbyastatins 1 and 3.68,69 These compounds were shown to induce similar morphological effects on cells, which thus indicated effects on actin polymerization as well.
Fig. 5.
Actin-targeting agents from marine cyanobacteria.
Lyngbyabellins70–73 and hectochlorin74 (Fig. 5) are structurally related modified cyclodepsipeptides characterized by a bisthiazole moiety and an unusual dichlorinated β-hydroxy acid. These compounds exhibited moderate antiproliferative activity, with low-micromolar IC50s against a variety of cell lines.70 Initial evidence for the effects of this class of compounds on actin came from morphological changes in treated cells, characterized by an extensively retracted cytoplasm and binucleated cells, concurrent with a disrupted microfilament network.70 The assembly of purified actin in the presence of hectochlorin was also monitored using light scattering experiments.74 Hectochlorin showed a dose-dependent effect in promoting actin polymerization, comparable to jasplakinolide.74 However, hectochlorin did not displace the binding of FITC-labeled phalloidin to actin polymer, indicating a distinct binding site from phalloidin.74 The MOA and target of the lyngbyabellin-class of compound were validated after it was purified as the bioactive component in a targeted screening for new actin modulators using high content image analysis.75 In this screening for actin-targeting compounds, the documented changes in the morphology of cancer cells following compound treatment were utilized as indicators of activity.75 HeLa cells treated with actin-targeting compounds showed decreased cytoplasmic area of up to 70% and an increase in distance between the centroid of the nucleus and centroid of the entire cell.75 Plotting the distance between centroids of nucleus and cell versus the area of the cytoplasm allowed for clustering of actin stabilizing and destabilizing compounds. Nuclear protrusion arising from actin-disruption by small molecules was first observed for the cyanobacterial compound tolytoxin.76 On this basis, the purified lyngbyabellin C was shown to be an actin stabilizer, and the cellular effects of lyngbyabellin C were accounted for by its weak inhibition of the polymerization of purified F-actin.75 This cell morphology-based screening also identified seven known actin targeting compounds: cytochalasin D, doliculide, jasplakinolide, latrunculin A, mycalolide B, seragamide A and swinholide, from a library of 400 purified natural products and, thereby, validated this screening platform.75 In addition, the cyanobacteria-derived compound bisebromoamide (Fig. 5) was also identified as a hit compound; likely an actin stabilizer.75 The morphological assay was validated by monitoring the assembly of purified actin in the presence of varying concentration of bisebromoamide. A concentration dependent effect on actin polymerization of pyrene labeled G-actin was observed with bisebromoamide treatment, together with inhibition of F-actin depolymerization.75 Synthesis of a bisebromoamide-fluorescein conjugate and monitoring of cellular localization in HeLa cells demonstrated the localization of bisebromoamide on actin filament.75
3.2 Nonclassical Anticancer Drug Targets
The advent of advanced molecular biology tools and the omics era has led not only to improved identification of cellular targets of small molecules but also in defining deregulated components of signaling pathways in cancer. It has become apparent that cancer cells are able to achieve their hallmark characteristics by mutations of proto-oncogenes to oncogenes leading to either increased expression or constitutively active signaling molecules such as enzymes, transcription factors, and cell surface receptors.77,78 As such, the inhibition of these overactive or upregulated enzymes has been recognized as a promising strategy for mitigating tumor growth. The differential expression of selected oncogenes in cancer compared to normal cells also allowed for the development of either selective small molecules or the design of magic bullets aimed at cancer cells, thereby giving improved success of the therapy and management of potential side effects. In addition, the loss-of-function of tumor suppressor genes is key to progression of malignancies, and reactivation of the expression of these genes has been recognized as bona fide targets for cancer therapy.77,78 Hence, the design of effective small molecule therapeutics for cancer that has distinct mechanisms of action from classical agents can also be associated with the improved understanding of the molecular basis of the disease. Several small molecules from cyanobacteria (Fig. 6) have been demonstrated to act via novel mechanisms and on different targets compared with classical anticancer agents.
Fig. 6.
Small molecules from marine cyanobacteria acting on non-classical cancer cell targets.
3.2.1 Secretory pathway
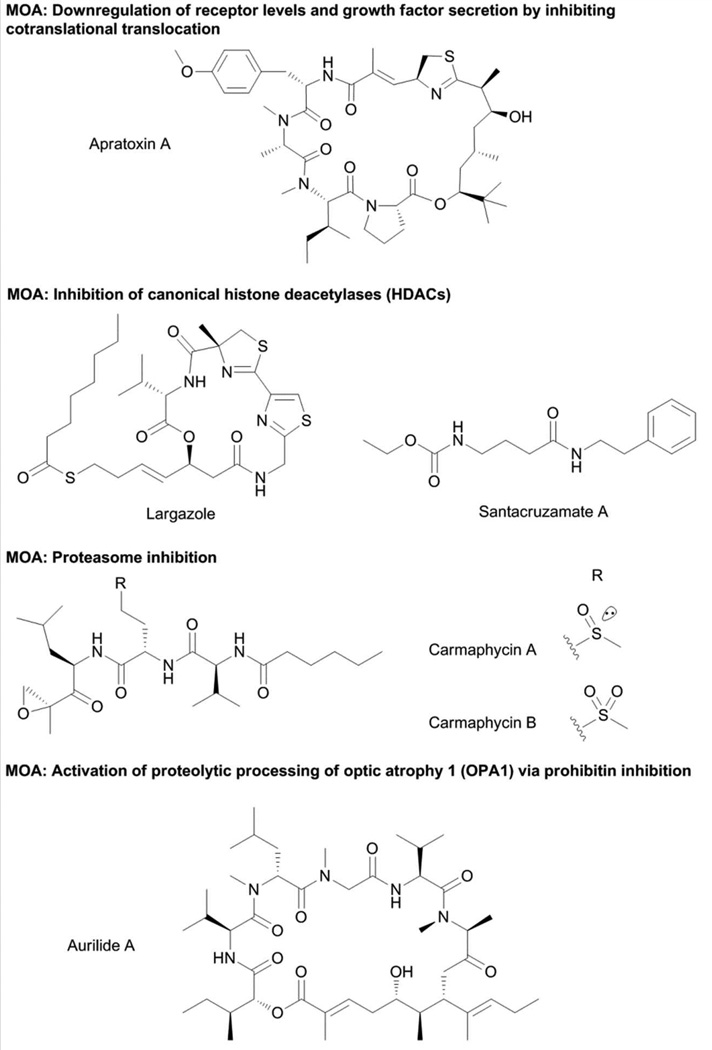
The apratoxins (Fig. 6) are a group of cyclodepsipeptides notably recognized by the presence of a contiguous pentapeptide chain, a modified Cys residue and a highly functionalized polyketide chain.79–84 The first member of this group of compounds, apratoxin A, showed potent in vitro antiproliferative activity against cancer cells.79 Apratoxin A did not affect the classical anticancer drug targets such as microfilaments, microtubules and topoisomerases, with the NCI-60 screening and COMPARE profile of apratoxin A indicating a novel MOA.79 Transcriptome analysis of HT29 cells treated with apratoxin A indicated changes in mRNA levels for more than 100 genes which include oncogenes, tumor suppressors, components of the cell cycle and stress response regulators.85 Genes that confer resistance to apratoxin A when overexpressed were interrogated using a library of 27,000 cDNA transiently transfected in U2OS cells. This genome-wide screening identified 46 cDNAs that attenuated the cytotoxic effects of apratoxin A through anti-apoptotic mechanisms, blockade of apratoxin A-dependent G1 cell cycle arrest and induction of apratoxin A-independent cell cycle arrest.85 A significant portion of the cDNAs conferring resistance to apratoxin A encoded for fibroblast growth factor receptor (FGFR) variants, some of which also have higher transcript levels in NCI-60 cell lines that were less susceptible to apratoxin A.85 Quantitative proteomics using iTRAQ labeling indicated that apratoxin A modulated the levels of a subset of proteins, specifically receptors or membrane-associated proteins and proteins that are localized in the endoplasmic reticulum.86 In addition, immunoblot analysis demonstrated that apratoxin A downregulated the levels of several cancer-associated receptors such as c-Met, Her2, PDGFR-β, and IGF1R-β.86 To elucidate the mechanism of apratoxin A-mediated downregulation of receptors, in vitro translation with or without microsomal membrane with [35S]methionine incorporation was performed and monitored by SDS-PAGE followed by autoradiography. Apratoxin A did not affect protein synthesis but rather inhibited glycosylation and also signal peptide-cleavage by preventing cotranslational translocation of proteins bound for the secretory pathway.86 Non-functional non-translocated receptors underwent proteasomal degradation instead. In a parallel study, apratoxin A-induced downregulation of membrane receptors was also suggested to occur as a result of demonstrated binding of an oxazoline analog of apratoxin A to HSP70, which in turn could promote HSP90 client protein (including receptor) degradation.87 The ability of apratoxin A to inhibit cotranslational translocation suggested the simultaneous downregulation of membrane receptors and inhibition of secretion of growth factors and other ligands.
Downregulation of receptor expression and secretion of corresponding ligands by apratoxin A proved to be a promising strategy to attenuate in vivo tumor growth in a murine colorectal tumor xenograft model. However, significant toxicity was observed with apratoxin A treatment and found to be irreversible.88 The design of new analogs of apratoxin A was then aimed to obtain maximum in vivo potency but without the irreversible toxicity exerted by apratoxin A. The knowledge of the MOA of apratoxin A was utilized in the design of new analogs. Comprehensive SAR information was obtained with monitoring of the antiproliferative activities against cancer cells and the levels of VEGF-A and c-MET.88 This corroborated that the biological activities of the apratoxin class of compounds are intricately related to their ability to reduce the expression of receptors and ligands which are crucial aspects in several of the clinical hallmarks of cancer. Importantly, in vivo studies suggested that the modified Cys unit is responsible for the irreversible toxicity of apratoxin A, indicating that this problem was a result of off-target activity rather than mechanism-based.88 The second generation apratoxin analog, apratoxin S4, showed the same effects as apratoxin A in cotranslational translocation, with potent in vivo activity in a colorectal xenograft model, but did not show irreversible toxicity.88 The ability to segregate the off-target effects and relevant pharmacological actions of apratoxin A through selective modifications on the core structure allows these potent small molecules to be further developed. Further structural modifications have led to an analog that is less prone to chemical and metabolic deactivation (apratoxin S8) and an even more potent analog (apratoxin S9).89
3.2.2 Histone deacetylases
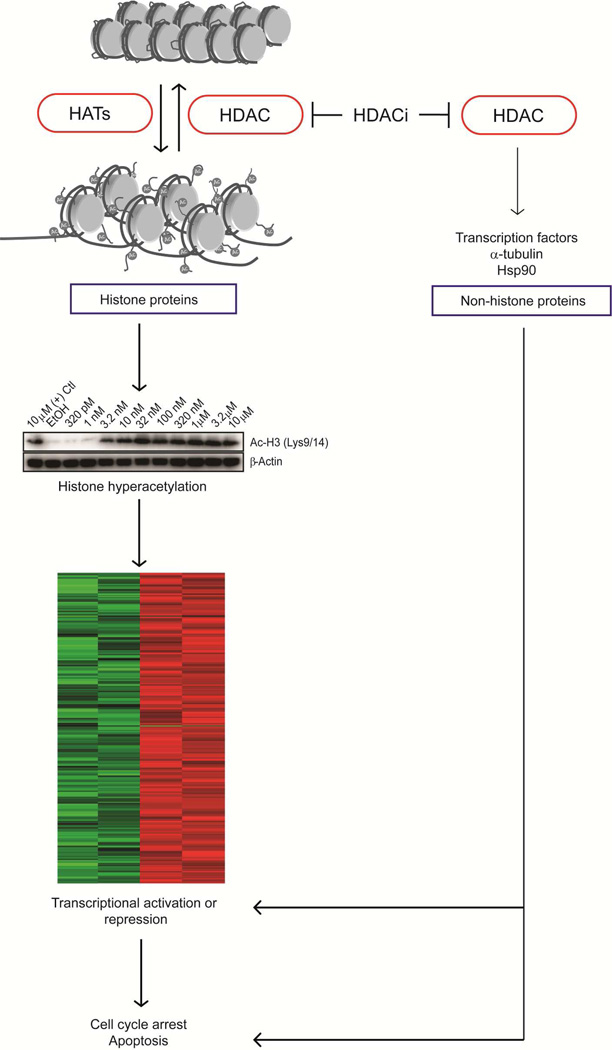
Histone deacetylases (HDACs), enzymes that modulate the acetylation of histones and nonhistone proteins, are divided into two groups, the canonical HDACs (classes I, II, IV) and class III sirtuins.90 Canonical HDACs utilize a catalytic Zn2+ residue and a His-Asp dyad system for the deacetylation reaction.90 On the other hand, sirtuins are NAD+-dependent enzymes, which have weak deacetylase activity and act mainly as ADP-ribosyltransferase.91 Modulation of histone acetylation neutralizes interactions between histone proteins and DNA of nucleosomes, thereby affecting higher-order chromatin structure and favoring the binding of transcription factors, ultimately leading to gene expression changes (Fig. 7).92 In addition, recent studies identified many non-histone proteins that are also subjected to acetylation and are likely to be substrates of canonical HDACs and SIRTs as well.10,93 Because of the well-documented cellular consequences of modulation of histone acetylation, pharmacological intervention targeting HDACs is being extensively explored for drug development.94 Two HDAC inhibitors have been approved for clinical use; the natural product romidepsin (FK228)95,96 and one natural product-like compound, vorinostat (SAHA).97 Combination therapy and non-cancer applications are also being pursued for HDAC inhibitors in the preclinical and clinical trial stages.93 The major challenge in HDAC inhibitor development is finding isoform-selective agents with the hope of minimizing undesired side effects of HDAC inhibitors and also defining the physiological roles of different HDAC isoforms.93
Fig. 7.
MOA of HDAC inhibitors. Cell cycle arrest and apoptosis mediated by HDAC inhibitors are in part due to alteration in gene expression arising from changes in acetylation levels of histone and non-histone proteins.
While several canonical HDAC inhibitors have been purified from terrestrial microbes such as trichostatins, FK228 and trapoxins, HDAC inhibition is so far a minor theme in marine microbes and macroorganisms.93 The sponge-derived psammaplin A and azumamides,93,98 marine fungus-derived microsporins99, and the cyanobacterial compounds largazole100,101 and santacruzamate A102 are the only known marine-sourced canonical HDAC inhibitors.
Largazole (Fig. 6) was purified from a Symploca sp. collection from Key Largo, Florida using a bioactivity-directed approach.101 The MOA of largazole is disguised by its prodrug characteristics, where activation by protein-assisted hydrolysis is required to liberate the bioactive species largazole thiol.103,104 Largazole thiol features a 3-hydroxy-7-mercaptohept-4-enoic acid moiety, a characteristic structural feature of the cyclic depsipeptides FK228, FR-901375,105 and spiruchostatins.106,107 This moiety is also disguised in FK228, FR-901375 and spiruchostatins as a disulfide, which requires glutathione-assisted reduction to liberate the bioactive species.95 FK228 was first shown to inhibit HDACs by similarity of its phenotypic effect with the known HDAC inhibitor trichostatin A in a screen for activators of SV40 promoter-dependent transcription.108 The structural similarity of the “warhead moiety” of largazole thiol and FK228 then indicated a similar direct target and MOA.103,104 Largazole was shown to selectivity inhibit class I HDACs, with comprehensive assessment of inhibitory activity against 12 purified human HDACs using fluorogenic HDAC substrates.100 The isoform selectivity of largazole is consistent with the cyclic depsipeptide HDAC inhibitor FK228, with largazole exhibiting slightly better potency.100 The molecular target of largazole was also confirmed by SAR studies that indicated the importance of the 3-hydroxy-7-mercaptohept-4-enoic acid moiety for biological activity.109 Molecular interactions of largazole with its target protein were also elegantly demonstrated by X-ray cocrystallization with HDAC8 at 2.14 Å resolution.110 Largazole binds to the HDAC active site and coordinates the active site Zn2+ residue of HDAC8, via the thiol moiety, present as thiolate and the macrocycle occupying the rim of the active site.110 Visualization of the molecular interactions of largazole with HDAC8 is the first ever reported for a cyclic depsipeptide HDAC inhibitor and, hence, provided critical insights into the binding modes of related compounds and will be valuable in the design of next generation HDAC inhibitors, thereby serving as an important molecular probe.110
The effects of the HDAC inhibitors largazole, FK228 and SAHA on the transcriptome was assessed using comparative microarray analysis of HCT116 cells treated with these compounds, leading to significant changes in gene expression of >800 genes.111 For example, these HDAC inhibitors significantly upregulated the expression of cell cycle inhibitor proteins p21, p19, p57 and p15 and a proapoptotic BCL2 protein variant (BCL2L11), and at the same time downregulated the levels of growth factor receptors (EGFR, HER2, MET) and also CDK6 and cyclin D1.111 In addition to its antiproliferative properties in vitro and in vivo, largazole also modulated the invasiveness of breast cancer cells through gene expression changes and non-histone mediated effects.112 Largazole was able to reverse the epigenetic silencing of E-cadherin expression in invasive breast cancer cells and to modify the composition of the E-cadherin complex.112 Largazole was shown to strongly induce E-cadherin expression (histone-mediated) and also increased the association of E-cadherin with γ-catenin to ensure proper cell membrane localization (non-histone mediated).112 These effects of largazole were a direct consequence of HDAC inhibition as trichostatin A and SAHA also displayed the same cellular effects, although largazole was more effective.112 Largazole’s anti-invasive properties were enhanced in combination with dexamethasone which blocked the production of the pro-invasive cleaved form of CDCP1 (CUB domain-containing protein 1), present in the E-cadherin complex, through upregulation of plasminogen activator inhibitor-1.112 Largazole and dexamethasone cooperated to induce E-cadherin localization in the plasma membrane.112
Combination therapy is also an active area of interest for HDAC inhibition. Another approach to design combination therapy is through exploring the combinatorial pharmacology utilized by marine cyanobacteria.48 In this regard, a combination of largazole and symplostatin 4 was employed to achieve cooperative effects in preventing the proliferation of human colorectal adenocarcinoma cells.48 It has been proposed that HDAC inhibitors sensitize cancer cells to the proapoptotic effects of microtubule disruptors and other agents.93
While largazole was initially purified based on its antiproliferative effects, the application of this small molecule has been expanded. Non-cancer therapeutic applications for HDAC inhibitors are also being actively explored. In the case of largazole, it was shown to also have bone-forming properties113 and cytoprotective effects against liver fibrosis.114 The in vivo osteogenic activity of largazole was attributed to its ability to stimulate bone formation and suppress bone resorption.113 These effects were mediated in part by largazole’s ability to modulate the expression of ALP, OPN and BMP-2, 4, 6, 7, and 9.113 The selectivity of the antiproliferative effects of largazole for transformed cells served as an advantage, as largazole did not exhibit any cytotoxic effects against murine pluripotent mesenchymal precursor C2C12 cells.114 The selective action of largazole was also observed in hepatic cells, where largazole increased the acetylation of histone H3 and H4 and also inhibited the proliferation of hepatic stellate cells, leading to reduction of liver fibrosis.114 The effects of largazole were shown to be a direct consequence of histone acetylation as a decrease in biological activity was observed with siRNA-mediated knockdown of HDACs 1–3. Downstream effects of HDAC inhibition in hepatic stellate cells include changes in expression of TGFβR2, VEGF, VEGFR, Smad2 and phosphorylated Akt.114 Efforts are underway to modulate the pharmacokinetics and pharmacodynamics of the natural product by designing largazole-based HDAC inhibitors with altered of prodrug properties.115 Furthermore, the class I HDAC selective largazole scaffold now serves as a template for the synthesis of isoform specific inhibitors.116
It must be noted, however, that HDAC inhibitors can oftentimes appear as promiscuous hits in screening, due to their ability to modulate gene expression. For example, psammaplin A was identified as a modulator of the Wnt signaling pathway.117 Rather than acting on the signaling pathway itself, the activity of psammaplin A was due to significant transcriptional activity.117 This was observed in the case of largazole in the screening of inhibitors for ubiquitin activating enzyme (E1) using mink lung epithelial cell line (Kip16) stably expressing the N-terminal GFP-p27 fusion.118 Largazole showed nanomolar potency for GFP-p27 stabilization, however, E1 inhibition was observed at high micromolar concentration.118
Another HDAC inhibitor recently purified from a Symploca-like marine cyanobacterium is santacruzamate A (Fig. 6), structurally reminiscent of the pan-selective HDAC inhibitor SAHA.102 This compound, while showing striking structural similarity with SAHA, was determined to be a class I selective HDAC inhibitor with potent activity at the sub-nanomolar range against HDACs 2,4 and 6.102 The HDAC inhibitory profile of santacruzamate A showed selectivity for certain class I over the tested class II HDACs.102 The antiproliferative activity of santacruzamate A is only modest with low-micromolar GI50s against HCT116 and HuT-78 cells and, depending on the cell type, showed 2–10-fold difference in activity compared to SAHA.102 The potent and selective inhibitory activity of santacruzamate A in cell-free systems provides a unique opportunity to explore the molecular determinants of selectivity in hydroxamate-type HDAC inhibitors which have been known to be pan-selective inhibitors.
While the pharmacophore for inhibitors of canonical HDACs and their cellular effects have been clearly established, these areas are less explored for sirtuins.93 The majority of sirtuin modulators have been derived from plants and agents targeting these proteins from marine sources have not been prevalent.119 From marine cyanobacteria, tanikolide dimer is the only SIRT inhibitor isolated to date.120 This compound was purified using target-based screening for SIRT2 inhibitors.120 Tanikolide dimer showed varying IC50s against SIRT2, from sub-nanomolar to low-micromolar, depending on the assay protocol employed.120 The activity of tanikolide dimer in the enzymatic assay, however, did not translate to potent inhibition of the growth of the human lung H-460 cancer cell line, consistent with other SIRT2 inhibitors. Thus, this SIRT inhibitor may find utility as a pharmacological probe and in non-malignant applications.120
3.2.3 Proteasome
The ubiquitin-proteasome pathway is one of the key mechanisms to regulate protein levels. Proteins tagged by polyubiquitination are subjected to proteolytic degradation by the proteasome.121 The 26S proteasome consists of a 20S proteolytic core and a 19S regulatory particle.121 The 20S proteolytic core is critical to degradation of protein substrates, containing two chymotrypsin-like, two trypsin-like and two caspase-like sites, while the 19S regulatory protein is responsible for substrate recognition and priming for proteolysis.121 The role of the proteasome in cancer was suggested by the high activity in rapidly dividing cells and the increased cell death associated with proteasome inhibition. The validation of the proteasome as a promising target for cancer therapy was fully realized with the approval of bortezomib for multiple myeloma.121,122
1H NMR- and bioactivity-guided purification of a Symploca collection yielded two potent antiproliferative agents, carmaphycins A and B (Fig. 6).123 These compounds are characterized by a tripeptide moiety fused to a hexanoic acid and α,β-epoxyketone on the N- and C-terminal ends, respectively. Insights into the MOA of the modified peptides carmaphycins A and B were obtained from the structural similarity of these compounds with the epoxomicin class of compounds.123 Carmaphycins and epoxomicins are characterized by an α,β-epoxyketone that has been demonstrated to be one of the key features in the latter for inhibiting the proteasome, forming a covalent bond with the catalytic Thr residue of the β5 subunit.121 Carmaphycins inhibited the chymotrypsin-like (β5 subunit) activity of the 20S proteasome, with comparable potency to epoxomicin, culminating in potent inhibition of the growth of cancer cells, particularly those which harbor KRAS/tp53 mutations.123 Using the X-ray crystal structure of the 20S proteasome derived from the complex with epoxomicins, the molecular interactions of carmaphycins were modeled.123 Extensive hydrogen bonding, van der Waals and solvent interactions were observed. This molecular dynamics simulation also provided insight into the role of the distinctive sulfoxide or sulfone moieties in carmaphycins, derived from the Met residue. This moiety appears to contribute to additional hydrogen bonding interactions with Gly23 residue of the 20S proteasome.123
There is much promise on the proteasome as a target of anticancer drugs, with the recent approval of the proteasome inhibitor, carfilzomib, for multiple myeloma.124 Carfilzomib was designed based on the natural product epoxomicin and highlights the importance of knowledge of the drug target to effectively design second-generation agents that are potent with good solubility and pharmacokinetic properties.124 The proteasome serves as an attractive target not just for cancer but other diseases as well, such as lupus nephritis, inflammation, reperfusion injury after stroke, infection and stimulation of bone and hair growth.121
3.2.4 Prohibitin
Aurilides125–127 (Fig. 6) and related compounds lagunamides,128–130 and kulokekahilides131,132 are potent antiproliferative agents that have nanomolar IC50s. Evaluation of aurilide B using the NCI-60 screening indicated potent antiproliferative activity with mean GI50 of 10 nM.126 Information about the direct target and MOA of this compound class was derived from chemical proteomics using an aurilide affinity probe.133 Aurilide was conjugated to a protease-cleavable polyproline linker with a biotin molecule tag and immobilized on neutravidin-agarose beads.133 Significantly enriched protein was observed in the membrane fraction and subsequent MS/MS analysis indicated this to be prohibitin 1 (PHB1).133 The results were also compared to a control probe, using the inactive analogue 6-epi-aurilide, which showed a decreased intensity of the protein band corresponding to PHB1.133 To validate the results of affinity purification, the response of HeLa cells to aurilide treatment was probed for both PHB1 overexpression and depletion. Stable cell lines that overexpressed PHB1 showed resistance to aurilide, while siRNA-mediated partial knockdown of PHB1 led to sensitivity.133 Comparison of the morphological changes in the mitochondria of PHB1 knockdown and aurilide-mediated PHB1 inhibition yielded the same phenotype, with the mitochondria appearing as fragmented.133 Immunoblotting experiments further demonstrated that PHB1 inhibition by aurilide A facilitates the proteolytic processing of the optic atrophy 1 (OPA1) protein, which in turn signals the initiation of apoptosis.133 Aurilide represents the first small molecule inhibitor of PHB1 and thus serves as an important pharmacological probe in elucidating the role of PHB1 and OPA1 in the initiation of apoptosis.133 In this case, the natural product aurilide enabled the discovery of a potentially novel anticancer drug target and also of a probe to understand the function of prohibitin and OPA1 in cancer progression.
3.2.5 Kinases
Several of the clinical hallmarks of cancer are products of deregulated kinase activity, either due to constitutive activity or upregulated expression.134 The phosphorylation of substrate proteins by kinases plays a key role in activation or protein inhibition as well as in nuclear translocation that culminates in gene expression.135 Kinases have been shown to act as oncogenes, or downstream effectors of transforming oncogenes and initiators of angiogenesis and metastasis.136 Among the cancer-associated kinases are the fusion protein Bcr-Abl, epidermal growth factor receptor (EGFR), vascular epidermal growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PDGFR).134,136 The many kinases and their involvement in critical signaling pathways have made them critical drug targets for cancers, immunological, neurological, metabolic and infectious diseases.136
Bisebromoamide and scytonemin are two cyanobacteria-derived compounds shown to target kinases and related enzymes. Bisebromoamide is a potent antiproliferative agent that bears unique structural features such as pivalic acid, methylthiazoline, and an unprecedented 2-(1-oxopropyl)pyrrolidine moieties.137 Bisebromoamide inhibited PDGF-initiated signaling in NRK cells and attenuated the phosphorylation of ERK.137 In addition to its effect on certain kinases, bisebromoamide was obtained as a screening hit for actin-targeting agents using high-content analysis for morphological changes in HeLa cells as a consequence of actin disruption.75 However, the relationship between the kinase inhibitory activity and actin destabilizing effects of bisebromoamide to the potent in vitro antiproliferative activity is not fully elucidated. The structural basis for bisebromoamide-mediated kinase inhibition has not been characterized.
A target-based screen for inhibitors of polo-like kinase 1 yielded the cyanobacterial pigment scytonemin.138 Polo-like kinase 1 has been demonstrated to have high expression in human tumors and is also an attractive drug target due to its involvement in cell cycle events such as mitosis, centrosome maturation, assembly of the bipolar spindle, separation of sister chromatids and exit from mitosis.139 Based on binding studies, scytonemin acts as both competitive and noncompetitive inhibitor of polo-like kinase 1. The inhibition of PLK1 through pharmacological modulation by a small molecule has proven to be an effective anticancer strategy as seen for the small molecule BI-2536.139
Additional profiling of scytonemin showed inhibitory effects on other kinases such as Myt1, checkpoint kinase 1, cyclin-dependent kinase 1/cyclin B and protein kinase Cβ2.138 As expected from the effects of scytonemin on kinases, the compound inhibited the proliferation of cells stimulated with growth factors, induced apoptosis but did not arrest cells at a specific stage, which may indicate multiple cellular targets for scytonemin, warranting further investigations.138
3.2.6 Metal chelation
Metal chelation is a possible mechanism of antiproliferative activity. Chelation of copper in particular has been actively explored for modulating angiogenesis. The copper chelating compounds penicillamine and tetrathiomolybdate have been evaluated in pre-clinical and clinical trials for anti-cancer and antiangiogenic activities.140,141 Critical in exploiting copper chelation as an antiangiogenic therapy is maintaining a low level of copper to block angiogenesis and simultaneously obtaining sufficient amount for critical copper-dependent processes to prevent clinical toxicity.142
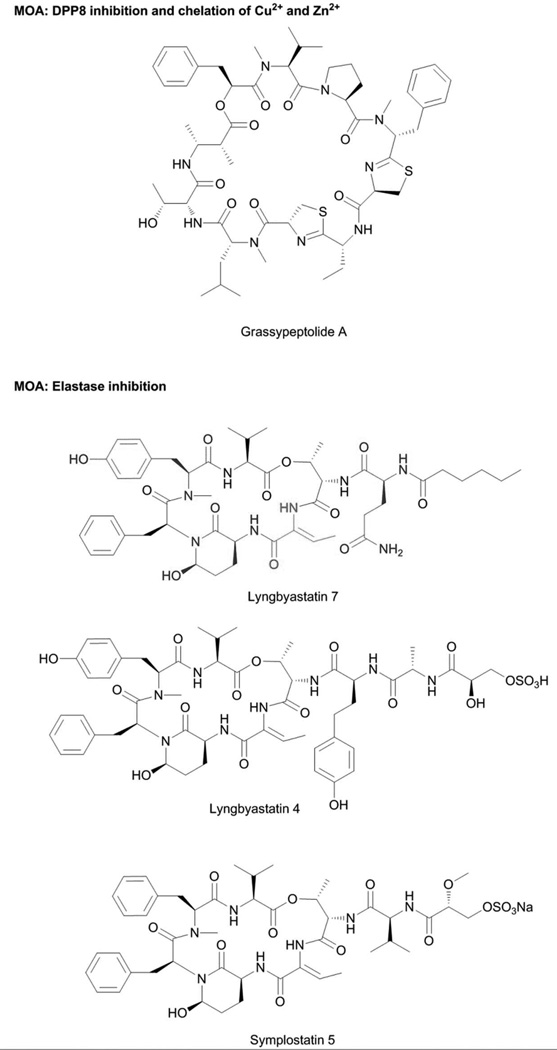
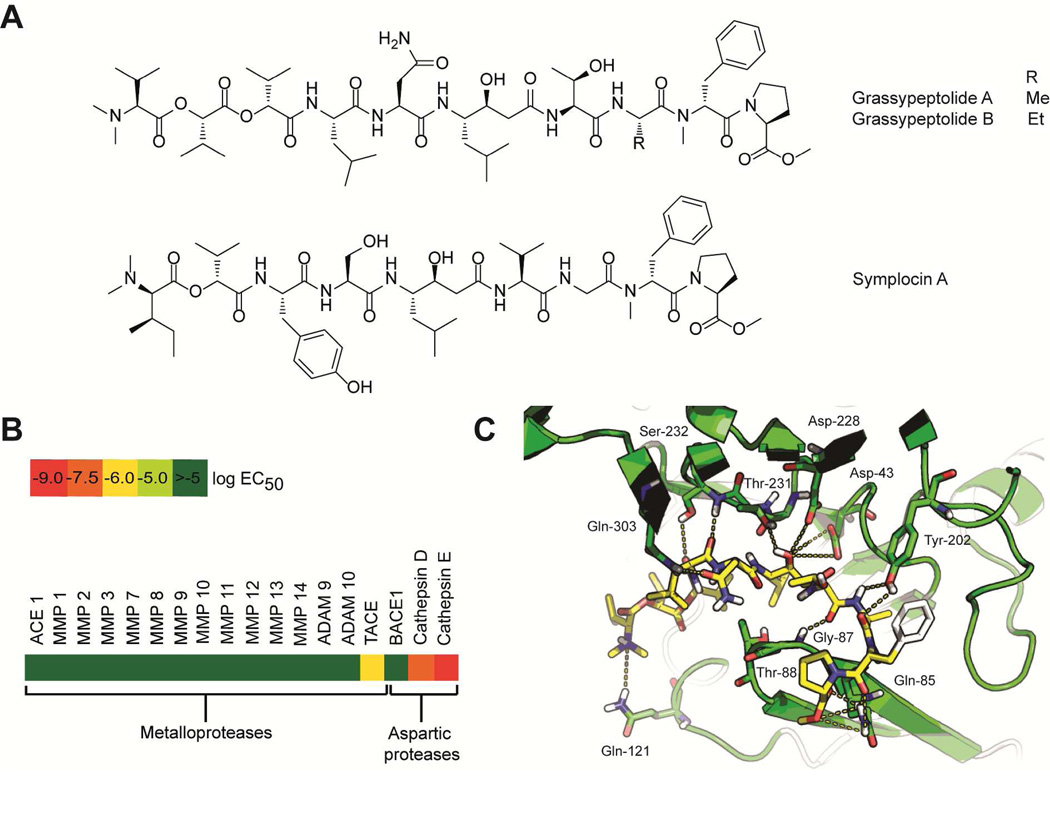
Grassypeptolides are a group of bis-thiazoline containing cyclic depsipeptides (Fig. 8) with antiproliferative activity against a variety of cancer cell lines, with sub-nanomolar to low-micromolar IC50s.143,144 Related to the grassypeptolides are the marine invertebrate-derived compounds patellamides A and C, ascidiacyclamide and lissoclinamide. Their antiproliferative activities were previously demonstrated to be due to the metal chelating properties, binding Cu2+ and Zn2+ via the tandem thiazole/oxazole or thiazoline/oxazoline moieties.145,146 In accord, the metal chelating activity of grassypeptolide A was assessed using mass spectrometry and circular dichroism (CD) measurements.144 Addition of 1 eq of Cu2+ caused changes in the CD spectrum of grassypeptolide A, and the [M-H+Cu]+ pseudomolecular ion was observed.144 No additional change was observed at higher equivalence ratios. Addition of 1 eq of Zn2+ caused a slight positive change in the CD spectrum of grassypeptolide A, similar to effects of lissoclinamide 10.144 Pseudomolecular ions corresponding to the Zn2+ adducts of grassypeptolide A were also observed. The contribution of metal binding to the antiproliferative properties of grassypeptolide has not been demonstrated and other pleiotropic effects have also been suggested to possibly play a role. SAR studies indicated the importance of the bisthiazoline moiety of grassypeptolides.147 Weak antiproliferative activity (IC50 in the high-micromolar range), was observed for grassypeptolide A analogs lacking the bis-thiazoline moiety.147 Grassypeptolide A was also shown to cause accumulation of cells at G0-G1 and apoptosis.147 An increased expression of the cell cycle inhibitor proteins p27 and p21 proteins was observed, with concomitant decrease in expression of antiapoptotic BCL-2 and BCL-xL.147 In addition, grassypeptolide A was also demonstrated to be a selective inhibitor of dipeptidyl peptidase 8 (DPP8), potentially linked to the observed cellular effects of lower IL-2 production and proliferation of activated T cells.148 This additional bioactivity indicates the potential of grassypeptolide as a probe to elucidate the functions of DPP8, a type of serine protease.148
Fig. 8.
Selective DPP8 and elastase inhibitors from marine cyanobacteria. The 2-amino-butenoic acid moiety (red) of modified cyanobacterial cyclodepsipeptides serves as the warhead in inhibiting elastase while other modified amino acid residues (blue) provides additional key interactions.
3.3 Protease Inhibition
3.3.1 Serine proteases
Serine protease inhibition is a major mechanistic theme, with more than 100 members in this type of compounds sourced from terrestrial, freshwater and marine cyanobacteria.19,52 The DPP8 inhibition by grassypeptolides148 is rather unique among cyanobacterial compounds. Most of these serine protease inhibitors are cyclic depsipeptides bearing the modified glutamic acid residue, 3-amino-6-hydroxypiperidone (Ahp) (Fig 8). They act as competitive inhibitors that mimic the endogenous substrates of serine proteases, but are not hydrolytically cleaved (Fig. 8).
The X-ray cocrystal structures of serine proteases were derived with compounds mostly from terrestrial cyanobacteria. The scyptolin-porcine pancreatic elastase,149 lyngbyastatin 7-porcine pancreatic elastase150 and A90720A-porcine pancreatic trypsin151 complexes provided insights into the molecular determinants of selectivity and potency, and corroborated with enzymatic assay results. The Ahp moiety and the adjacent residue on the N-terminal side are central to the biological activity, with the latter occupying the enzyme active site. The residue on the N-terminal side of Ahp determines specificity; the presence of a basic residue (e.g., Arg in A90720A) provides selectivity for inhibiting trypsin, while a hydrophobic residue at this position (e.g., Phe in micropeptin T-20) gives preferential inhibition for chymotrypsin. Small, nonpolar residues such as Thr in scyptolin or 2-amino-2-butenoic acid (Abu) in lyngbyastatin 7 are critical for inhibition of elastase and chymotrypsin, although more potent inhibition is observed for the former (Fig. 8).152
The marine cyanobacteria-derived members of this compound class bear the modified Thr residue, Abu moiety, at this position and are among the most potent elastase inhibitors. The structure-activity relationship studies on the compounds symplostatins 5–10 together with the X-ray cocrystal complex of lyngbyastatin 7-porcine pancreatic elastase provided key insights on other structural features that are critical for potent elastase inhibition (Figs. 8 and 9).150 Having a polar functional group on residues that modify the N-terminus of the cyclodepsipeptide scaffold together with an N-Me-Tyr moiety on the macrocycle provides significant improvements to biological activity.150
Fig. 9.
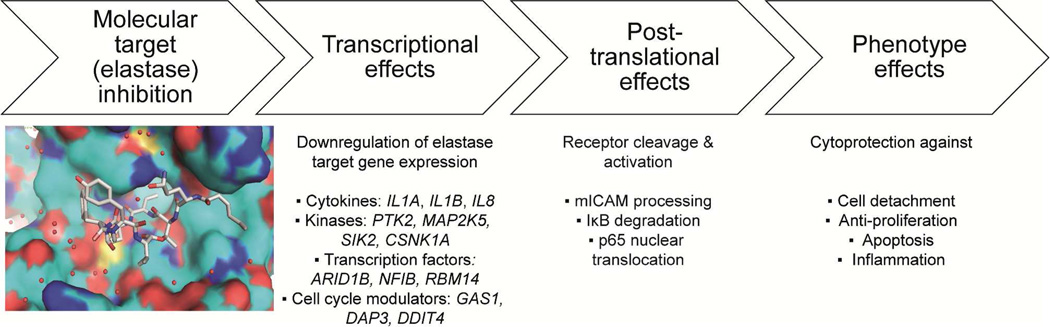
MOA of elastase inhibitors from marine cyanobacteria. Elastase inhibitors from marine cyanobacteria modulate the transcriptional and post-translational effects of elastase in bronchial epithelial cells leading to significant cytoprotection from elastase-induced cell detachment and anti-proliferation.
Comprehensive protease screening using a panel of 68 proteases and 26 serine proteases for lyngbyastatin 7 and symplostatin 5, respectively, revealed that these compounds are not promiscuous inhibitors and inhibit only a subset of serine proteases with elastase being most potently inhibited, followed by bovine chymotrypsin.150 With the information that members of the Abu-containing cyclic depsipeptides indeed are selective and potent elastase inhibitors, these compounds were utilized to modulate the cellular effects of elastase on bronchial epithelial cells. Elastase is a key player involved in the progression of chronic obstructive pulmonary disease (COPD), which is a major health concern due to the lack of effective therapeutics and surging patient populations.153
Bronchial epithelial cells were stimulated with elastase together with either solvent control or the model compound symplostatin 5 (Fig. 8). Elastase treated cells showed changes in morphology, adhesion and growth, due in part to global transcript changes, upregulation of caspase activity, NF-κB nuclear translocation and cleavage of intercellular adhesion molecule-1 (ICAM-1).150 Elastase caused significant upregulation of the transcript levels of pro-inflammatory cytokines IL1A, IL1B and IL8, together with transcription factors and components of the spliceosome and cell cycle (Fig. 9).150 Symplostatin 5 was able to attenuate the cellular effects of elastase providing cytoprotection to bronchial epithelial cells and alleviating several of the hallmarks of COPD such as inflammation, detachment and cell death (Fig. 9).150 Symplostatin 5 promises to possess a wide therapeutic window since it did not cause any cytotoxic effects to bronchial epithelial cells and had almost no transcriptional effects on cells that have not been stimulated with elastase. Aside from demonstrating the pharmacological utility of symplostatin 5 and related compounds as small molecule therapeutics, monitoring of the effects of elastase with an elastase-selective inhibitor provided insights into the cellular consequences of the proteolytic activity of elastase on bronchial epithelial cells. The comprehensive and global assessment of the effects of elastase on transcript levels indicates that elastase modulates the expression of a plethora of genes, with several of these having a yet-to-be determined relationship.
3.3.2 Aspartic proteases
Three modified linear peptides, grassystatins A–C (Fig. 10A), were purified from a Lyngbya cf. confervoides collection.154 These compounds bear a Leu-derived γ-amino-β-hydroxy acid, previously described in the broad-spectrum aspartic protease inhibitor pepstatin A (Fig. 10A).154 The structural similarity between grassystatins and pepstatin A provided insights into the possible molecular target of the former as aspartic proteases.154 Grassystatins A–C potently inhibited the aspartic proteases cathepsin D and E, but did not affect other members of this protease family such as renin and BACE1 (Fig. 10B).154 Validation of the protease screening results indeed showed that grassystatins are potent cathepsins D and E inhibitors with pico- to nanomolar IC50s, with preferential targeting of cathepsin E (Fig. 10B). Molecular docking studies of grassystatins A and C with cathepsins D and E were performed in order to visualize the molecular interactions of these inhibitors and, in addition, compare the interactions with that of pepstatin A to explain the differences in potency and selectivity (Fig. 10C).154 Grassystatins A and B both bear L-Asn, while grassystatin C has N-Me-L-Gln that occupies the P2 site, in contrast, to L-Val of pepstatin A.154 Molecular docking studies indicated that a basic residue at P2 confers selectivity for cathepsin E. The potent inhibitory activity of grassystatin A was demonstrated to be due to the optimum length and complementarity of the functional groups of grassystatin A with the amino acid residues of cathepsin E in the binding pocket, based on the molecular docking studies (Fig. 10C). The N-Me-L-Asn unit of grassystatin A also contributes hydrogen bonding interactions with Gln303, while the O-Me-Pro residue forms extensive hydrogen bonding interactions with Gln85.154
Fig. 10.
Structure and bioactivity of cathepsin E inhibitors. A. Cathepsin E inhibitors from marine cyanobacteria. B. Selectivity profile of grassystatin A against metalloproteases and aspartic proteases. Grassystatin A did not inhibit members of other families of proteases such as serine and cysteine proteases, dipeptidyl/tripeptidyl peptidases and cysteine carboxypeptidases. C. Key molecular interactions between grassystatin A and cathepsin E. Reprinted with permission from J. C. Kwan, E. A. Eksioglu, C. Liu, V. J. Paul and H. Luesch, J. Med. Chem., 2009, 52, 5732–5747. Copyright 2009 American Chemical Society.
Although structurally related, cathepsins D and E have distinct cellular functions and, as such, it is important to have selective inhibitors that can distinguish between these two proteases. Among other things, cathepsin D is involved in programmed cell death, while cathepsin E has been implicated in the breakdown of antigenic peptides that leads to T cell proliferation and consequent increase in pro-inflammatory cytokines, indicating involvement in autoimmune diseases.155 Grassystatin A showed potent and selective inhibition of cathepsin E; thus its effects on antigen presentation were evaluated by monitoring antigen presenting cells in human peripheral blood mononuclear cells stimulated with tetanus toxin C-fragment and in a mixed lymphocyte reaction of dendritic cells propagated in the CD4+ T cells, TTc and phorbol myristate acetate.154 Grassystatin A significantly lowered the proliferation of T cells in response to the exogenous antigen tetanus toxin C-fragment in human peripheral blood mononuclear cells. Furthermore, grassystatin A also decreased the proliferation of T cells in the autologous mixed lymphocyte reaction and significantly reduced the levels of the pro-inflammatory mediators IL-17 and IFN-γ.154 The potent and selective cathepsin E-inhibitory activity of grassystatin A, combined with cell permeability and cellular activity, is key to its use as a molecular probe to characterize the functions of cathepsin E in pathways of autoimmune disease. In this light, the synthesis of grassystatin A was reported as well as its utility as a probe for the role of cathepsin E in MHC Class II-dependent antigen processing.156 Inhibition of cathepsin E, via grassystatin A treatment, did not show any relevant role in ovalbumin antigen processing and peptide presentation, which was distinct from studies with other aspartic protease inhibitors.156 Similarly, the related statine-bearing compound, symplocin A (Fig. 10A), from a Symploca sp. collection also demonstrated selective and potent inhibitory activity against cathepsin E.157
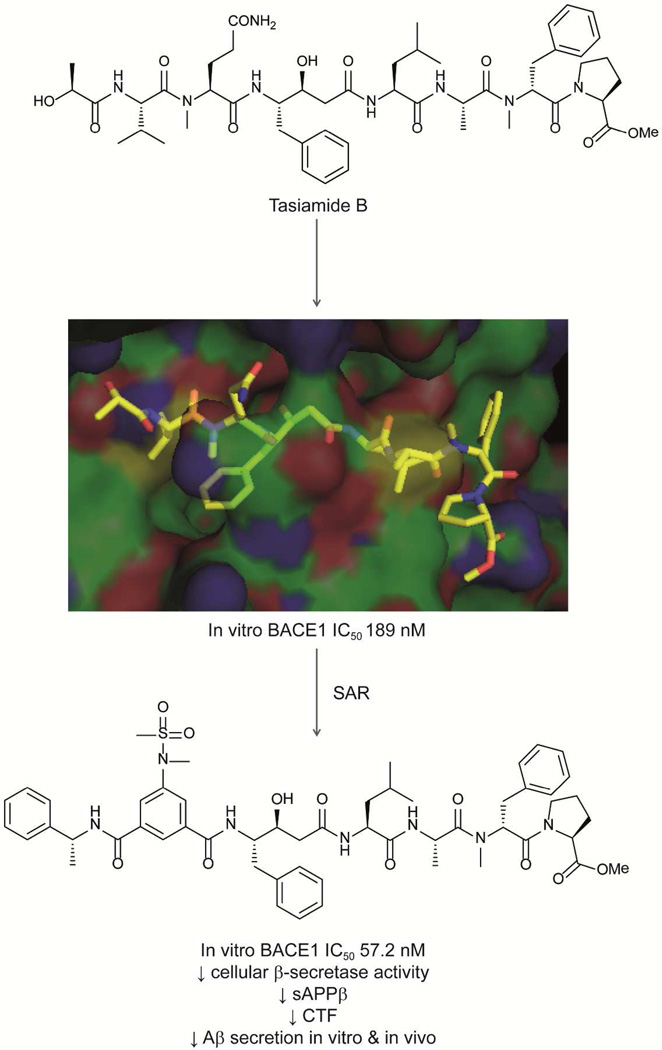
Like the grassystatins, the phenylstatine-bearing peptide tasiamide B158,159 (Fig. 11) also exhibited aspartic protease inhibitory activity but with a different selectivity profile.160 Tasiamide B inhibited β-secretase 1 (BACE1) and cathepsins D and E with nanomolar IC50s.160 The selectivity of these compounds was probed using X-ray cocrystallization of tasiamide B and BACE1.160 Tasiamide B occupies the binding groove between the C-terminal and N-terminal lobes of BACE1, and creates hydrogen bonding interactions with the residues of BACE1 including Gly34, Pro70, Thr72, Gln73, Tyr178, Gly230 and Thr232, in addition to 30 hydrophobic interactions with the enzyme.160 Based on the SAR results for the proteolytic activity of tasiamide B and its analogs against cathepsins D and E, the selectivity can be fine-tuned by modifying the residues on both the C- and N-termini of the molecule.160
Fig. 11.
Design of new BACE1 inhibitor based on the cyanobacterial compound tasiamide B.
BACE1 is the key β-secretase involved in the proteolytic processing of the amyloid precursor protein to generate Aβ peptides linked to the formation of plaques in the brain, one of the important factors implicated in the pathogenesis of Alzheimer’s disease. To probe the cellular consequences of tasiamide B and analogs on BACE1 inhibition, the levels of Aβ, Aβ40 and Aβ42 peptides in vitro and in vivo were monitored.160 Tasiamide B reduced sAPPβ secretion in stably transfected H4 cells by approximately 50% at 10 µM concentration and also caused a significant reduction in the levels of total Aβ, Aβ40 and Aβ42 in CHO 2B7 cells, with an approximate IC50 of 10 µM.160 With tasiamide B showing significant cellular effects relating to β-amyloid production as a consequence of BACE1 inhibition, this compound then served as an important prototype in designing more potent inhibitors of BACE1 with improved cellular activity. Since the P2 site has been demonstrated to be important for inhibition of aspartic proteases, synthetic analogs of tasiamide B were designed to incorporate modifications to the moiety occupying this site, while maintaining the central phenylstatine moiety and the C-terminus portion of tasiamide B.160 The most potent analogs with cellular activity that were designed bear an isophthalic acid moiety which contributes to increased hydrogen bonding and hydrophobic interactions with BACE1 to achieve higher potency (Fig. 11).160 Compared with tasiamide B, these hybrid compounds more potently reduced secreted sAPPβ, total Aβ, Aβ40 and Aβ42 levels.160 The increased cellular activity for the tertiary (N-methyl) sulfonamide (Fig. 11) translated into in vivo activity. This compound demonstrated sufficient pharmacokinetic stability and blood-brain barrier penetration and together with the potent inhibitory activity against BACE1, resulted in significant reduction by 23% of Aβ40 levels in CF-1 mice treated with a 30 mg/kg dose (Fig. 11).160
3.4 Voltage-gated Ion Channel Modulation
Modulation of the activity of ion channels is also a prominent theme in the MOA of small molecules from marine cyanobacteria (Fig. 12).19 The MOA of these compounds was probed using small molecule agonists and antagonists of ion channels and downstream effector proteins. Among the most widely studied ion channel modulators from cyanobacteria is antillatoxin A (Fig. 12), an ichthytoxic compound, discovered by screening a library of cyanobacterial extracts for sodium channel modulators using mouse neuroblastoma cells.161–163 Antillatoxin A showed a 10-fold higher potency compared to its analog antillatoxin B in activating voltage gated sodium channels (VGSCs).163 Further neurochemical and pharmacological approaches were utilized to elucidate the MOA of antillatoxin A. Antillatoxin A caused neuronal loss in cerebellar granule cells, which was prevented by either tetrodotoxin or the NMDA receptor antagonists MK-801 and dextrorphan, reminiscent of the effects of brevetoxin and domoic acid, and suggested that the neurotoxic effects of antillatoxin A are mediated by VGSCs.164 The effects of antillatoxin A on the influx of Ca2+ were also examined by monitoring fluo-3, a fluorescence indicator of intracellular [Ca2+]. Antillatoxin A increased Ca2+ levels and this was abrogated by pretreatment of cells with tetrodotoxin. In order to probe for the interaction of antillatoxin A on VGSCs, the binding of [3H]batrachotoxin to VGSCs was monitored in the presence of antillatoxin A.164 A concentration-dependent stimulation of [3H]batrachotoxin binding was observed with antillatoxin A. A synergistic effect was found with a combination of antillatoxin A and brevetoxin, but not with the sea anemone toxin or deltamethrin. The increase in binding of [3H]batrachotoxin was due to the allosteric modulation mediated by antillatoxin A. These data indicated that antillatoxin A is unlikely to bind to the neurotoxin sites 1, 2, 3, 5 and 7 on sodium channels and may either bind on a distinct site or at the neurotoxin site 4.164 The pharmacological properties and selectivity of antillatoxin A was further evaluated using cells that heterologously express rNav1.2, rNav1.4 or rNav1.5 α-subunits. [Na+] influx was monitored using the Na+ binding dye benzofuran isophthalate.165 Antillatoxin A induced Na+ influx in all three cell types, and there were no significant differences in the efficacy of Na+ influx in the three cell types. The pharmacological profile of antillatoxin A-induced Na+ influx was distinct from that of other neurotoxins that bind to sites 2 and 5 of VGSCs. The cellular consequences of antillatoxin A-mediated Na+ influx included neurite outgrowth in immature cerebrocortical neurons.166 The signaling pathway modulated by antillatoxin A leading to neuritogenesis was probed using chemical inhibitors and by monitoring changes in neurite outgrowth and Na+ and Ca2+ levels. Antillatoxin A activated VGSCs, leading to an increase in Na+ levels. This was accompanied by an activation of the Src kinase family, leading to potentiation of NMDAR function, which culminated in Ca2+ influx, engagement of Ca2+/calmodulin-dependent protein kinase and eventually neurite outgrowth.166
Fig. 12.
Structures of modulators of voltage-gated ion channels from marine cyanobacteria.
Another class of neurotoxins from marine cyanobacteria is the hoiamide family, unique lipopeptides distinguished by contiguous heterocyclic ring systems consisting of two methylated thiazolines and one thiazole ring.167–169 Using a bioactivity-directed approach, hoiamide A (Fig. 12) was purified as the active principle, causing a dose-dependent and rapid influx of [Na+] in neocortical neurons.167 Hoiamide A, however, produced a maximum Na+ influx less than the full VGSC agonist batrachotoxin, suggesting that hoiamide A is a partial agonist.167 The MOA of hoiamide A was interrogated using the VGSC inhibitor tetrodotoxin, NMDA receptor antagonist MK-801 and AMPA receptor antagonist NBQX, monitoring for hoiamide A-induced elevation in [Na+] in neocortical neurons.167 Tetrodotoxin A and MK-801, but not NBQX, caused a significant decrease in hoiamide A-induced Na+ influx and an increase in maximum response of Na+ influx with pyrethroid and brevetoxin 3 cotreatment.167 This indicated a positive allosteric interaction between the hoiamide A binding site and neurotoxin sites 5 and 7, and suggested that hoiamide A occupies neurotoxin site 2.167 Hoiamide A inhibited the specific binding of [3H]batrachotoxin on neurotoxin site 2, which provided direct evidence for the interaction of hoiamide A with VGSC, and also reduced the maximum Na+ influx in response to batrachotoxin, which is consistent with the reported interaction between a full and partial agonist. These results suggested that hoiamide A and batrachotoxin interact in a mutually exclusive manner with a common recognition site on VGSC.167 The structural similarity between hoiamides A and B suggested that the latter also exhibited similar pharmacological effects. Further assays with hoiamide B indeed showed increased [Na+] and suppression in Ca2+ oscillations.168 The effects of both hoiamides A and B on Ca2+ oscillations were of higher potency compared to the effects on Na+ influx, and were not related to the effects on VGSCs, suggesting that hoiamides A and B may have additional molecular target leading to the observed pharmacological effects in Na+ influx and Ca2+ oscillations.168
The neurotoxin kalkitoxin (Fig. 12), an unusual thiazoline-bearing lipid, was originally purified as the cytotoxic component using brine shrimp and fish toxicity.170 The neurotoxic effect of kalkitoxin was demonstrated in primary cultures of rat neurons; this cellular effect of kalkitoxin was alleviated by cotreatment with an NMDA receptor antagonist.162 Initial assessment indicated that kalkitoxin is a potent VGSC inhibitor, with comparable potency as saxitoxin in neuro-2a cells. Kalkitoxin inhibited veratridine-induced neurotoxicity and [Ca2+] release and decreased [3H]batrachotoxin incorporation in cells cotreated with deltamethrin, which suggested that kalkitoxin and pyrethroid binding sites may be allosterically coupled.171 Additional studies on kalkitoxin, as well as other neuroactive compounds purified from marine cyanobacteria (Fig. 12) including the macrolide palmyrolide,172 modified peptide alotamide173 and the lipopeptide jamaicamides A–C,174 are needed to establish the detailed MOA.
4 Mechanisms of Action and Direct Targets of Biologically Active Cyanobacteria Metabolites in Microbial and Protozoal Pathogens
The discovery of novel drug targets for antiprotozoal agents and antibiotics is of great interest particularly in tackling drug resistance. Small molecules that have unique molecular targets and mechanisms of action compared with currently available drugs can provide insight into alternative pathways that may be exploited to combat drug resistant strains of bacterial pathogens and protozoa.175–177 Equally important is finding new molecular targets in bacteria and protozoa that are not utilized by their mammalian host, thereby giving the possibility of an improved selectivity window and minimizing undesirable side effects.
4.1 Falcipains
The modified linear peptide symplostatin 4 (Fig. 13), also referred to as gallinamide A, was independently purified from Symploca and Schizothrix by the Luesch and Linington groups, respectively.48,178 Synthesis of symplostatin 4/gallinamide A and isomers indicated that these two compounds are indeed identical.179 Symplostatin 4 was demonstrated to induce G2 cell cycle arrest at high-micromolar concentration, which is related to the microtubule disrupting effects as evidenced by immunofluorescence staining of tubulin in HeLa cells.48 In addition, symplostatin 4 was purified as the antimalarial constituent using a bioactivity-directed fractionation of a Schizothrix collection.178 Symplostatin 4 and synthetic analogs showed potent activity against drug resistant and wild-type strains of Plasmodium falciparum.179,180 Interestingly, symplostatin 4 did not cause lysis of red blood cells, even at high-micromolar concentration.181 Clues to the mechanism of antiplasmodial activity of symplostatin 4 were obtained from the distinct food vacuole phenotype of P. falciparum observed following symplostatin 4 treatment.181 Giemsa stained ring stage parasites treated with symplostatin 4 showed a red swollen food vacuole phenotype.181 This phenotype is characteristic, arising from accumulation of nondigested hemoglobin or oligopeptides in the food vacuole due to inhibition of proteases involved in this pathway, and thus delineated the possible cellular targets of symplostatin 4 in P. falciparum.181 A rhodamine (Rh) labeled molecule was prepared to visualize target proteins in the intact schizont.181 Following protein isolation, SDS-PAGE analysis showed two bands in the 28 kDa molecular weight range to be strongly labeled by symplostatin 4.181 The formation of the food vacuole phenotype, together with the bands observed with treatment of Rh-labeled symplostatin 4 and the importance of the methylmethoxypyrrolinone moiety in symplostatin 4 suggested its possible role in the inhibition of one or more types of falcipains, which are plasmodial papain-like Cys proteases.181 Symplostatin 4 was shown to target falcipains 2/2’ and 3 using a competitive activity-based protein profiling (ABPP) experiment employing Cy5-DCG-04 as the activity based probe for papain-like Cys proteases.181 Symplostatin 4 was further demonstrated to be an irreversible inhibitor of falcipains 2 and 3, through analysis of the substrate turnover using recombinant falcipains.181 The effects of symplostatin 4 on other Cys proteases were also evaluated; cathepsin L was identified as the predominant mammalian cellular target.181 A follow up study indicated that symplostatin 4 is a covalent inhibitor of cathepsin L, using the vinyl amino acid derived moiety as the warhead.182 Symplostatin 4 has an IC50 of 5.0 nM against mammalian cathepsin L,182 although the direct consequence of this activity in mammalian cells treated with symplostatin 4 is not evident at this point. Hence, determination of off-target cellular effects of cathepsin L inhibition is warranted. The selective inhibition of symplostatin 4 on falcipains established this compound as a useful pharmacological probe and also suggests an alternative therapeutic target in combating the Plasmodium spp. parasites.
Fig. 13.
Protozoal parasite targeting compounds from marine cyanobacteria and their MOA.
4.2 Glycosomes
The linear lipopentapeptide almiramides A–C (Fig. 13) were isolated from a Lyngbya majuscula collection as the active principle against the parasite Leishmania donovania, the causative agent of leishmaniasis.183 The close relation of this parasite to other kinetoplastid parasites such as Trypanosoma brucei prompted its evaluation for additional biological activity.184 Almiramides A–C and related synthetic analogs showed low micromolar IC50s against both Leishmania sp. and T. b. brucei, with the majority of the compounds having greater than 10-fold selectivity for the parasites than mammalian cells.184 The MOA of almiramides against T. brucei was interrogated using an almiramide C-biotinylated probe, photoaffinity probe and affinity-resin conjugated probe. The use of complementary techniques to search for the direct target of almiramides addressed the limitations of each technique and thereby provided greater reliability of the protein hits from affinity purification.184 The biotinylated probe was constructed using a commercially available biotin reporter tag with a primary amine terminus that was coupled to an almiramide C analog bearing a terminal carboxylic acid moiety.184 Incubation of the biotinylated probe with cell lysates of T. brucei yielded 38 possible protein targets.184 The affinity resin on the other hand was prepared using a 15 Å PEG linker and a 6-aminohexanol-capped affi-gel resin and identified 12 proteins that directly bind to almiramide.184 The photoaffinity probe was prepared like the biotinylated probe, except for the addition of a benzophenone moiety. It relied on the generation of short-lived carbene species following irradiation of the biotinylated probe and cell lysates with UV, and identified 73 possible protein targets.184 Integration of the results of the three methods identified seven common candidate proteins, including β-tubulin, elongation factor 1-α, and five glycosomal proteins: glycosomal membrane protein, glycerol kinase, glyceraldehyde 3-phosphate dehydrogenase, Gim 5A protein and orotidine 5-phosphate decarboxylase.184 β-Tubulin and elongation factor 1-α were suggested to be false-positive targets because of their cellular abundance, and almiramides are then likely to affect the glycosomes of T. brucei, which is the sole energy machinery of these parasites.184 Almiramides exert a novel mechanism of inhibition of T. brucei, are the first small molecules shown to inhibit glycosomal function, and are an attractive chemotherapeutic target since glycosomes have no mammalian homologs.184 Validation of the direct target of almiramides, however, proved to be difficult due to limitations in expressing glycosomal-encoding genes.184
4.3 Quorum Sensing
Small molecule inhibitors of bacterial quorum sensing were also isolated from marine cyanobacteria, with several of these compounds showing structural similarities to acylhomoserine lactones (AHLs) used by Gram-negative bacteria to regulate quorum sensing. AHLs can bind to intercellular receptor proteins which leads to activation of gene expression pathways relevant to biofilm formation, virulence, luminescence and other quorum sensing-dependent phenotypes.185 The bioactivity of quorum sensing inhibitors was demonstrated through transcript and protein level monitoring of relevant quorum sensing genes such as lasB and phzG1, pyocyanin pigment production in Pseudomonas aeruginosa, and measurement of quorum sensing-dependent phenotype such as bioluminescence in Vibrio harveyi.186 Quorum sensing may be regulated by small molecules through competition for the AHL binding site, downregulation of expression of quorum sensing genes or regulation of the quorum sensing repressor RsaL.186
Using a bioactivity-directed isolation, malyngolide (Fig. 14) was purified as the anti-quorum sensing component.187 Malyngolide, at a sublethal concentration, acts as an indirect quorum sensing inhibitor, preventing the response of lasR+lasI+luxCDABE reporter pSB1075 in the presence of N-oxo-dodecanoyl-L-homoserine lactone (C6-HSL).187 Malyngolide, however, did not affect the quorum sensing-dependent phenotype of Agrobacterium tumanifaciens reporter, in the presence or absence of C6-HSL stimulation, suggesting that malyngolide does not act as a direct agonist or antagonist of quorum sensing.187 To further validate the effects of malyngolide in attenuating bacterial quorum sensing, the production of elastase, which is controlled by 3-oxo C12-HSL and LasR, was monitored. Malyngolide inhibited elastase production, while the exact MOA of malyngolide is yet to be fully elucidated.187 Accordingly, the structural relatedness of the tumonoic acids to AHLs prompted their evaluation for quorum sensing inhibition. Tumonoic acids E–H modulated the quorum sensing-dependent bioluminescence in V. harveyi, with tumonoic acid F being the most potent.188
Fig. 14.
Structures of quorum sensing inhibitors from marine cyanobacteria.
The similarity of the cyclopropane “tagged” fatty acid lyngbyoic acid (Fig. 14) with structures of natural AHL disrupters prompted investigations of its effects on quorum sensing.189 Lyngbyoic acid was evaluated in three E. coli reporter strains (pSB401, pSB536 and pSB1075), in the presence or absence of cognate AHLs. These reporters encode for different transcriptional activator proteins (R proteins) and, consequently, differ in their response to cognate AHLs. Lyngbyoic acid had the greatest inhibitory activity in the pSB1075 reporter system in the presence of 3-oxo-C12-AHL, while decreasing the background luminescence for all three reporters in the absence of AHLs.189 Using the pTIM5319 reporter which is similar to pSB1075 but lacked the AHL-binding site, lyngbyoic acid also reduced background luminescence, indicating that the effects of lyngbyoic acid on quorum sensing depend on neither the cognate AHL nor the AHL-binding domain of the receptor.189 In the pTIM505pTIM5211 reporter, which lacks the transcriptional repressor rsaL, 3-oxo-C12-HSL was able to compete with lyngbyoic acid, indicating that the effects of lyngbyoic acid are mediated by both AHL-binding domain-dependent and independent mechanisms. Unlike the related dodecanoic acid, lyngbyoic acid decreased pyocyanin and elastase production in wild-type P. aeruginosa cultures, due in part to downregulation of lasB and phzG1 transcript levels but also directly inhibited elastase.189
Analysis of the global changes in the transcriptome of P. aeruginosa PAO1 treated with lyngbyoic acid using microarray analysis revealed 969 upregulated genes and 887 downregulated genes in response to treatment.189 Relevant genes in pyocyanin production, secreted enzymes and rhamnolipid production were significantly downregulated, together with iron-regulated and biofilm-relevant genes being differentially expressed as well.189 The results of the transcriptome analysis of lyngbyoic acid-treated PAO1 indicated that this compound may serve as a useful molecular tool to probe for process adaptation of P. aeruginosa in cystic fibrosis patients and to elucidate the role of fatty acids as pathway modulators.189
5 Multiple Modes of Inhibition Mediated by Biologically Active Cyanobacteria Metabolites
The modified fatty acid esters honaucins A–C and pitinoic acids A–C (Fig. 15) represent a class of cyanobacterial compounds that consists of two structurally distinct simple carbon-chain components fused by an ester linkage. Presumably upon hydrolysis of the ester bond, these “modular” compounds deliver two different molecules that have distinct biological activities and, hence, display bifunctional activities. Honaucin A consists of a (S)-3-hydroxy-γ-butyrolactone and 4-chlorocrotonic acid, while honaucins B and C both bear a 3,4-di-O-substituted butanoic acid.190 Pitinoic acid B, on the other hand, consists of pitinoic acid A (5-methylene decanoic acid) and pitinoic acid C (5-chloro-2-hydroxy-pent-4-enoic acid). Pitinoic acids A and C were also purified as individual components of the crude extract.191
Fig. 15.
Bifunctional molecules from marine cyanobacteria and the MOA pitinoic acid B and the corresponding component fatty acids.
The structural similarities of honaucins A–C with microbial signaling small molecules prompted their evaluation as bacterial quorum sensing inhibitors, dose-dependently inhibiting bioluminescence, a quorum sensing-dependent phenotype in V. harveyi BB120.190 Honaucins were further evaluated by monitoring GFP-fluorescence arising from the activation of the LuxR receptor via the exogenous autoinducer 3-oxo-hexanoyl homoserine lactone in Escherichia coli JB525.190 Testing of synthetic analogs of honaucins confirmed the importance of the γ-butyrolactone ring, the position and configuration of the OH group on the γ-butyrolactone moiety, presence and position of the double bond, carbon chain length and nature of the halogen.190 Further evaluation of the natural and synthetic honaucins indicated that these compounds can inhibit lipopolysaccharide (LPS) mediated nitric oxide production in RAW macrophage cells, and the same structural features for the quorum sensing modulation were deemed essential for this biological activity in mammalian cells.190 Evaluation of the mechanism of inhibition of nitric oxide production indicated downregulation of the mRNA expression of several important inflammatory mediators such as TNF-α, IL-1β, IL-6 and iNOS by honaucin A.190
The structural similarity between honaucins and pitinoic acid B prompted the evaluation of the latter for anti-inflammatory effects using LPS-stimulated THP cells.191 Ester pitinoic acid B and its alcohol component, pitinoic acid C, downregulated the expression of several pro-inflammatory interleukins (IL-1β, IL-6 and IL-8) as well as TNF-α.191 Pitinoic acid A was also evaluated for its quorum sensing inhibitory activity due to the precedence of such effects of related fatty acids such as lyngbyoic acid.191 Pitinoic acid A decreased the transcript level of lasB and pyocyanin biosynthetic member phzG1 and, in accord, decreased elastase and pyocyanin levels in culture supernatants of P. aeruginosa.191
Symplostatin 4 is another example of a bifunctional molecule that has both mammalian and protozoal targets. Instead of delivering two different molecules upon metabolic activation, two distinct regions of the molecule bind to different targets. The N-terminus of dolastatin 10 related compounds has been shown to be critical to modulating tubulin polymerization based on SAR results, suggesting that the N-terminus of symplostatin 4 is key to the observed antiproliferative and tubulin disrupting activity.48 In addition, the pentenoic acid-derived moiety in the C-terminus of symplostatin 4 is detrimental to the antiproliferative activity, as observed for analogs of dolastatin 10 bearing the same modification. Based on the SAR for symplostatin 4 against P. falciparum, it is clear that the unusual methyl-methoxypyrrolinone moiety in the C-terminus of the molecule is central to targeting the falcipains of this protozoal parasite by forming a covalent adduct.181 The α,β-unsaturated system in the C-terminus of symplostatin 4 is determinant for the formation of a covalent bond with a Cys residue of cathepsin L.182
The presence of distinct moieties in cyanobacterial small molecules that can be liberated upon biotransformation gives rise to bifunctional biological activities and may represent a prodrug strategy employed by these primitive organisms. Prodrug strategies in general are quite commonly utilized by Nature.192 Also, the presence of defined regions of the molecule that gives rise to biological activities toward mammalian cancer cells and bacterial or protozoan pathogens offers the opportunity for modulating potential off-target effects and improving the selectivity index.
6 Perspective
Marine cyanobacteria continue to be a rich source of structurally diverse and potent pharmacological agents. The potential utilities of these small molecules are now better realized with the establishment of the MOA and identification of molecular targets. Improvements in bioactivity screening technologies, chemical genomics and chemical proteomics techniques have further strengthened the value of cyanobacteria as source organisms for drug discovery and chemical biology. The elucidation of the mechanisms of action and targets of cyanobacterial small molecule has allowed for the discovery of novel molecular targets that offers a novel way for disease intervention. The targets of cyanobacterial small molecules extend well beyond commonly reported drug targets for mammalian, protozoal and bacterial systems, which may be exploited for tackling drug resistance and minimizing side effects. For example, aurilides represent the first small molecule modulators of prohibitin and, as such, aurilides will become a valuable chemical tool to elucidate the role of this protein in signaling networks related to cancer, and may potentially represent a novel approach in combating malignancies. Largazole, among the most potent and selective HDAC inhibitors to date, has also emerged as a tool to visualize the molecular interactions of depsipeptides with HDACs. This discovery will aid in the development of the next generation of cyclodepsipeptide-based inhibitors. The discovery of the molecular target of largazole has also widened the prospect for this cyanobacterial molecule. Aside from cancer-related applications, largazole is now also being pursued for other diseases where gene expression changes may be beneficial. The elucidation of the MOA has also benefited the design of new analogs of cyanobacterial compounds that possess improved potency and selectivity. Tasiamide B, which targets the important BACE1 enzyme, has yielded new analogs with potent in vitro and in vivo activity and pharmacokinetic stability. The antiprotozoal compounds symplostatin 4 and almiramides have given insights into new molecular targets for malaria and trypanosomiasis, respectively. The mechanisms of action of these antiprotozoal compounds provide a possible new route in tackling protozoal parasites with potentially fewer side effects to the human host. More importantly, deciphering the MOA also allowed for weeding out potential off-target effects of cyanobacterial small molecules. This has been beneficial for the potent antiproliferative agent apratoxin A and has enabled the design of new analogs without the irreversible toxicity that has limited the development of this small molecule. The elucidation of the bioactivity, SAR and mechanisms of action of the dolastatin 10 family of compounds highlights the privileged scaffold of these cyanobacterial metabolites. The primitive source organism of these compounds utilizes the same basic skeleton to target multiple diseases and cellular targets in cancer and malaria, with minimal overlap presumably through a switching mechanism. The tubulin disrupting and anticancer activity of the dolastatin 10 family of compounds relies on the N-terminal moieties for potent bioactivity and abrogated by the introduction of a double bond on the C-terminus. In contrast, the antiplasmodial properties of the dolastatin 10 family rely on the modified amino acid residues on the C-terminus.
The relevance of the MOA and direct targets of antiproliferative natural products to cancer is also related to a better understanding of the molecular basis of the disease. The molecular mechanisms of the unintended side effects of several promising cyanobacterial small molecules are poorly defined and would be of interest to trigger the development of these compounds as effective therapeutics in the future. In all, natural products discoveries offer a wider research prospective with the elucidation of the MOA and target. In the coming years, the development of new platforms in screening technologies, next-generation sequencing and use of model organisms such as S. cerevisiae, C. elegans and D. rerio will pave the way for the discovery of new small molecules from marine cyanobacteria and their pharmacological effects. As cyanobacterial natural products have shown exemplary activities towards their protein targets, it is warranted to explore cyanobacteria to discover small molecules against poorly understood diseases to find molecular probes and effective therapeutics as well.
Fig. 3.
MOA of tubulin-targeting agents. Tubulin-disrupting compounds from marine cyanobacteria cause significant cellular microtubule depolymerization, leading to G2 cell cycle arrest and cell death.
Acknowledgements
Cyanobacteria-based natural products research in the authors’ laboratory is supported by the National Institutes of Health, NCI grants R01CA172310 and R01CA138544, the Bankhead-Coley Cancer Research Program, grants 4BB07 and 3BF03, and the James & Esther King Biomedical Research Program, grant 3KF04. LASR acknowledges support from the University of the Philippines (UP) Balik PhD Program, the UP Marine Science Institute, UP Office of the Vice-Chancellor for Research and Development and the Philippine Council for Health Research and Development.
Biographies

Lilibeth A. Salvador-Reyes earned her Bachelor and Master’s degree in Chemistry from the University of the Philippines Diliman. She joined the Luesch laboratory in 2008 and received her Ph.D. in Pharmaceutical Sciences - Medicinal Chemistry at the University of Florida in 2013. Lilibeth is currently an Assistant Professor at the Marine Science Institute, University of the Philippines Diliman. Her research interests include chemical ecology, symbiosis and discovery and development of antibiotics and anticancer agents from marine organisms.

Hendrik Luesch received his Diplom in Chemistry at the University of Siegen (Germany) in 1997. He studied marine natural products chemistry at the University of Hawaii at Manoa and obtained his Ph.D. with Professor Richard E. Moore in 2002. He undertook three years of postdoctoral studies as an Irving S. Sigal Fellow at The Scripps Research Institute with Professor Peter G. Schultz in functional genomics. Since 2005 he is faculty at the University of Florida and currently Associate Professor of Medicinal Chemistry, leading a multidisciplinary marine natural products drug discovery and development program.
Footnotes
Conflict of Interest
HL is a co-founder of Oceanyx Pharmaceuticals, Inc., which has been licensing various patents and patent applications related to cyanobacterial compounds discussed in this review.
References
- 1.Newman DJ, Cragg GM. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong F, Cheung AK, Huang SA. Annu. Rev. Pharmacol. Toxicol. 2012;52:57–78. doi: 10.1146/annurev-pharmtox-010611-134639. [DOI] [PubMed] [Google Scholar]
- 3.Hurco O. Neurochem. Int. 2012;61:892–898. doi: 10.1016/j.neuint.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Ziegler S, Pries V, Hedberg C, Waldmann H. Angew. Chem. Int. Ed. 2013;52:2744–2794. doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]
- 5.Breinbauer R, Vetter IR, Waldmann H. Angew. Chem. Int. Ed. 2002;41:2878–2890. doi: 10.1002/1521-3773(20020816)41:16<2878::AID-ANIE2878>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 6.Carlson EE. ACS Chem. Biol. 2010;5:639–653. doi: 10.1021/cb100105c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taunton J, Collins JL, Schreiber SL. J. Am. Chem. Soc. 1996;118:10412–10422. [Google Scholar]
- 8.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Proc. Natl. Acad. Sci. USA. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang B, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Bolden J, Peart MJ, Johnstone RW. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins AL, Groom CR. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 12.Muda M, McKenna S. Drug Discov. Today. 2004;1:55–59. doi: 10.1016/j.ddtec.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Ridley CP, Bergquist PR, Harper MK, Faulkner DJ, Hooper JN, Haygood MG. Chem. Biol. 2005;12:397–406. doi: 10.1016/j.chembiol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat. Chem. Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 15.Luesch H, Harrigan GG, Goetz G, Horgen FD. Curr. Med. Chem. 2002;9:1791–1806. doi: 10.2174/0929867023369051. [DOI] [PubMed] [Google Scholar]
- 16.Gerwick WH, Moore BS. Chem. Biol. 2012;19:85–98. doi: 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senter PD, Sievers EL. Nat. Biotechnol. 2012;30:631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 18.Tan LT. Drug Discov. Today. 2013;18:863–871. doi: 10.1016/j.drudis.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Tan LT. J. Appl. Phycol. 2010;22:659–676. [Google Scholar]
- 20.Tan LT. Phytochemistry (Elsevier) 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Shoemaker RH. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 22.Bai RL, Paull KD, Herald CL, Malspeis L, Pettit GR, Hamel E. J. Biol. Chem. 1991;25:15882–15889. [PubMed] [Google Scholar]
- 23.Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, Scherf U, Lee JK, Reinhold WO, Weinstein JN, Mesirov JP, Lander ES, Brown PO, Golub TR. Proc. Natl. Acad. Sci. USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Nat. Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 25.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, Scudiero DA, Eisen MB, Sausville EA, Pommier Y, Botstein D, Brown PO, Weinstein JN. Nat. Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 26.Futamura Y, Kawatani M, Kazami S, Tanaka K, Muroi M, Shimizu T, Tomita T, Watanabe N, Osada H. Chem. Biol. 2012;19:1620–1630. doi: 10.1016/j.chembiol.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Lamb J. Nat. Rev. Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 28.Wacker S, Houghtaling B, Elemento O, Kapoor T. Nat. Chem. Biol. 2012;8:235–237. doi: 10.1038/nchembio.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luesch H. Mol. Biosyst. 2006;2:609–620. doi: 10.1039/b609384a. [DOI] [PubMed] [Google Scholar]
- 30.Sato S, Murata A, Shirakawa T, Uesugi M. Chem. Biol. 2010;17:616–623. doi: 10.1016/j.chembiol.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Rix U, Superti-Furga G. Nat. Chem. Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 32.Futamura Y, Muroi M, Osada H. Mol. Biosyst. 2013;9:897–914. doi: 10.1039/c2mb25468a. [DOI] [PubMed] [Google Scholar]
- 33.Schenone M, Dančík V, Wagner BK, Clemons PA. Nat. Chem. Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo F. Proc. Natl. Acad. Sci. USA. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito SY. Prog. Mol. Subcell. Biol. 2009;46:187–219. doi: 10.1007/978-3-540-87895-7_7. [DOI] [PubMed] [Google Scholar]
- 36.Dumontet C, Jordan MA. Nat. Rev. Drug Discov. 2010;9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan MA, Wilson L. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 38.Jordan MA, Wilson L. Curr. Opin. Cell Biol. 1998;10:123–130. doi: 10.1016/s0955-0674(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 39.Bai R, Pettit G, Hamel E. Biochem. Pharmacol. 1990;39:1941–1949. doi: 10.1016/0006-2952(90)90613-p. [DOI] [PubMed] [Google Scholar]
- 40.Pettit GR, Kamano Y, Herald CL, Dufresne C, Cerny RL, Herald DL, Schmidt JM, Kizu H. J. Am. Chem. Soc. 1989;111:5015–5017. [Google Scholar]
- 41.Harrigan GG, Luesch H, Yoshida WY, Moore RE, Nagle DG, Paul VJ, Mooberry SL, Corbett TH, Valeriote FA. J. Nat. Prod. 1998;61:1075–1077. doi: 10.1021/np980321c. [DOI] [PubMed] [Google Scholar]
- 42.Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. J. Nat. Prod. 2001;64:907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 43.Mooberry SL, Leal RM, Tinley TL, Luesch H, Moore RE, Corbett TH. Int. J. Cancer. 2003;104:512–521. doi: 10.1002/ijc.10982. [DOI] [PubMed] [Google Scholar]
- 44.Mitra A, Sept D. Biochemistry. 2004;43:13955–13962. doi: 10.1021/bi0487387. [DOI] [PubMed] [Google Scholar]
- 45.Vaishampayan U, Glode M, Du W, Kraft A, Hudes G, Wright J, Hussain M. Clin. Cancer Res. 2000;6:4205–4208. [PubMed] [Google Scholar]
- 46.Deng C, Pan B, O’Connor OA. Nat. Rev. Drug Discov. 2011;11:19–20. [Google Scholar]
- 47.Luesch H, Yoshida WY, Moore RE, Paul VJ, Mooberry SL, Corbett TH. J. Nat. Prod. 2002;65:16–20. doi: 10.1021/np010317s. [DOI] [PubMed] [Google Scholar]
- 48.Taori K, Liu Y, Paul VJ, Luesch H. ChemBioChem. 2009;10:1634–1639. doi: 10.1002/cbic.200900192. [DOI] [PubMed] [Google Scholar]
- 49.Horgen FD, Kazmierski EB, Westenburg HE, Yoshida WY, Scheuer PJ. J. Nat. Prod. 2002;65:487–491. doi: 10.1021/np010560r. [DOI] [PubMed] [Google Scholar]
- 50.Simmons TL, McPhail KL, Ortega-Barria E, Mooberry SL, Gerwick WH. Tetrahedron Lett. 2006;47:3387–3390. [Google Scholar]
- 51.Golakoti T, Ogino J, Heltzel CE, Husebo TL, Jensen CM, Larsen LK, Patterson GML, Moore RE, Mooberry SL, Corbett TH, Valeriote FA. J. Am. Chem. Soc. 1995;117:12030–12049. [Google Scholar]
- 52.Chlipala GE, Mo S, Orjala J. Curr. Drug Targets. 2011;12:1654–1673. doi: 10.2174/138945011798109455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trimurtulu G, Ohtani I, Patterson GML, Moore RE, Corbett TH, Valeriote FA, Demchiko L. J. Am. Chem. Soc. 1994;116:4729–4737. [Google Scholar]
- 54.Kobayashi M, Aoki S, Ohyabu N, Kurosu M, Wang W, Kitagawa I. Tetrahedron Lett. 1994;35:7969–7972. [Google Scholar]
- 55.Smith CD, Zhang X, Mooberry SL, Patterson GM, Moore RE. Cancer Res. 1994;54:3779–3784. [PubMed] [Google Scholar]
- 56.Morita K, Koiso Y, Hashimoto Y, Kobayashi M, Wang W, Ohyabu N, Iwasaki S. Biol. Pharm. Bull. 1997;20:171–174. doi: 10.1248/bpb.20.171. [DOI] [PubMed] [Google Scholar]
- 57.Panda D, Ananthnarayan V, Larson G, Shih C, Jordan MA, Wilson L. Biochemistry. 2000;39:14121–14127. doi: 10.1021/bi0010827. [DOI] [PubMed] [Google Scholar]
- 58.Edelman MJ, Gandara DR, Hausner P, Israel V, Thornton D, DeSanto J, Doyle LA. Lung Cancer. 2003;39:197–199. doi: 10.1016/s0169-5002(02)00511-1. [DOI] [PubMed] [Google Scholar]
- 59.D’Agostino G, del Campo J, Mellado B, Izquierdo MA, Minarik T, Cirri L, Marini L, Perez-Gracia JL, Scambia G. Int. J. Gynecol. Cancer. 2006;16:71–76. doi: 10.1111/j.1525-1438.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 60.Gerwick WH, Proteau PJ, Nagle DG, Hamel E, Blokhin A, Slate DL. J. Org. Chem. 1994;59:1243–1245. [Google Scholar]
- 61.Verdier-Pinard P, Sitachitta N, Rossi JV, Sackett DL, Gerwick WH, Hamel E. Arch. Biochem. Biophys. 1999;370:51–58. doi: 10.1006/abbi.1999.1363. [DOI] [PubMed] [Google Scholar]
- 62.Dominguez R, Holmes KC. Annu. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cooper JA. J. Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettit GR, Kamano Y, Herald CL, Fujii Y, Kizu H, Boyd RR, Boettner FE, Doubek DL, Schmidt JM, Chapuis J, Michel C. Tetrahedron. 1993;49:9151–9170. [Google Scholar]
- 65.Bai R, Verdier-Pinard P, Gangwar S, Stessman CC, McClure KJ, Sausville EA, Pettit GR, Bates RB, Hamel E. Mol. Pharmacol. 2001;59:462–469. doi: 10.1124/mol.59.3.462. [DOI] [PubMed] [Google Scholar]
- 66.Oda T, Crane ZD, Dicus CW, Sufi BA, Bates RB. J. Mol. Biol. 2003;328:319–324. doi: 10.1016/s0022-2836(03)00306-1. [DOI] [PubMed] [Google Scholar]
- 67.Williams DE, Burgoyne DL, Rettig SJ, Andersen RJ, Fathi-Afshar ZR, Allen TM. J. Nat. Prod. 1993;56:545–551. [Google Scholar]
- 68.Williams PG, Moore RE, Paul VJ. J. Nat. Prod. 2003;66:1356–1363. doi: 10.1021/np0302145. [DOI] [PubMed] [Google Scholar]
- 69.Harrigan GG, Yoshida WY, Moore RE, Nagle DG, Park PU, Biggs J, Paul VJ, Mooberry SL, Corbett TH, Valeriote FA. J. Nat. Prod. 1998;61:1221–1225. doi: 10.1021/np9801211. [DOI] [PubMed] [Google Scholar]
- 70.Luesch H, Yoshida WY, Moore RE, Paul VJ, Mooberry SL. J. Nat. Prod. 2000;63:611–615. doi: 10.1021/np990543q. [DOI] [PubMed] [Google Scholar]
- 71.Luesch H, Yoshida WY, Moore RE, Paul VJ. J. Nat. Prod. 2000;63:1437–1439. doi: 10.1021/np000104n. [DOI] [PubMed] [Google Scholar]
- 72.Han B, McPhail KL, Gross H, Goeger DE, Mooberry SL, Gerwick WH. Tetrahedron. 2005;61:11723–11729. [Google Scholar]
- 73.Matthew S, Salvador LA, Schupp PJ, Paul VJ, Luesch H. J. Nat. Prod. 2010;73:1544–1552. doi: 10.1021/np1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marquez BL, Watts KS, Yokochi A, Roberts MA, Verdier-Pinard P, Jimenez JI, Hamel E, Scheuer PJ, Gerwick WH. J. Nat. Prod. 2002;65:866–871. doi: 10.1021/np0106283. [DOI] [PubMed] [Google Scholar]
- 75.Sumiya E, Shimogawa H, Sasaki H, Tsutsumi M, Yoshita K, Ojika M, Suenaga K, Uesugi M. ACS Chem. Biol. 2011;6:425–431. doi: 10.1021/cb1003459. [DOI] [PubMed] [Google Scholar]
- 76.Patterson GM, Smith CD, Kimura LH, Britton BA, Carmeli S. Cell Motil. Cytoskeleton. 1993;24:39–48. doi: 10.1002/cm.970240105. [DOI] [PubMed] [Google Scholar]
- 77.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 78.Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 79.Luesch H, Yoshida WY, Moore RE, Paul VJ, Corbett TH. J. Am. Chem. Soc. 2001;123:5418–5423. doi: 10.1021/ja010453j. [DOI] [PubMed] [Google Scholar]
- 80.Luesch H, Yoshida WY, Moore RE, Paul VJ. Tetrahedron. 2002;58:7959–7966. [Google Scholar]
- 81.Luesch H, Yoshida WY, Moore RE, Paul VJ. Bioorg. Med. Chem. 2002;10:1973–1978. doi: 10.1016/s0968-0896(02)00014-7. [DOI] [PubMed] [Google Scholar]
- 82.Matthew S, Schupp PJ, Luesch H. J. Nat. Prod. 2008;71:1113–1116. doi: 10.1021/np700717s. [DOI] [PubMed] [Google Scholar]
- 83.Gutierrez M, Suyama TL, Engene N, Wingerd JS, Matainaho T, Gerwick WH. J. Nat. Prod. 2008;71:1099–1103. doi: 10.1021/np800121a. [DOI] [PubMed] [Google Scholar]
- 84.Tidgewell K, Engene N, Byrum T, Media J, Doi T, Valeriote FA, Gerwick WH. ChemBioChem. 2010;11:1458–1466. doi: 10.1002/cbic.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luesch H, Chanda SK, Raya RM, DeJesus PD, Orth AP, Walker JR, Izpisua Belmonte JC, Schultz PG. Nat. Chem. Biol. 2006;2:158–167. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- 86.Liu Y, Law BK, Luesch H. Mol. Pharmacol. 2009;76:91–104. doi: 10.1124/mol.109.056085. [DOI] [PubMed] [Google Scholar]
- 87.Shen S, Zhang P, Lovchik MA, Li Y, Tang L, Chen Z, Zeng R, Ma D, Yuan J, Yu Q. J. Cell Biol. 2009;185:629–639. doi: 10.1083/jcb.200810183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen QY, Liu Y, Luesch H. ACS Med. Chem. Lett. 2011;2:861–865. doi: 10.1021/ml200176m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen QY, Liu Y, Cai W, Luesch H. J. Med. Chem. 2014;57:3011–3029. doi: 10.1021/jm4019965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith BC, Denu JM. Biochim. Biophys. Acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith BC, Hallows WC, Denu JM. Chem. Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 93.Salvador LA, Luesch H. In: Natural Products and Cancer Drug Discovery, Cancer Drug Discovery and Development. Koehn FE, editor. New York: Springer; 2013. pp. 59–94. [Google Scholar]
- 94.Ganesan A, Nolan L, Crabb SJ, Packham G. Curr. Cancer Drug Targets. 2009;9:963–981. doi: 10.2174/156800909790192428. [DOI] [PubMed] [Google Scholar]
- 95.Furumai R, Matsuyama A, Kobashi N, Lee K, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, Horinouchi S. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- 96.Lech-Maranda E, Robak E, Korycka A, Robak T. Mini-Rev. Med. Chem. 2007;7:1062–1069. doi: 10.2174/138955707782110178. [DOI] [PubMed] [Google Scholar]
- 97.Marks PA, Breslow R. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 98.Pina IC, Gautschi JT, Wang G, Sanders ML, Schmitz FJ, France D, Cornell-Kennon S, Sambucetti LC, Remiszewski SW, Perez LB, Bair KW, Crews P. J. Org. Chem. 2003;68:3866–3873. doi: 10.1021/jo034248t. [DOI] [PubMed] [Google Scholar]
- 99.Gu W, Cueto M, Jensen PR, Fenical W, Silverman RB. Tetrahedron. 2007;63:6535–6541. [Google Scholar]
- 100.Hong J, Luesch H. Nat. Prod. Rep. 2012;29:449–456. doi: 10.1039/c2np00066k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Taori K, Paul VJ, Luesch H. J. Am. Chem. Soc. 2008;130:1806–1807. doi: 10.1021/ja7110064. [DOI] [PubMed] [Google Scholar]
- 102.Pavlik C, Wong C, Ononye S, Lopez DD, Engene N, McPhail KL, Gerwick WH, Balunas MJ. J. Nat. Prod. 2013;76:2026–2033. doi: 10.1021/np400198r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ying Y, Taori K, Kim H, Hong J, Luesch H. J. Am. Chem. Soc. 2008;130:8455–8459. doi: 10.1021/ja8013727. [DOI] [PubMed] [Google Scholar]
- 104.Bowers A, West N, Taunton J, Schreiber SL, Bradner JE, Williams RM. J. Am. Chem. Soc. 2008;130:11219–11222. doi: 10.1021/ja8033763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen Y, Gambs C, Abe Y, Wentworth PJ, Janda KD. J. Org. Chem. 2003;68:8902–8905. doi: 10.1021/jo034765b. [DOI] [PubMed] [Google Scholar]
- 106.Masuoka Y, Nagai A, Shin-ya K, Furihata K, Nagai K, Suzuki K-, Hayakawa Y, Seto H. Tetrahedron Lett. 2001;42:41–44. [Google Scholar]
- 107.Salvador L, Luesch H. Curr. Drug Targets. 2012;13:1029–1047. doi: 10.2174/138945012802008973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S. Expt. Cell. Res. 1998;241:126–133. doi: 10.1006/excr.1998.4027. [DOI] [PubMed] [Google Scholar]
- 109.Ying Y, Liu Y, Byeon SR, Kim H, Luesch H, Hong J. Org. Lett. 2008;10:4021–4024. doi: 10.1021/ol801532s. [DOI] [PubMed] [Google Scholar]
- 110.Cole KE, Dowling DP, Boone MA, Phillips AJ, Christianson DW. J. Am. Chem. Soc. 2011;133:12474–12477. doi: 10.1021/ja205972n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y, Salvador LA, Byeon S, Ying Y, Kwan JC, Law BK, Hong J, Luesch H. J. Pharmacol. Exp. Ther. 2010;335:351–361. doi: 10.1124/jpet.110.172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Law ME, Corsino PE, Jahn SC, Davis BJ, Chen S, Patel B, Pham K, Lu J, Sheppard B, Nørgaard P, Hong J, Higgins P, Kim JS, Luesch H, Law BK. Oncogene. 2013;32:1316–1329. doi: 10.1038/onc.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee SU, Kwak HB, Pi SH, You HK, Byeon SR, Ying Y, Luesch H, Hong J, Kim SH. ACS Med. Chem. Lett. 2011;2:248–251. doi: 10.1021/ml1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu Y, Wang Z, Wang J, Lam W, Kwong S, Li F, Friedman SL, Zhou S, Ren Q, Xu Z, Wang X, Ji L, Tang S, Zhang H, Lui EL, Ye T. Liver Int. 2013;33:504–515. doi: 10.1111/liv.12034. [DOI] [PubMed] [Google Scholar]
- 115.Salvador LA, Park H, Al-Awadhi FH, Liu Y, Kim B, Zeller SL, Chen Q, Hong J, Luesch H. ACS Med. Chem. Lett. 2014;5:905–910. doi: 10.1021/ml500170r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim B, Park H, Salvador LA, Serrano PE, Kwan JC, Zeller SL, Chen QY, Ryu S, Liu Y, Byeon S, Luesch H, Hong J. Bioorg. Med. Chem. Lett. 2014;24:3728–3731. doi: 10.1016/j.bmcl.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCulloch MWB, Coombs GS, Banerjee N, Bugni TS, Cannon KM, Harper MK, Veltri CA, Virshup DM, Ireland CM. Bioorg. Med. Chem. 2009;17:2189–2198. doi: 10.1016/j.bmc.2008.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ungermannova D, Parker SJ, Nasveschuk CG, Wang W, Quade B, Zhang G, Kuchta RD, Phillips AJ, Liu X. PLoS One. 2012;7:e29208. doi: 10.1371/journal.pone.0029208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Balcerczyk A, Pirola L. Biofactors. 2010;36:383–393. doi: 10.1002/biof.112. [DOI] [PubMed] [Google Scholar]
- 120.Gutierrez M, Andrianasolo EH, Shin WK, Goeger DE, Yokochi A, Schemies J, Jung M, France D, Cornell-Kennon S, Lee E, Gerwick WH. J. Org. Chem. 2009;74:5267–5275. doi: 10.1021/jo900578j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kisselev AF, van der Linden WA, Overkleeft HS. Chem. Biol. 2012;19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nalepa G, Rolfe M, Harper JW. Nat. Rev. Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 123.Pereira AR, Kale AJ, Fenley AT, Byrum T, Debonsi HM, Gilson MK, Valeriote FA, Moore BS, Gerwick WH. ChemBioChem. 2012;13:810–817. doi: 10.1002/cbic.201200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim KB, Crews CM. Nat. Prod. Rep. 2013;30:600–604. doi: 10.1039/c3np20126k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suenaga K, Mutou T, Shibata T, Itoh T, Kigoshi H, Yamada K. Tetrahedron Lett. 1996;37:6771–6774. [Google Scholar]
- 126.Suenaga K, Mutou T, Shibata T, Itoh T, Fujita T, Takada N, Hayamizu K, Takagi M, Irifune T, Kigoshi H, Yamada K. Tetrahedron. 2004;60:8509–8527. [Google Scholar]
- 127.Han B, Gross H, Goeger DE, Mooberry SL, Gerwick WH. J. Nat. Prod. 2006;69:572–575. doi: 10.1021/np0503911. [DOI] [PubMed] [Google Scholar]
- 128.Tripathi A, Puddick J, Prinsep MR, Rottmann M, Tan LT. J. Nat. Prod. 2010;73:1810–1814. doi: 10.1021/np100442x. [DOI] [PubMed] [Google Scholar]
- 129.Tripathi A, Puddick J, Prinsep MR, Rottmann M, Chan KP, Chen DY, Tan LT. Phytochemistry (Elsevier) 2011;72:2369–2375. doi: 10.1016/j.phytochem.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 130.Dai L, Chen B, Lei H, Wang Z, Liu Y, Xu Z, Ye T. Chem. Commun. 2012;48:8697–8699. doi: 10.1039/c2cc34187e. [DOI] [PubMed] [Google Scholar]
- 131.Kimura J, Takada Y, Inayoshi T, Nakao Y, Goetz G, Yoshida WY, Scheuer PJ. J. Org. Chem. 2002;67:1760–1767. doi: 10.1021/jo010176z. [DOI] [PubMed] [Google Scholar]
- 132.Nakao Y, Yoshida WY, Takada Y, Kimura J, Yang L, Mooberry SL, Scheuer PJ. J. Nat. Prod. 2004;67:1332–1340. doi: 10.1021/np049949f. [DOI] [PubMed] [Google Scholar]
- 133.Sato S, Murata A, Orihara T, Shirakawa T, Suenaga K, Kigoshi H, Uesugi M. Chem. Biol. 2011;18:131–139. doi: 10.1016/j.chembiol.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 134.Zhang J, Yang PL, Gray NS. Nat. Rev. Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rauch J, Volinsky N, Romano D, Kolch W. Cell. Commun. Signal. 2011;9:23. doi: 10.1186/1478-811X-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lahiry P, Torkamani A, Schork NJ, Hegele RA. Nat. Rev. Genet. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 137.Teruya T, Sasaki H, Fukazawa H, Suenaga K. Org. Lett. 2009;11:5062–5065. doi: 10.1021/ol9020546. [DOI] [PubMed] [Google Scholar]
- 138.Stevenson CS, Capper EA, Roshak AK, Marquez B, Eichman C, Jackson JR, Mattern M, Gerwick WH, Jacobs RS, Marshall LA. J. Pharmacol. Exp. Ther. 2002;303:858–866. doi: 10.1124/jpet.102.036350. [DOI] [PubMed] [Google Scholar]
- 139.Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krssák M, Gürtler U, Garin-Chesa P, Lieb S, Quant J, Grauert M, Adolf GR, Kraut N, Peters JM, Rettig WJ. Curr. Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 140.Antoniades V, Sioga A, Dietrich EM, Meditskou S, Ekonomou L, Antoniades K. Med. Hypotheses. 2013;81:1159–1163. doi: 10.1016/j.mehy.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 141.Brewer GJ, Dick RD, Grover DK, LeClaire V, Tseng M, Wicha M, Pienta K, Redman BG, Jahan T, Sondak VK, Strawderman M, LeCarpentier G, Merajver SD. Clin. Cancer Res. 2000;6:1–10. [PubMed] [Google Scholar]
- 142.Antoniades V, Sioga A, Dietrich EM, Meditskou S, Ekonomou L, Antoniades K. Med. Hypotheses. 2013;81:1159–1163. doi: 10.1016/j.mehy.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 143.Kwan JC, Rocca JR, Abboud KA, Paul VJ, Luesch H. Org. Lett. 2008;10:789–792. doi: 10.1021/ol702946d. [DOI] [PubMed] [Google Scholar]
- 144.Kwan JC, Ratnayake R, Abboud KA, Paul VJ, Luesch H. J. Org. Chem. 2010;75:8012–8023. doi: 10.1021/jo1013564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morris LA, Milne BF, Thompson GS, Jaspars M. J. Chem. Soc. Perkin Trans. 2002;2:1072–1075. [Google Scholar]
- 146.Latifi R, Bagherzadeh M, Milne BF, Jaspars M, de Visser SP. J. Inorg. Biochem. 2008;102:2171–2178. doi: 10.1016/j.jinorgbio.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 147.Liu H, Liu Y, Wang Z, Xing X, Maguire AR, Luesch H, Zhang H, Xu Z, Ye T. Chem. Eur. J. 2013;19:6774–6784. doi: 10.1002/chem.201203667. [DOI] [PubMed] [Google Scholar]
- 148.Kwan JC, Liu Y, Ratnayake R, Hatano R, Kuribara A, Morimoto C, Ohnuma K, Paul VJ, Ye T, Luesch H. ChemBioChem. 2014;15:799–804. doi: 10.1002/cbic.201300762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Matern U, Schelberger C, Jelakovic S, Weckesser J, Schulz GE. Chem. Biol. 2003;10:997–1001. doi: 10.1016/j.chembiol.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 150.Salvador LA, Taori K, Biggs JS, Jakoncic J, Ostrov DA, Paul VJ, Luesch H. J. Med. Chem. 2013;56:1276–1290. doi: 10.1021/jm3017305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee AY, Smitka TA, Bonjouklian R, Clardy J. Chem. Biol. 1994;1:113–117. doi: 10.1016/1074-5521(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 152.Taori K, Matthew S, Rocca JR, Paul VJ, Luesch H. J. Nat. Prod. 2007;70:1593–1600. doi: 10.1021/np0702436. [DOI] [PubMed] [Google Scholar]
- 153.Barnes PJ, Stockley RA. Eur. Respir. J. 2005;25:1084–1106. doi: 10.1183/09031936.05.00139104. [DOI] [PubMed] [Google Scholar]
- 154.Kwan JC, Eksioglu EA, Liu C, Paul VJ, Luesch H. J. Med. Chem. 2009;52:5732–5747. doi: 10.1021/jm9009394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Conus S, Simon HU. Swiss Med. Wkly. 2010;140:w13042. doi: 10.4414/smw.2010.13042. [DOI] [PubMed] [Google Scholar]
- 156.Yang S, Zhang W, Ding N, Lo J, Liu Y, Clare-Salzler MJ, Luesch H, Li Y. Bioorg. Med. Chem. 2012;20:4774–4780. doi: 10.1016/j.bmc.2012.05.077. [DOI] [PubMed] [Google Scholar]
- 157.Molinski TF, Reynolds KA, Morinaka BI. J. Nat. Prod. 2012;75:425–431. doi: 10.1021/np200861n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Williams PG, Yoshida WY, Moore RE, Paul VJ. J. Nat. Prod. 2003;66:1006–1009. doi: 10.1021/np030114z. [DOI] [PubMed] [Google Scholar]
- 159.Sun T, Zhang W, Zong C, Wang P, Li Y. J. Pept. Sci. 2010;16:364–374. doi: 10.1002/psc.1254. [DOI] [PubMed] [Google Scholar]
- 160.Liu Y, Zhang W, Li L, Salvador LA, Chen T, Chen W, Felsenstein KM, Ladd TB, Price AR, Golde TE, He J, Xu Y, Li Y, Luesch H. J. Med. Chem. 2012;55:10749–10765. doi: 10.1021/jm301630s. [DOI] [PubMed] [Google Scholar]
- 161.Orjala J, Nagle DG, Hsu VL, Gerwick WH. J. Am. Chem. Soc. 1995;117:8281–8282. [Google Scholar]
- 162.Berman FW, Gerwick WH, Murray TF. Toxicon. 1999;37:1645–1648. doi: 10.1016/s0041-0101(99)00108-7. [DOI] [PubMed] [Google Scholar]
- 163.Nogle LM, Okino T, Gerwick WH. J. Nat. Prod. 2001;64:983–985. doi: 10.1021/np010107f. [DOI] [PubMed] [Google Scholar]
- 164.Li WI, Berman FW, Okino T, Yokokawa F, Shioiri T, Gerwick WH, Murray TF. Proc. Natl. Acad. Sci. USA. 2001;98:7599–7604. doi: 10.1073/pnas.121085898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cao Z, Gerwick WH, Murray TF. BMC Neuroscience. 2010;11:154–166. doi: 10.1186/1471-2202-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jabba SV, Prakash A, Dravid SM, Gerwick WH, Murray TF. J. Pharmacol. Expt. Ther. 2010;332:698–709. doi: 10.1124/jpet.109.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pereira A, Cao Z, Murray TF, Gerwick WH. Chem. Biol. 2009;16:893–906. doi: 10.1016/j.chembiol.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Choi H, Pereira AR, Cao Z, Shuman CF, Engene N, Byrum T, Matainaho T, Murray TF, Mangoni A, Gerwick WH. J. Nat. Prod. 2010;73:1411–1421. doi: 10.1021/np100468n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Malloy KL, Choi H, Fiorilla C, Valeriote FA, Matainaho T, Gerwick WH. Bioorg. Med. Chem. Lett. 2012;22:683–688. doi: 10.1016/j.bmcl.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu M, Okino T, Nogle LM, Marquez BL, Williamson RT, Sitachitta N, Berman FW, Murray TF, McGough K, Jacobs R, Colsen K, Asano T, Yokokawa F, Shioiri T, Gerwick WH. J. Am. Chem. Soc. 2000;122:12041–12042. [Google Scholar]
- 171.LePage KT, Goeger D, Yokokawa F, Asano T, Shioiri T, Gerwick WH, Murray TF. Toxicol. Lett. 2005;158:133–139. doi: 10.1016/j.toxlet.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 172.Pereira AR, Cao Z, Engene N, Soria-Mercado IE, Murray TF, Gerwick WH. Org. Lett. 2010;12:4490–4493. doi: 10.1021/ol101752n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Soria-Mercado IE, Pereira A, Cao Z, Murray TF, Gerwick WH. Org. Lett. 2009;11:4704–4707. doi: 10.1021/ol901438b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem. Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 175.Müller J, Hemphill A. Int. Rev. Cell Mol. Biol. 2013;301:359–401. doi: 10.1016/B978-0-12-407704-1.00007-5. [DOI] [PubMed] [Google Scholar]
- 176.Wilkinson SR, Kelly JM. Expert Rev. Mol. Med. 2009;11:e31. doi: 10.1017/S1462399409001252. [DOI] [PubMed] [Google Scholar]
- 177.Hughes D. Nat. Rev. Genet. 2003;4:432–441. doi: 10.1038/nrg1084. [DOI] [PubMed] [Google Scholar]
- 178.Linington RG, Clark BR, Trimble EE, Almanza A, Urena L, Kyle DE, Gerwick WH. J. Nat. Prod. 2009;72:14–17. doi: 10.1021/np8003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Conroy T, Guo JT, Linington RG, Hunt NH, Payne RJ. Chem. Eur. J. 2011;17:13544–13552. doi: 10.1002/chem.201102538. [DOI] [PubMed] [Google Scholar]
- 180.Conroy T, Guo JT, Hunt NH, Payne RJ. Org. Lett. 2010;12:5576–5579. doi: 10.1021/ol1024663. [DOI] [PubMed] [Google Scholar]
- 181.Stolze SC, Deu E, Kaschani F, Li N, Florea BI, Richau KH, Colby T, van der Hoorn RA, Overkleeft HS, Bogyo M, Kaiser M. Chem. Biol. 2012;19:1546–1555. doi: 10.1016/j.chembiol.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Miller B, Friedman AJ, Choi H, Hogan J, McCammon JA, Hook V, Gerwick WH. J. Nat. Prod. 2014;77:92–99. doi: 10.1021/np400727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sanchez LM, Lopez D, Vesely BA, Della Togna G, Gerwick WH, Kyle DE, Linington RG. J. Med. Chem. 2010;53:4187–4197. doi: 10.1021/jm100265s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sanchez LM, Knudsen GM, Helbig C, De Muylder G, Mascuch SM, Mackey ZB, Gerwick L, Clayton C, McKerrow JH, Linington RG. J. Nat. Prod. 2013;76:630–641. doi: 10.1021/np300834q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Miller MB, Bassler BL. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 186.Kalia VC. Biotech. Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 187.Dobretsov S, Teplitski M, Alagely A, Gunasekera SP, Paul VJ. Environ. Microbiol. Rep. 2010;2:739–744. doi: 10.1111/j.1758-2229.2010.00169.x. [DOI] [PubMed] [Google Scholar]
- 188.Clark BR, Engene N, Teasdale ME, Rowley DC, Matainaho T, Valeriote FA, Gerwick WH. J. Nat. Prod. 2008;71:1530–1537. doi: 10.1021/np800088a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kwan JC, Meickle T, Ladwa D, Teplitski M, Paul VJ, Luesch H. Mol. Biosyst. 2011;7:1205–1216. doi: 10.1039/c0mb00180e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Choi H, Mascuch SJ, Villa FA, Byrum T, Teasdale ME, Smith JE, Preskitt LB, Rowley DC, Gerwick L, Gerwick WH. Chem. Biol. 2012;19:589–598. doi: 10.1016/j.chembiol.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Montaser R, Paul VJ, Luesch H. Org. Lett. 2013;15:4050–4053. doi: 10.1021/ol401396u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kwan JC, Luesch H. Chem. Eur. J. 2010;16:13020–13029. doi: 10.1002/chem.201001562. [DOI] [PMC free article] [PubMed] [Google Scholar]