Abstract
Cryptococcus neoformans is an opportunistic human fungal pathogen and can undergo both bisexual and unisexual mating. Despite the fact that one mating type is dispensable for unisexual mating, the two sexual cycles share surprisingly similar features. Both mating cycles are affected by similar environmental factors and regulated by the same pheromone response pathway. Recombination takes place during unisexual reproduction in a fashion similar to bisexual reproduction and can both admix pre-existing genetic diversity and also generate diversity de novo just like bisexual reproduction. These common features may allow the unisexual life cycle to provide phenotypic and genotypic plasticity for the natural Cryptococcus population, which is predominantly α mating type, and to avoid Muller’s ratchet. The morphological transition from yeast to hyphal growth during both bisexual and unisexual mating may provide increased opportunities for outcrossing and the ability to forage for nutrients at a distance. The unisexual life cycle is a key evolutionary factor for Cryptococcus as a highly successful global fungal pathogen.
Keywords: Unisexual mating, Recombination, de novo genetic diversity, Muller’s ratchet, Nutrient foraging, Filamentation
1. Introduction
Sex is ubiquitous throughout biology. Numerous fungal species were long considered clonal or asexual, for example, the human fungal pathogens Candida albicans and Aspergillus fumigatus. However, bioinformatic studies of whole genome sequences revealed the existence of both opposite alleles of the mating-type locus (MAT), and population genetics studies revealed genetic recombination within the population. These advances first suggested sexual reproduction is an integral life cycle feature for these and other fungal species that have been thought to be asexual (Butler et al., 2009; Ene and Bennett, 2014; Heitman, 2006).
Sexual identity in fungi is defined by the mating type locus (MAT), which contains genes that regulate cell identity and sexual reproduction (Ni et al., 2011). Mating can be either heterothallic, involving mating between partners harboring different mating type alleles with high DNA sequence dissimilarity, or homothallic, which involves sexual reproduction of solo cultured individual isolates. Homothallism can also involve selfing, or same-sex mating/unisexual reproduction. Heterothallism and homothallism in fungi have been reviewed in detail (Lee et al., 2010; Ni et al., 2011). Table 1 lists some classic examples for different types of heterothallic and homothallic mating in fungi.
Table 1.
Examples of fungal mating strategies.
| Phylum | Species | Reference | |
|---|---|---|---|
| Heterothallism | |||
| Bipolar | Ascomycota |
Aspergillus spp. (A. fumigatus; A. oryzae) |
(Galagan et al., 2005) |
| Candida albicans | (Hull and Johnson, 1999) | ||
| Neurospora crassa | (Metzenberg and Glass, 1990) | ||
| Basidiomycota | Cryptococcus neoformans | (Hsueh et al., 2011b) | |
| Ustilago hordei | (Lee et al., 1999) | ||
| Tetrapolar | Basidiomycota | Coprinopsis cinerea | (Casselton and Kues, 2007) |
| Cryptococcus heveanensis | (Metin et al., 2010) | ||
| Ustilago maydis | (Spellig et al., 1994) | ||
| Homothallism | |||
| Pseudohomothallism | Ascomycota | Neurospora tetrasperma | (Merino et al., 1996) |
| Podospora anserina | (Picard et al., 1991) | ||
| Mating type switching |
Ascomycota | Kluyveromyces lactis | (Butler et al., 2004) |
| Saccharomyces cerevisiae | (Herskowitz et al., 1992) | ||
| Schizosaccharomyces pombe | (Nielsen and Egel, 2007) | ||
| Basidiomycota | Agrocybe aegerita | (Labarere and Noel, 1992) | |
| Two fused or linked opposite MAT loci |
Ascomycota | Cochliobolus spp. | (Yun et al., 1999) |
| Didymella zeae-maydis | (Yun et al., 2013) | ||
|
Neurospora spp. (N. pannonica; N. terricola) |
(Poggeler, 1999) | ||
| Stemphylium spp. | (Inderbitzin et al., 2005) | ||
| Sordaria macrospora | (Poggeler, 1999) | ||
| Two unlinked opposite MAT loci |
Ascomycota | Aspergillus nidulans | (Galagan et al., 2005) |
| Neosartorya fischeri | (Rydholm et al., 2007) | ||
| One MAT locus | Ascomycota | Candida albicans | (Alby et al., 2009) |
|
Neurospora spp. (N. africana; N. dodgei; N. galapagonensis; N. lineolata) |
(Poggeler, 1999) | ||
| Stemphylium spp. | (Inderbitzin et al., 2005) | ||
| Basidiomycota | Cryptococcus neoformans | (Lin et al., 2005) | |
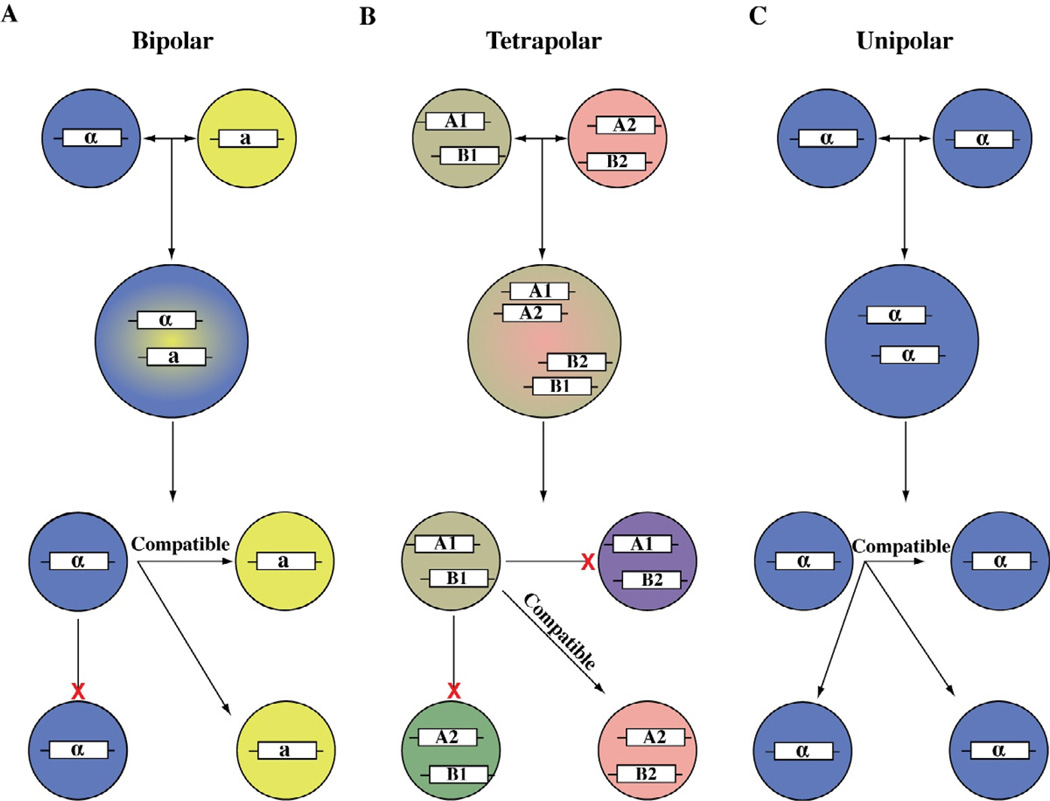
Heterothallic mating occurs either in a bipolar or a tetrapolar system. The bipolar mating system contains a single bi-allelic MAT locus that controls mating between partners of opposite mating types, such as the genetic model Neurospora crassa, the prominent human fungal pathogenic basidiomycetes Cryptococcus gattii and C. neoformans, and the ascomycetes Candida albicans and Aspergillus fumigatus (Galagan et al., 2005; Hsueh et al., 2011b; Hull and Johnson, 1999; Metzenberg and Glass, 1990). Many basidiomycetes, such as Coprinopsis cinerea, Cryptococcus heveanensis, and Ustilago maydis exhibit tetrapolar mating systems which contain two unlinked, multiallelic MAT loci that control mating between partners of different mating types that differ at both MAT loci (Casselton and Kues, 2007; Metin et al., 2010; Spellig et al., 1994). Tetrapolar mating systems restrict inbreeding more than bipolar mating systems because they reduce the chances of mating between the progeny to 25% versus 50% in a bipolar mating system (Figure 1) (Ene and Bennett, 2014). Homothallism involves sexual reproduction of solo isolates without a partner and there are at least five different types of homothallism in different fungal species, including pseudohomothallism, mating type switching, two fused or linked MAT loci, two unlinked MAT loci, and one MAT locus (Ni et al., 2011). Pseudohomothallism occurs in Neurospora tetrasperma and Podospora anserina, and refers to selfing of spores that contain two nuclei of different mating types (Merino et al., 1996; Picard et al., 1991). Mating type switching refers to a daughter cell mating with the mother cell that has converted its mating type through endonuclease facilitated recombination in response to DNA double-strand breaks, and it occurs in Kluyveromyces lactis, Saccharomyces cerevisiae, and Schizosaccharomyces pombe (Butler et al., 2004; Herskowitz et al., 1992; Nielsen and Egel, 2007). Even though pseudohomothallism and mating type switching facilitate mating within the same cell, or among cells from an asexually derived colony, mating still occurs between two nuclei of the opposite mating types. In certain fungal species, both alleles of the MAT locus are present in one nucleus enabling the fungus to be self-fertile. The MAT loci can be fused or linked on one chromosome as in the ascomycetes Cochliobolus spp., Stemphylium spp., and Neurospora spp (Inderbitzin et al., 2005; Poggeler, 1999; Yun et al., 1999), or unlinked as in the ascomycetes Aspergillus nidulans and Neosartorya fischeri (Galagan et al., 2005; Rydholm et al., 2007). Finally, in four Neurospora spp. (N. africana, N. dodgei, N. galapagonensis, and N. lineolata) and a group of Stemphylium spp., only one MAT locus is present in the genome and yet these species are self-fertile and bona fide homothallic isolates (Glass and Smith, 1994; Inderbitzin et al., 2005; Poggeler, 1999).
Figure 1.
A) Bipolar mating type systems encode mating type determinants at one locus. Alleles for the MAT locus must be different for heterothallic mating to occur and diploids to be formed. The resulting spores are able to mate with 50% of their siblings in a tetrad. B) Mating type determinants are encoded at two unlinked loci in tetrapolar mating systems. Cells must carry different alleles at both MAT loci for mating to occur and spores can only mate with 25% of their sibling meiotic products. C) C. neoformans has a unisexual cycle, in addition, to the standard bipolar heterothallic mating cycle. α cells are able to mate with other α cells to produce diploids that are homozygous at the MAT loci. These diploids are able to sporulate and produce inter-fertile spores.
Homothallism has also been reported in two bipolar heterothallic human fungal pathogens, Candida albicans and Cryptococcus neoformans. Alby et al. (Alby et al., 2009) reported that when the α pheromone protease Bar1 is lost in C. albicans a cells, the α pheromone produced by a cells can promote a-a mating. In addition, in the presence of α cells, two a cells can mate efficiently under the influence of α cell expressed α pheromone. During C. albicans mating, two diploid nuclei fuse to form a tetraploid nucleus, which then undergoes a parasexual cycle and returns to the diploid or aneuploid state through stochastic chromosome loss. The monokaryotic fruiting behavior reported in the basidiomycete Cryptococcus neoformans was first thought to be an asexual reproduction event, and to be restricted to isolates of only the α mating type (Lin et al., 2006; Tscharke et al., 2003; Wickes et al., 1996). Subsequently, isolates of a mating type were identified that also produce hyphae and spores and this process was shown to be a quantitative trait in which the α allele of the MAT locus enhances hyphal growth (Lin et al., 2006; Tscharke et al., 2003; Wickes et al., 1996). Lin et al. (Lin et al., 2005) showed that there is a high frequency of recombination among monokaryotic fruiting progeny and deletion of the meiosis specific recombinase Dmc1 impaired monokaryotic sporulation, which provides evidence that monokaryotic fruiting is a sexual cycle involving cells of the same mating type, and thus C. neoformans is capable of homothallic mating. An insertional mutagenesis study further showed that Spo11, an enzyme that drives meiotic recombination by inducing DNA double-strand breaks, is required for sporulation during monokaryotic fruiting, which further verified the existence of a unisexual life cycle in C. neoformans (Feretzaki and Heitman, 2013a; Lin et al., 2005).
It is interesting to note that C. neoformans has a bipolar mating system and can undergo robust a-α heterothallic mating, and yet there is an extant parallel unisexual life cycle. The potential significance of unisexual mating has been highlighted for the Cryptococcus spp, specifically serotype A C. neoformans var. grubii and serotype D C. neoformans var. neoformans, by the following key findings: 1) Environmental and clinical isolates of C. neoformans are predominantly α mating type (Kwon-Chung and Bennett, 1978), with one documented exception of a Cryptococcus population in sub-Saharan Africa, which has a more balanced ratio of a and α cells (Litvintseva et al., 2003); 2) Mating of C. neoformans has not been observed in nature, but C. neoformans can readily mate both bisexually and unisexually under laboratory conditions, including media containing pigeon guano and live plants (Kwon-Chung, 1976a; Nielsen et al., 2007; Xue et al., 2007); 3) Approximately 8% of natural isolates tested are diploids for Cryptococcus spp, and the reported diploids include α/α diploids (fusion products of two serotype A α cells), αADα diploids (fusion products of serotype A and serotype D α cells), and hybrid AD diploids of opposite mating types (Cogliati et al., 2001; Lengeler et al., 2001; Lin et al., 2007; Lin et al., 2009). Taken together, these reports suggest unisexual reproduction plays a pivotal role in the life cycles, and possibly also the virulence cycles, of the globally prevalent human pathogenic Cryptococcus species. In this review, we take a deeper look into the consequences and the biological impacts of α-α unisexual mating in C. neoformans in comparison with traditional bisexual mating.
2. Environmental factors and genetic circuits for C. neoformans sexual cycles
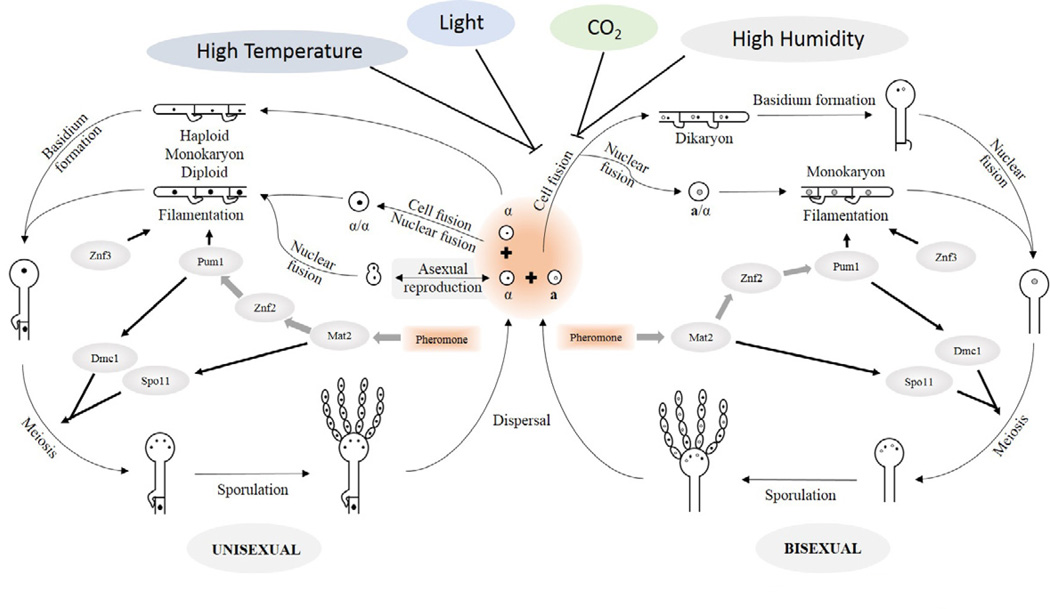
During the typical C. neoformans a-α bisexual mating, cells of opposite mating type (a and α) fuse to form a zygote, which undergoes a transition to filamentous growth. Parental nuclei remain separate in the dikaryotic hyphae. Clamp cell fusion with the neighboring hyphal compartment allows inheritance of both parental nuclei in each hyphal cell. The tip of the hyphae eventually enlarges and forms a basidium, within which nuclear fusion occurs and one round of meiosis produces four meiotic products. Repeated rounds of mitosis from these meiotic products produce nuclei that are packed into spores, and subsequently bud from the surface of the basidium to form four chains of basidiospores (Figure 2) (Idnurm, 2010; Kwon-Chung, 1975; Kwon-Chung, 1976a; Kwon-Chung, 1976b; Lin, 2009). Under conditions unsuitable for hyphal growth, dikaryotic hyphae produce diploid yeast cells by nuclear fusion. When conditions become favorable for hyphal development, diploid yeast cells can reenter the sexual cycle and form monokaryotic hyphae (Figure 2) (Sia et al., 2000).
Figure 2.
The life cycle of Cryptococcus neoformans, and environmental factors and genetic circuits that govern C. neoformans bisexual and unisexual reproduction.
During C. neoformans unisexual mating, either nuclear fusion following cell fusion between two α cells, nuclear fusion between mother and daughter nuclei, or nuclear endoreplication at different stages of the unisexual cycle can establish the α-α diploid state. Clamp cells of the monokaryotic hyphae do not fuse with the adjacent hyphal compartment, and the tip of the hyphae gives rise to the basidium. Meiosis and mitosis then take place to generate haploid spores (Figure 2) (Feretzaki and Heitman, 2013b; Lin et al., 2005; Wickes et al., 1996). Both environmental factors and genetic circuits govern diploidization (morphological transition from yeast cell to hyphal growth), meiosis, mitosis, and sporulation during both sexual cycles in C. neoformans. These regulatory mechanisms have been reviewed in detail (Kozubowski and Heitman, 2012; Lin, 2009; Wang and Lin, 2011). Here we focus on highlights of the regulatory mechanisms and recent discoveries.
Environmental cues for bisexual and unisexual life cycles exhibit a high degree of overlap. Factors that can stimulate cell-cell fusion and hyphal growth in both bisexual and unisexual mating include: ambient temperature, nitrogen limitation, dehydrated substrates, darkness, presence of mating pheromones, inositol, the plant hormone auxin (indole-3-acetic acid), and copper ions (Hull and Heitman, 2002; Kent et al., 2008; Kozubowski and Heitman, 2012; Lin, 2009; Rutherford et al., 2008; Schwartz and Staib, 1980; Torres-Guererro and Edman, 1994; Walton et al., 2005; Wang and Lin, 2011; Xue et al., 2007). Environmental conditions that inhibit bisexual mating also inhibit unisexual mating, such as high temperature, high levels of CO2, high humidity, and continuous white light exposure (Figure 2) (Bahn et al., 2005; Sia et al., 2000; Wang et al., 2012a; Xue et al., 2007). The mechanisms by which these environmental factors impact the sexual cycles often lie within genetic regulatory circuits. For example, copper ions up-regulate pheromone expression and promote mating (Kent et al., 2008), and continuous light activates the Bwc1/2 photoreceptor which negatively regulates mating specific gene expression (Idnurm and Heitman, 2005; Lu et al., 2005; Yeh et al., 2009).
The pheromone response pathway regulates both bisexual mating and unisexual mating (Figure 2) (Reviewed by Kozubowski and Heitman, 2012; Wang and Lin, 2011). The Cpk1 mitogen activated protein kinase (MAPK) pathway is the conserved yeast pheromone response pathway in C. neoformans. Mutation of components in this pathway blocks or impairs both bisexual and unisexual mating, such as deletion of Mfα (α pheromone), Ste11/Ste7/Cpk1 (core components of the Cpk1 MAPK pathway), and Mat2 (transcription factor for the Cpk1 MAPK pathway) (Davidson et al., 2003; Hsueh et al., 2009; Lin et al., 2010; Shen et al., 2002; Yue et al., 1999). On the other hand, overexpression of these components enhances sexual development (Hsueh et al., 2007; Hsueh et al., 2009; Shen et al., 2002; Yue et al., 1999). Besides the canonical pheromone response pathway, other factors have been identified as common regulators for both bisexual and unisexual mating, including two zinc finger transcription factors Znf2 and Znf3 (Feretzaki and Heitman, 2013a; Lin et al., 2010). Znf2 was later identified as the master regulator for hyphal morphogenesis (Wang et al., 2012a). Transcriptomic studies showed that Znf2 directs the yeast-hyphal transition through the Pumilio-family protein Pum1 during mating. Pum1 temporally up-regulates expression of Dmc1, a meiotic recombinase required for sporulation, during mating (Figure 2) (Wang et al., 2014). The pheromone response pathway positively regulates Znf2 expression during mating; however, overexpression of Znf2 can drive the yeast-hyphal transition through an adhesion protein, Cfl1, independent of mating (Lin et al., 2010; Wang et al., 2013a). Znf3 promotes cell fusion and pheromone production for both bisexual and unisexual cycles, and it also negatively regulates expression of transposable elements, which suggests that Znf3 may function in transposon silencing during mating (Feretzaki and Heitman, 2013a).
Despite the similarity in the environmental regulation and the genetic circuits between bisexual and unisexual mating, evidence has accumulated to suggest unisexual mating is more plastic. gpa2Δ gpa3Δ double mutants lacking two G protein α subunits are defective in cell fusion and bisexual mating; however, the mutant can undergo robust self-filamentation, suggesting cell fusion can be separate from unisexual reproduction (Hsueh et al., 2007). It is also worth noting that a transcriptomic study of a highly self-fertile strain showed that a cell fusion gene, PRM1, is highly upregulated during unisexual reproduction (Lin et al., 2010). Orthologs of Prm1 in Saccharomyces cerevisiae and Neurospora crassa contribute to plasma membrane fusion during mating (Aguilar et al., 2007; Fleissner et al., 2009). It is possible that Prm1 is a fortuitous target given that the Cpk1 MAPK pathway is shared between bisexual and unisexual reproduction. Deletion of the STE6 gene encoding a pheromone transporter severely affects cell fusion and filamentation during bisexual mating, yet unisexual mating is not affected (Hsueh and Shen, 2005). The differences between these two different sexual cycles suggest other pathways, independent of or in addition to the upstream pheromone response pathway, also regulate unisexual mating.
3. Recombination during bisexual and unisexual reproduction
Morphologically, α-α unisexual reproduction resembles a-α bisexual reproduction, although the initiation of unisexual reproduction can be either via fusion of two α cells, or by endoreplication of the nucleus of an α cell that establishes a dikaryon/diploid state. α-α unisexual reproduction, like a-α bisexual reproduction, requires meiotic genes (e.g. DMC1 and SPO11), demonstrating that unisexual reproduction also involves meiosis (Feretzaki and Heitman, 2013a; Lin et al., 2005; Wang et al., 2014). Additionally, our recent studies show that meiotic recombination in a-α bisexual and α-α unisexual reproduction occurs in a similar fashion (unpublished data). Specifically, we found that between the two modes of sexual reproduction: 1) the numbers of crossovers along chromosomes have comparable distributions; 2) the chromosomal average recombination frequencies (physical distance/genetic distance, kb/cM) are similar and comparable to those that have been previously reported (Marra et al., 2004); and 3) recombination hotspots, such as those flanking the MAT locus during a-α bisexual reproduction (Hsueh et al., 2006), also operate during α-α unisexual reproduction.
Extensive sequence divergence and chromosomal rearrangements are present between the two MAT alleles, a and α, which have been hypothesized to pose physical barriers for crossing over within the MAT locus during a-α bisexual reproduction. Indeed, in a study of more than 150 meiotic progeny from a-α bisexual reproduction of serotype D C. neoformans, no crossover has been detected within the MAT locus. However, gene conversion does occur within the MAT locus around a GC-rich intergenic region between the RPO41 and BSP2 genes (Sun et al., 2012). Interestingly, two recombination hot spots that flank the MAT locus are also located within GC-rich regions. During α-α unisexual reproduction, the two MATα alleles align with each other during meiosis, and the physical constraints on recombination posed by sequence divergence and chromosomal rearrangements are no longer present. However, the lack of physical constraints on recombination does not result in elevated crossover during α-α unisexual reproduction. Among 156 analyzed unisexual meiotic progeny, only one crossover event occurred within the MAT locus (unpublished data). This frequency of crossover within the MAT locus is considerably lower than expected given the size of MAT and the average recombination frequency during α-α unisexual reproduction. A possible explanation could be that the two recombination hot spots that flank the MAT locus cause crossover interference within MAT.
In C. neoformans, recombination can occur not only during sexual reproduction in the form of meiotic recombination, but also during vegetative growth in the form of mitotic recombination. During a-α bisexual mating, it is generally accepted that nuclear fusion does not occur until shortly before meiosis in the basidium, which limits the opportunities for mitotic recombination. In a recent study of progeny from AD hybridization, the authors found that more than four haplotypes are present among progeny dissected from a single basidium, which contradicts the principle that one meiotic event can produce a maximum of four genotypes in each basidium. The authors proposed that, post-meiotic mitotic recombination within aneuploid/diploid basidiospores is the reason for the observation of greater than four haplotypes during AD hybridization (Vogan et al., 2013). During both a-α bisexual and α-α unisexual reproduction in serotype D C. neoformans, we also found evidence suggesting that mitotic recombination occurs at a fairly high frequency, and in this case, most likely after karyogamy and prior to meiosis. Specifically, we found that among progeny from α-α unisexual reproduction, certain chromosomal regions exhibited highly skewed allele inheritance, while among progeny from a-α bisexual reproduction, alleles from one of the two parents at certain chromosomal regions were absent in the progeny recovered from a single basidium. Although our observation could be the result of gene conversion events during meiosis, the fact that the regions where alleles from one parent are absent among progeny cluster together and encompass large chromosomal regions suggest pre-meiotic mitotic recombination is the likely cause of loss of heterozygosity.
In addition, mitotic recombination breakpoints are enriched in certain chromosomal regions, suggesting the presence of fragile sites in the C. neoformans genome that are vulnerable to mitotic recombination, similar to those recently reported in Saccharomyces cerevisiae (Song et al., 2014). Additionally, some of these potential fragile sites overlap with meiotic recombination hot spots that have been previously identified, indicating certain characteristics of these chromosomal regions, such as a high GC content, might make them prone to induction of both meiotic and mitotic recombination (Hsueh et al., 2006).
In both a-α bisexual and α-α unisexual reproduction, nuclear fusion can occur immediately following cell fusion, resulting in heterozygous diploid cells. These diploid cells produce monokaryotic hyphae with unfused clamp cells, before the formation of the basidium and the onset of meiosis. Thus, ample opportunities are present for mitotic recombination to occur during both modes of sexual mating in C. neoformans (Hsueh et al., 2011a).
Maintaining a certain level of recombination during sexual reproduction is critical for proper meiosis. In hybridization between serotypes A and D, elevated sequence divergence and the presence of chromosomal rearrangements serve to repress meiotic recombination, and as a consequence, meiosis is compromised and the majority of hybrid progeny are aneuploid (Sun and Xu, 2007; Sun and Xu, 2009; Vogan et al., 2013). On the other hand, repression of meiotic recombination at certain chromosomal regions could also be beneficial, such as the MAT locus, where crossing over within could result in abnormal chromosomes due to chromosomal rearrangements between the opposite alleles, or non-functional MAT alleles by breaking up co-adapted alleles. This could be one of the reasons why recombination hot spots are present at both edges of the MAT locus and function in both bisexual and unisexual mating, as crossover interference exerted by these hot spots may further repress recombination within MAT, and thus ensure the inheritance of intact MAT alleles.
4. Sex as a diversity generator
Sexual reproduction is a key contributor to the generation of diversity in both genotype and phenotype, an adaptive function in response to environmental variations. In the Cryptococcus species complex, recombination takes place during both bisexual and unisexual mating. The advantages of canonical bisexual mating can be attributed to the admixture of preexisting diversity and generation of novel genotypes, while the role of unisex is substantially more puzzling. Unlike large multicellular eukaryotes that are obligately sexual, Cryptococcus can produce offspring without going through meiosis, so the advantages of a unisexual cycle that would clearly promote inbreeding are not immediately clear, especially given the energetic and opportunity costs of sex. The role of unisex may be explained by the ability of the sexual cycle in Cryptococcus to generate de novo diversity, through access to the diploid state and generation of aneuploidy during sex. Additionally there are a number of other potential hypotheses as to how unisex may play a role in generating diversity, including co-opting the sexual machinery to produce polyploid Titan cells, enable spread of cytoplasmic elements, and stimulate RNAi pathways operating during the sexual cycle.
4.1 Sex contributes to admixture of preexisting diversity in the population
First, sex serves a key role in the reassortment of alleles, during both unisexual (Lin et al., 2005) and bisexual reproduction. One key role of sexual reproduction, the elimination of deleterious alleles via Muller’s ratchet, will be discussed in more detail below. Sex can also play a critical role in the development of virulence through recombination between two diverse lineages, as occurred in Toxoplasma (Grigg et al., 2001). In Cryptococcus, the ability of the sexual cycle to mediate transmission of virulence characteristics between lineages under laboratory conditions has been tested. In crosses between VGII C. gattii isolates, hypervirulence traits contributed by an outbreak VGII parent were easily transmitted (Voelz et al., 2013). In contrast, hypervirulence traits were not easily transferred across a species barrier between the VGII and VGIII C. gattii lineages.
Both bisex and unisex are important in Cryptococcus because of the paucity of a isolates in the environment. Outside of a hypothesized ancestral origin in Botswana (Litvintseva et al., 2011), the Cryptococcus neoformans var. grubii group is largely clonal globally, with a preponderance of MATα isolates (Kwon-Chung and Bennett, 1978). Thus unisex may allow access to some of the inherent advantages of the sexual cycle. Alternatively, in some regions, like Sub-Saharan Africa, a isolates are present at a much higher frequency and population genetics analyses have revealed hallmarks of mating likely to involve canonical bisexual reproduction (Litvintseva et al., 2003; Litvintseva et al., 2006). In addition, recombination has been observed at the population level in two subtypes of VGIII C. gattii in the California area (Byrnes et al., 2011 and unpublished data). One of these subtypes, VGIIIb, is also characterized by a substantial number of a isolates, while the other is primarily comprised of α isolates (Byrnes et al., 2011). Consequently, it is possible that both bisexual and unisexual cycles are acting to admix these populations within the California area.
Sexual reproduction has also been implicated, at the population level, in the development of pathogenic clonal clusters in VGII Cryptococcus gattii, which is the etiological agent currently responsible for an ongoing outbreak in the Pacific Northwest of the United States. Early analysis employing multilocus sequence typing (MLST) suggested that unisexual mating may have contributed to the development of the virulent VGIIa group that constituted the major portion of the outbreak, as approximately 50% of MLST markers were shared between the major VGIIa and the minor VGIIb components of the outbreak (Fraser et al., 2005). More recent work employing whole genome sequencing of multiple VGII isolates suggests that at least the highly virulent and clonal VGIIc group found in the Pacific Northwest may be the product of sexual crosses between preexisting VGII lineages not involved in the outbreak (Billmyre et al., 2014; Engelthaler et al., 2014). In addition, the VGII subtype as a whole appears to have undergone substantial admixture through sexual reproduction during its proposed emergence from South America (Hagen et al., 2013). In summary, the sexual cycle operates to recombine preexisting diversity on the population level, even in an organism with a skewed mating type ratio and a preponderance of clonal lineages.
4.2 Sex contributes to de novo diversity
Sex can also contribute not just by mixing preexisting diversity, but also by creating de novo diversity, not previously represented in the population. One mechanism by which this can occur is through the production of not only haploid progeny, but also diploid intermediates. This contributes diversity either by accessing the diploid state or by masking deleterious recessive traits in the diploid hybrids. In Saccharomyces, the diploid state is advantageous under some conditions and disadvantageous under others (Zörgö et al., 2013). Diploids occur frequently in the C. neoformans population, with approximately 8% of natural isolates tested being diploid. The majority of these cases were the result of autodiploidization or self-fusion between two cells of the same mating type (Lin et al., 2009). Alternatively, intervarietal αADα diploid hybrids have also been identified. The hybridization of two diverse genomes allowed transgressive phenotypes for both thermotolerance and UV resistance in these hybrids (Lin et al., 2007). Both of these diploid states provide advantages through access to the diploid state, but outcrossing between the Cryptococcus varieties also allows access to hybrid vigor.
In addition, sexual reproduction contributes to the generation of de novo diversity through the production of aneuploid progeny. Aneuploidy is typically thought of as deleterious, especially in the case of multicellular eukaryotes like humans, where it can result in dramatic defects, including Down’s syndrome or embryonic lethality. However, in limited circumstances, even multicellular organisms occasionally utilize aneuploid cells within specific developmental niches, like the fly hindgut (Fox and Duronio, 2013). Likewise, recent work in S. cerevisiae has shown that in unicellular eukaryotes, aneuploidy can provide a selective advantage under the appropriate conditions (Pavelka et al., 2010; Torres et al., 2007). In C. neoformans, aneuploid progeny with altered phenotypes were identified after both bisexual and unisexual mating at a frequency of approximately 4% (Ni et al., 2013). Aneuploidy has previously been observed as a mechanism of drug resistance during normal mitotic growth on fluconazole (Sionov et al., 2010), and not surprisingly, both increases and decreases in fluconazole sensitivity can be observed in aneuploid progeny produced without fluconazole selection by the sexual cycle (Ni et al., 2013). Aneuploidy can also contribute to variation in virulence factor expression, such as melanin production, in C. neoformans (Hu et al., 2011). Notably, both bisexual and unisexual mating produce aneuploid progeny at similar frequencies. In addition, variations in chromosomal structure are present during both sexual reproductions, including segmental deletions (Ni et al., 2013). This suggests that the process of sex may be inherently mutagenic and, like the parasexual cycle of Candida albicans, create diversity at a high rate through error-prone reduction of ploidy (Forche et al., 2008). Thus, the unisexual cycle, in the absence of preexisting diversity to admix, can inherently generate diversity de novo.
4.3 Alternative possible roles for unisexual reproduction
In addition to the known roles for bisexual and unisexual mating in generating diversity de novo, there are a number of potential roles yet to be explored in detail in Cryptococcus. These include ploidy increases and reduction in Titan cells, spread of cytoplasmic elements, and transposon silencing by RNAi pathways during the sexual cycles.
Sex may be involved in ploidy variation during both the generation of polyploidy Titan cells and the production of cells derived from Titan cells. Titan cells have been reviewed in detail (Zaragoza and Nielsen, 2013), but briefly, stimulation of Ste3a can induce the production of Titan cells (Okagaki et al., 2010). In addition, these Titan cells produce haploid or nearly haploid aneuploid progeny, despite containing 8n or even higher chromosome numbers (Nielsen, 2014). Together with the induction by Ste3a, this raises the question of whether ploidy reduction in Titan cells is sexual, or possibly parasexual, in nature. Studies may be limited by a lack of genomic variation to determine recombination, but tests with deletions of known meiotic components may be illuminating. In either case, the process appears to at least employ some elements of the sexual machinery to accomplish substantial changes in both ploidy and phenotype.
Recent work in S. cerevisiae suggests sex can result in advantageous selection independent of genomic diversity, but instead by transmission of cytoplasmic elements, like dsRNA viruses (Edwards et al., 2014) or prions (Suzuki et al., 2012). Interspecies mating may cross species barriers, and usually results in abortive mating, yet cytoplasmic elements are able to be transmitted between the mating pairs. Crosses between clonal individuals within a population could allow the transmission of an established prion into other cells. Cytoplasmic viruses are present in other basidiomycetes including Ustilago (Drinnenberg et al., 2011), but have not been observed to this point in Cryptococcus. Studies of prions and mycoviruses in Cryptococcus may illuminate an unseen biological impact for both the bisexual and unisexual life cycles.
Sex also induces silencing of endogenous transposons and transgenes in Cryptococcus. C. neoformans undergoes two distinct transgene dependent silencing processes, one during normal mitotic growth, called MIS (mitotic induced silencing) (Wang et al., 2012b), and one during the sexual cycle, called SIS (sex induced silencing) (Wang et al., 2010). Both processes function to repress expression of transposons, either at a basal level during vegetative growth (Dumesic et al., 2014; Wang et al., 2012b), or at a much higher level under mating conditions (Wang et al., 2010). The key difference between these two processes is that SIS occurs at a much higher frequency, which is in line with the increased expression of transposons and elevated levels of RNAi components under mating conditions. Deletion of RNAi components allows increased expression and movement of transposable elements, which can have direct functional consequences, such as development of FK506 resistance (Wang et al., 2010). Notably, SIS occurs during both bisexual and unisexual mating (Wang et al., 2013b). In addition, the VGII subtype of C. gattii has conspicuously lost its RNAi pathway (Billmyre et al., 2013; D’Souza et al., 2011; Wang et al., 2010). This suggests that the role of transposons in generating diversity may be amplified in the VGII lineage, as the control mechanism used in C. neoformans is absent. Furthermore, transposons are a key component of the centromere in Cryptococcus (Janbon et al., 2014). Loss of RNAi in VGII may therefore amplify the generation of aneuploidy during the sexual cycle in Cryptococcus, either through changes in chromatin structure in the centromeres, like in Schizosaccharomyces pombe (Volpe et al., 2003), or simply by allowing more rapid transposon movement to change the otherwise conserved structure of the centromeres.
5. Reversing Muller’s ratchet
In the Cryptococcus species complex, it is clear that unisexual mating is mechanistically similarly to traditional bisexual mating, however, it does not explain the evolution of a parallel unisexual cycle. Here we explore the significance of unisexual mating in Cryptococcus from an evolutionary perspective.
It is puzzling that most eukaryotes reproduce sexually during their life cycle when considering the two-fold cost of sex (Smith, 1978; Williams, 1975). Each daughter in a sexual system requires contributions from two parents while a single asexual mother is sufficient to produce daughters mitotically. This is typically thought of as a cost of providing non-reproducing males, but hermaphroditic mating systems have similar evolutionary costs (Lloyd, 1988). A sexually reproducing organism’s genome is diluted every generation. Every allele will be passed on to every daughter during clonal division but each allele has only a 50% chance to be passed on during sexual reproduction. Homothallic reproductive strategies such as unisex may have initially evolved, in part, to avoid the two-fold cost of modern sex.
The ubiquity of sex must be the result of significant advantages to counterbalance the costs of sex (Bell, 1982; Williams, 1975). Hermann Muller first recognized the fundamental challenge facing asexually dividing populations; their genomes would gradually accumulate moderately deleterious mutations without a means to purge them, a process since termed Muller’s ratchet (Felsenstein, 1974; Haldane, 1937; Muller, 1932; Muller, 1964). Mutations occur during both cell division and growth, eroding the fitness of individuals and of the population. Natural selection works to remove strongly deleterious alleles from the population, but weakly deleterious alleles can increase in frequency through genetic drift or linkage to a beneficial allele. In an asexual population, eventually every individual will carry at least one harmful mutation, and the best class of genome will be lost. Sexual reproduction allows organisms to use recombination to reassort alleles and purge the genome of these deleterious mutations to recreate a best class of genome. Selection is not sufficient to maintain fitness without the ability to recombine and reassort chromosomes to purge mildly deleterious mutations.
Genetic drift is thought to play a central role in the rate at which a population accumulates deleterious mutations or the speed that Muller’s ratchet turns (Bell, 1988; Charlesworth and Charlesworth, 1998; Gessler, 1995; Haigh, 1978). The population of C. neoformans is globally distributed and large, leading some to hypothesize that the impact of genetic drift and Muller’s ratchet is small. However, the population of C. neoformans is highly structured and we have only begun to investigate the population history. There are four distinct serotypes, based on capsular agglutination, in the species complex containing C. neoformans and C. gattii (Belay et al., 1996). Interserotype diploid hybrids are found in both clinical and environmental samples, but while these diploids display hybrid vigor, the spores produced rarely germinate, indicating significant barriers to gene flow (Lengeler et al., 2001). In addition, there are multiple molecular types in each serotype, with both cosmopolitan and geographically isolated types. The cosmopolitan molecular types, found worldwide, are thought to have larger populations but appear to be largely clonal expansions. There is little, if any, genetic exchange between molecular types and there is even significant population structure within some molecular types (Brandt et al., 1996; Bui et al., 2008; Campbell et al., 2005; Casali et al., 2003; Franzot et al., 1997; Halliday and Carter, 2003; Jain et al., 2005; Litvintseva et al., 2003; Litvintseva and Mitchell, 2012; Ngamskulrungroj et al., 2009; Nishikawa et al., 2003). This suggests that C. neoformans may not be one large, global, interbreeding population but numerous, local, genetically isolated populations of unknown size. If C. neoformans represents many smaller populations, the effects of genetic drift and Muller’s ratchet on its population will be much larger than previously considered. Unisexual reproduction may be of particular importance to those isolated populations with few or no a alleles and restricted opportunities to access the bisexual reproductive pathway.
Both bisexual and unisexual cycles utilize the same post cell-fusion meiotic and recombinational pathways (Bui et al., 2008; Feretzaki and Heitman, 2013a; Lin et al., 2005). Therefore, unisexual reproduction, like bisexual reproduction, is hypothesized to allow Cryptococcus populations to avoid Muller’s ratchet, which remains to be validated experimentally.
6. Foraging for mating partners and nutrients
The transition from haploid to diploid brings about a large number of transcriptional and morphological changes. These changes can be environmentally dependent, or specific to the life cycle of a species. Under limiting growth conditions, diploid cells are generally larger, increasing the ratio of volume to surface area. In many populations of C. neoformans, the frequency of the MATa alleles is low enough that the unisexual cycle is likely the dominant pathway to instigate these morphological transitions. The change in surface area changes the number of transporters and receptors the cell can utilize while a change in volume can affect the protein content and number of organelles. Changes in the ratio of these components interacts with specific environmental conditions which pose environmental advantages to either diploids or haploids (Zörgö et al., 2013).
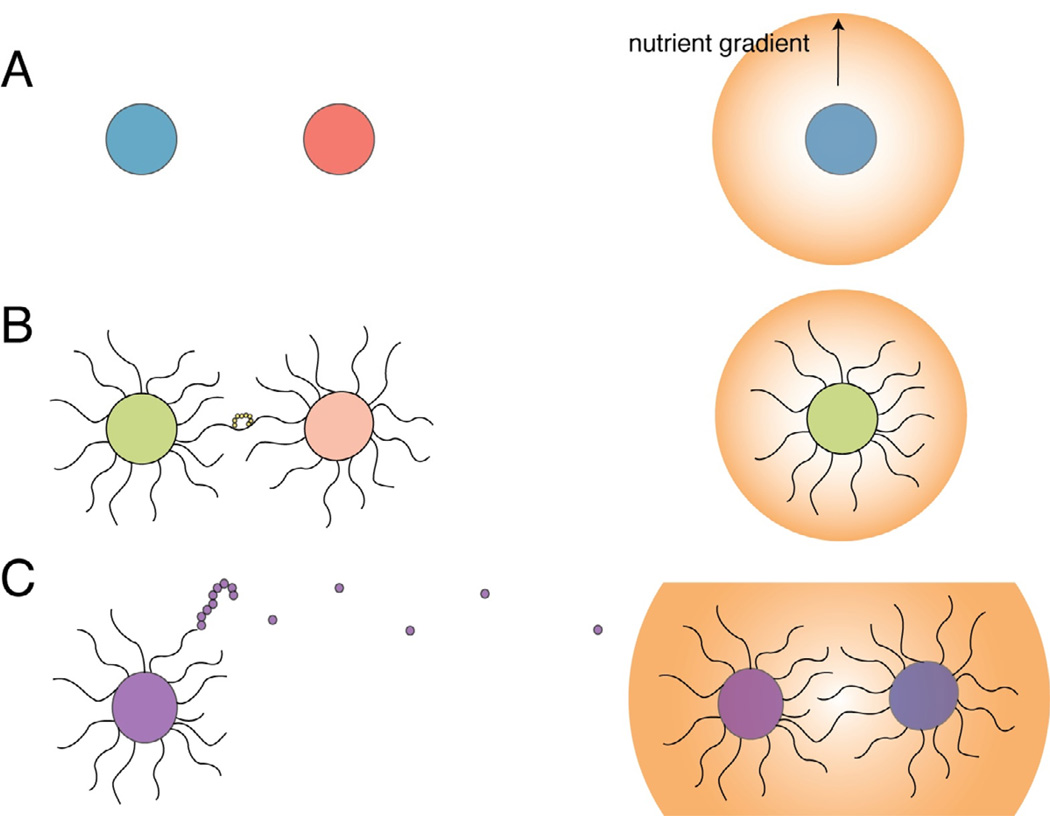
For the Cryptococcus species complex, the biological impacts of its sexual cycles are manifested through its morphological transition from yeast to hyphal growth (Roach et al., 2014). C. neoformans grows mitotically as a haploid yeast and produces hyphae, basidia, diploid cells, and spores during the unisexual or bisexual cycles. These morphologically differentiated cell types provide plasticity for environmental adaptation. For example, C. neoformans can employ the hyphae produced by the unisexual cycle to explore the environment and forage for nutrients (Phadke et al., 2013), and spores can be easily dispersed widely by wind or water and are resistant to adverse environmental conditions (Alvarez and Casadevall, 2006; Botts et al., 2009). The exploration of the environment may also have other beneficial effects such as bringing mating ready cells into contact with distant mating partners, either through the production of hyphae to explore the local environment or the production of spores for wider dispersal. Thus, filamentation during sexual life cycles can increase the number and frequency of outcrossing by enabling foraging for mates, akin to courtship in S. cerevisiae (Figure 3) (Jackson and Hartwell, 1990a; Jackson and Hartwell, 1990b).
Figure 3.
Hyphal growth during both bisexual and unisexual reproduction provides advantages in foraging for mating partners and nutrients. A) Asexually dividing Cryptococcus cells are unable to undergo the dimorphic switch. They grow as a yeast colony unable to reach potential mating partners or distant nutrient sources. B) During bisexual and unisexual reproduction Cryptococcus undergoes hyphal growth. The hyphae are able to explore the environment, bringing them into contact with mating partners or new nutrient sources. C) Unisexual and bisexual mating produce spores, in addition to hyphae, and these spores are able to be easily dispersed by the wind or water to colonize new environments, bringing them into contact with both mating partners and nutrients.
The yeast-hyphal transition was once considered a hallmark for Cryptococcus mating; however, evidence has shown that there is also a sex-independent form of thermally activated hyphal growth (Fu et al., 2013). Bilateral mating between two znf2Δ α cells failed to initiate the morphological transition despite the fact that diploid cells can be easily detected (Wang and Lin, 2011). It is understood now that Znf2 functions as a master regulator for Cryptococcus filamentation (Wang et al., 2012a). Znf2 is regulated by the pheromone pathway during mating and can induce two different types of hyphal cells, aerial hyphae that can sporulate, and invasive hyphae (Wang et al., 2012a). Invasive hyphae usually concentrate at the periphery of the mating colony and can invade solid agar medium, extend on the surface, and expand into the air (Wang et al., 2012a). Overexpression of Znf2 under mating suppressive conditions revealed that invasive hyphal growth is regulated by Znf2 through an adhesion protein, Cfl1 (Wang et al., 2013a; Wang et al., 2014). Though it is not clear whether this pathway can be activated independent of sex in nature, it does suggest Cryptococcus may have a pathway analogous to yeast pseudohyphal differentiation in response to nutrient limitation (Cullen and Sprague, 2012). Interestingly, nutrient deprivation does promote pseudohyphal growth in Cryptococcus (Lee et al., 2012; Lin, 2009). Nonetheless, both bisexual and unisexual mating produce hyphae, which may provide fitness benefits by enabling hyphal cells to forage for nutrients in places yeast colonies are unable to reach.
Highlights.
Similar environmental factors and genetic circuits regulate both unisexual and bisexual mating in Cryptococcus neoformans.
Recombination happens in a similar fashion during unisexual and bisexual mating.
Unisexual mating can generate phenotypic diversity de novo.
Filamentous growth during sexual reproduction allows Cryptococcus colonies to forage for mating partners and nutrients.
Summary points.
Unisexual and bisexual mating are controlled by similar environmental factors and genetic circuits in Cryptococcus neoformans.
Recombination happens in a similar fashion during unisexual and bisexual mating. Recombination hotspots for bisexual mating also function during unisexual mating. MAT loci recombination is also repressed during unisexual mating despite no or limited sequence divergence.
Unisexual mating can generate phenotypic diversity de novo, like bisexual mating. Unisexual mating can also enable a predominantly α Cryptococcus population to avoid Muller’s ratchet.
Filamentous growth during both unisexual mating and bisexual mating may provide fitness benefits by increasing opportunities for outcrossing and allowing Cryptococcus colonies to forage for distant nutrients.
Acknowledgements
Studies reviewed here were supported in part by NIH/NIAID R37 MERIT Award AI39115-17 and R01 AI50113-10.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar PS, Engel A, Walter P. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Mol. Biol. Cell. 2007;18:547–556. doi: 10.1091/mbc.E06-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alby K, Schaefer D, Bennett RJ. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature. 2009;460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Cox GM, Perfect JR, Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr. Biol. 2005;15:2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Belay T, Cherniak R, O’Neill EB, Kozel TR. Serotyping of Cryptococcus neoformans by dot enzyme assay. J. Clin. Microbiol. 1996;34:466–470. doi: 10.1128/jcm.34.2.466-470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. The Evolution and Genetics of Sexuality. Los Angeles, CA: University of California Press; 1982. The Masterpiece of Nature. [Google Scholar]
- Bell G. Recombination and the immortality of the germ line. J. Evol. Biol. 1988;1:67–82. [Google Scholar]
- Billmyre RB, Calo S, Feretzaki M, Wang X, Heitman J. RNAi function, diversity, and loss in the fungal kingdom. Chromosome Research. 2013;21:561–572. doi: 10.1007/s10577-013-9388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre RB, Croll D, Li W, Mieczkowski P, Carter DA, Cuomo CA, Kronstad JW, Heitman J. Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. mBio. 2014;5:e01494–14. doi: 10.1128/mBio.01494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot. Cell. 2009;8:595–605. doi: 10.1128/EC.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt ME, Hutwagner LC, Klug LA, Baughman WS, Rimland D, Graviss EA, Hamill RJ, Thomas C, Pappas PG, Reingold AL, Pinner RW. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T, Lin X, Malik R, Heitman J, Carter DA. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, α mating type populations. Eukaryot. Cell. 2008;7:1771–1780. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe KH. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, Dietrich FS, May RC, Chaturvedi S, Chatuverdi S, Chaturvedi V, Chatuverdi V, Heitman J. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 2011;7:e1002205. doi: 10.1371/journal.ppat.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot. Cell. 2005;4:1403–1409. doi: 10.1128/EC.4.8.1403-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali AK, Goulart Lc, Rosa e Silva LvK, Silva ÂM, Amaral AA, Alves SH, Schrank A, Meyer W, Vainstein MH. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. FEMS Yeast Research. 2003;3:405–415. doi: 10.1016/S1567-1356(03)00038-2. [DOI] [PubMed] [Google Scholar]
- Casselton LA, Kues U. The origin of multiple mating types in the model mushrooms Coprinopsis cinerea and Schizophyllum commune. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in Fungi: Molecular Determination and Evolutionary Implications. Washington, D.C.: ASM Press; 2007. pp. 283–300. [Google Scholar]
- Charlesworth B, Charlesworth D. Some evolutionary consequences of deleterious mutations. Genetica. 1998;102–103:3–19. [PubMed] [Google Scholar]
- Cogliati M, Esposto MC, Clarke DL, Wickes BL, Viviani MA. Origin of Cryptococcus neoformans var. neoformans diploid strains. J. Clin. Microbiol. 2001;39:3889–3894. doi: 10.1128/JCM.39.11.3889-3894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr. The regulation of filamentous growth in yeast. Genetics. 2012;190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, Lee N, Zeilmaker T, Sawkins J, McVicker G, Shah S, Gnerre S, Griggs A, Zeng Q, Bartlett K, Li W, Wang X, Heitman J, Stajich JE, Fraser JA, Meyer W, Carter D, Schein J, Krzywinski M, Kwon-Chung KJ, Varma A, Wang J, Brunham R, Fyfe M, Ouellette BFF, Siddiqui A, Marra M, Jones S, Holt R, Birren BW, Galagan JE, Cuomo CA. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio. 2011;2:e00342–10. doi: 10.1128/mBio.00342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 2003;49:469–485. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic PA, Natarajan P, Chen C, Drinnenberg IA, Schiller BJ, Thompson J, Moresco JJ, Yates JR, III, Bartel DP, Madhani HD. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2014;152:957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MD, Symbor-Nagrabska A, Dollard L, Gifford DK, Fink GR. Interactions between chromosomal and nonchromosomal elements reveal missing heritability. Proc. Natl. Acad. Sci. USA. 2014;111:7719–7722. doi: 10.1073/pnas.1407126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene IV, Bennett RJ. The cryptic sexual strategies of human fungal pathogens. Nat. Rev. Microbiol. 2014;12:239–251. doi: 10.1038/nrmicro3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler DM, Hicks ND, Gillece JD, Roe CC, Schupp JM, Driebe EM, Gilgado F, Carriconde F, Trilles L, Firacative C, Ngamskulrungroj P, Castañeda E, Lazera MDS, Melhem MSC, Pérez-Bercoff A, Huttley G, Sorrell TC, Voelz K, May RC, Fisher MC, Thompson GR, Lockhart SR, Keim P, Meyer W. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio. 2014;5:e01464–14. doi: 10.1128/mBio.01464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Heitman J. Genetic circuits that govern bisexual and unisexual reproduction in Cryptococcus neoformans. PLoS Genet. 2013a;9:e1003688. doi: 10.1371/journal.pgen.1003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Heitman J. Unisexual reproduction drives evolution of eukaryotic microbial pathogens. PLoS Pathog. 2013b;9:e1003674. doi: 10.1371/journal.ppat.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner A, Diamond S, Glass NL. The Saccharomyces cerevisiae PRM1 homolog in Neurospora crassa is involved in vegetative and sexual cell fusion events but also has postfertilization functions. Genetics. 2009;181:497–510. doi: 10.1534/genetics.108.096149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzot SP, Hamdan JS, Currie BP, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: Evidence for both local genetic differences and a global clonal population structure. J. Clin. Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- Fu J, Morris IR, Wickes BL. The production of monokaryotic hyphae by Cryptococcus neoformans can be induced by high temperature arrest of the cell cycle and is independent of same-sex mating. PLoS Pathog. 2013;9:e1003335. doi: 10.1371/journal.ppat.1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae . Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gessler DDG. The constraints of finite size in asexual populations and the rate of the ratchet. Genetics Research. 1995;66:241–253. doi: 10.1017/s0016672300034686. [DOI] [PubMed] [Google Scholar]
- Glass NL, Smith ML. Structure and function of a mating-type gene from the homothallic species Neurospora africana . Mol. Gen. Genet. 1994;244:401–409. doi: 10.1007/BF00286692. [DOI] [PubMed] [Google Scholar]
- Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science. 2001;294:161–165. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
- Hagen F, Ceresini PC, Polacheck I, Ma H, van Nieuwerburgh F, Gabaldón T, Kagan S, Pursall ER, Hoogveld HL, van Iersel LJJ, Klau GW, Kelk SM, Stougie L, Bartlett KH, Voelz K, Pryszcz LP, Castañeda E, Lazera M, Meyer W, Deforce D, Meis JF, May RC, Klaassen CHW, Boekhout T. Ancient dispersal of the human fungal pathogen Cryptococcus gattii from the Amazon rainforest. PLoS One. 2013;8:e71148. doi: 10.1371/journal.pone.0071148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh J. The accumulation of deleterious genes in a population—Muller’s ratchet. Theoretical Population Biology. 1978;14:251–267. doi: 10.1016/0040-5809(78)90027-8. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. The effect of variation of fitness. The American Naturalist. 1937;71:337–349. [Google Scholar]
- Halliday CL, Carter DA. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 2003;41:703–711. doi: 10.1128/JCM.41.2.703-711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr. Biol. 2006;16:R711–R725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- Herskowitz I, Rine J, Strathern J. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae . In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces, Volume 2, Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laborotory Press; 1992. pp. 583–656. [Google Scholar]
- Hsueh YP, Idnurm A, Heitman J. Recombination hotspots flank the Cryptococcus mating-type locus: implications for the evolution of a fungal sex chromosome. PLoS Genet. 2006;2:e184. doi: 10.1371/journal.pgen.0020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Lin X, Kwon-Chung KJ, Heitman J. Sexual reproduction of Cryptococcus . In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, editors. Cryptococcus: From Human Pathogen to Model Yeast. Washington, D.C.: ASM Press; 2011a. p. 81. [Google Scholar]
- Hsueh YP, Metin B, Findley K, Rodriguez-Carres M, Heitman J. The mating-type locus of Cryptococcus: evolution of gene clusters governing sex determination and sexual reproduction from the phylogenomic perspective. In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, editors. Cryptococcus: From Human Pathogen to Model Yeast. ASM Press; Washington, D.C: 2011b. pp. 139–149. [Google Scholar]
- Hsueh YP, Shen WC. A homolog of Ste6, the a-factor transporter in Saccharomyces cerevisiae, is required for mating but not for monokaryotic fruiting in Cryptococcus neoformans . Eukaryot. Cell. 2005;4:147–155. doi: 10.1128/EC.4.1.147-155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. G protein signaling governing cell fate decisions involves opposing Gα subunits in Cryptococcus neoformans . Mol. Biol. Cell. 2007;18:3237–3249. doi: 10.1091/mbc.E07-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans . EMBO J. 2009;28:1220–1233. doi: 10.1038/emboj.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wang J, Choi J, Jung WH, Liu I, Litvintseva AP, Bicanic T, Aurora R, Mitchell TG, Perfect JR, Kronstad JW. Variation in chromosome copy number influences the virulence of Cryptococcus neoformans and occurs in isolates from AIDS patients. BMC Genomics. 2011;12:526. doi: 10.1186/1471-2164-12-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 2002;36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans . Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- Idnurm A. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans . Genetics. 2010;185:153–163. doi: 10.1534/genetics.109.113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. Lateral transfer of mating system in Stemphylium . Proc. Natl. Acad. Sci. USA. 2005;102:11390–11395. doi: 10.1073/pnas.0501918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Hartwell LH. Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 1990a;63:1039–1051. doi: 10.1016/0092-8674(90)90507-b. [DOI] [PubMed] [Google Scholar]
- Jackson CL, Hartwell LH. Courtship in Saccharomyces cerevisiae: an early cell-cell interaction during mating. Mol. Cell. Biol. 1990b;10:2202–2213. doi: 10.1128/mcb.10.5.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Wickes BL, Keller SM, Fu J, Casadevall A, Jain P, Ragan MA, Banerjee U, Fries BC. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J. Clin. Microbiol. 2005;43:5733–5742. doi: 10.1128/JCM.43.11.5733-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, Chatterjee G, Mullapudi N, Hon C-C, Billmyre RB, Brunel F, Bahn Y-S, Chen W, Chen Y, Chow EWL, Coppée J-Y, Floyd-Averette A, Gaillardin C, Gerik KJ, Goldberg J, Gonzalez-Hilarion S, Gujja S, Hamlin JL, Hsueh Y-P, Ianiri G, Jones S, Kodira CD, Kozubowski L, Lam W, Marra M, Mesner LD, Mieczkowski PA, Moyrand F, Nielsen K, Proux C, Rossignol T, Schein JE, Sun S, Wollschlaeger C, Wood IA, Zeng Q, Neuvéglise C, Newlon CS, Perfect JR, Lodge JK, Idnurm A, Stajich JE, Kronstad JW, Sanyal K, Heitman J, Fraser JA, Cuomo CA, Dietrich FS. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014;10:e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent CR, Ortiz-Bermudez P, Giles SS, Hull CM. Formulation of a defined V8 medium for induction of sexual development of Cryptococcus neoformans . Appl. Environ. Microbiol. 2008;74:6248–6253. doi: 10.1128/AEM.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Heitman J. Profiling a killer, the development of Cryptococcus neoformans . FEMS Microbiol. Rev. 2012;36:78–94. doi: 10.1111/j.1574-6976.2011.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans . Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans . Mycologia. 1976a;68:821–833. [PubMed] [Google Scholar]
- Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976b;68:943–946. [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 1978;108:337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Phadke S, Sun S, Heitman J. Pseudohyphal growth of Cryptococcus neoformans is a reversible dimorphic transition in response to ammonium that requires Amt1 and Amt2 ammonium permeases. Eukaryot. Cell. 2012;11:1391–1398. doi: 10.1128/EC.00242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Cox GM, Heitman J. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 2001;69:115–122. doi: 10.1128/IAI.69.1.115-122.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect. Genet. Evol. 2009;9:401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG, Heitman J. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATα allele enhances filamentation. PLoS Genet. 2006;2:e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans . Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans . PLoS Genet. 2010;6:e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–1990. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Carbone I, Rossouw J, Thakur R, Govender NP, Mitchell TG. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS One. 2011;6:e19688. doi: 10.1371/journal.pone.0019688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in Sub-Saharan Africa. Eukaryot. Cell. 2003;2:1162–1168. doi: 10.1128/EC.2.6.1162-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Mitchell TG. Population genetic analyses reveal the African origin and strain variation of Cryptococcus neoformans var. grubii . PLoS Pathog. 2012;8:e1002495. doi: 10.1371/journal.ppat.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd DG. Benefits and costs of biparental and uniparental reproduction in plants. In: Michod RE, Levin BR, editors. The Evolution of Sex. Sunderland, MA: Sinauer Associates; 1988. pp. 233–252. [Google Scholar]
- Lu YK, Sun KH, Shen WC. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans . Mol. Microbiol. 2005;56:480–491. doi: 10.1111/j.1365-2958.2005.04549.x. [DOI] [PubMed] [Google Scholar]
- Marra RE, Huang JC, Fung E, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans) Genetics. 2004;167:619–631. doi: 10.1534/genetics.103.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino ST, Nelson MA, Jacobson DJ, Natvig DO. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma . Genetics. 1996;143:789–799. doi: 10.1093/genetics/143.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metin B, Findley K, Heitman J. The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genet. 2010;6:e1000961. doi: 10.1371/journal.pgen.1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg RL, Glass NL. Mating type and mating strategies in Neurospora . Bioessays. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Some genetic aspects of sex. The American Naturalist. 1932;66:118–138. [Google Scholar]
- Muller HJ. The relation of recombination to mutational advance. Mutation Research. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, Mitchell TG, Vainstein MH, Meyer W. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One. 2009;4:e5862. doi: 10.1371/journal.pone.0005862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans . PLoS Biol. 2013;11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Sun S, Wang X, Heitman J. Sex in fungi. Annu. Rev. Genet. 2011;45:405–430. doi: 10.1146/annurev-genet-110410-132536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K. 9th International Conference on Cryptococcus and Cryptococcosis. Vol. 57. The Netherlands: Mycoses, Amsterdam; 2014. The role of titan cells during infection; p. 11. [DOI] [PubMed] [Google Scholar]
- Nielsen K, De Obaldia AL, Heitman J. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot. Cell. 2007;6:949–959. doi: 10.1128/EC.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O, Egel R. The mat genes of Schizosaccharomyces pombe: expression, homothallic switch, and silencing. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in Fungi: Molecular Determination and Evolutionary Implications. Washington, D.C.: ASM Press; 2007. pp. 143–157. [Google Scholar]
- Nishikawa MM, Lazera MS, Barbosa GG, Trilles L, Balassiano BR, Macedo RCL, Bezerra CCF, Pérez MA, Cardarelli P, Wanke B. Serotyping of 467 Cryptococcus neoformans isolates from clinical and environmental sources in Brazil: analysis of host and regional patterns. J. Clin. Microbiol. 2003;41:73–77. doi: 10.1128/JCM.41.1.73-77.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadke SS, Feretzaki M, Heitman J. Unisexual reproduction enhances fungal competitiveness by promoting habitat exploration via hyphal growth and sporulation. Eukaryot. Cell. 2013;12:1155–1159. doi: 10.1128/EC.00147-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Debuchy R, Coppin E. Cloning the mating types of the heterothallic fungus Podospora anserina: developmental features of haploid transformants carrying both mating types. Genetics. 1991;128:539–547. doi: 10.1093/genetics/128.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggeler S. Phylogenetic relationships between mating-type sequences from homothallic and heterothallic ascomycetes. Curr. Genet. 1999;36:222–231. doi: 10.1007/s002940050494. [DOI] [PubMed] [Google Scholar]
- Roach KC, Feretzaki M, Sun S, Heitman J. Unisexual reproduction. Adv. Genet. 2014;85:257–307. doi: 10.1016/B978-0-12-800271-1.00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford JC, Lin X, Nielsen K, Heitman J. Amt2 permease is required to induce ammonium-responsive invasive growth and mating in Cryptococcus neoformans . Eukaryot. Cell. 2008;7:237–246. doi: 10.1128/EC.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydholm C, Dyer PS, Lutzoni F. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri . Eukaryot. Cell. 2007;6:868–874. doi: 10.1128/EC.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Staib F. Effect of purines on the formation of the perfect state of Cryptococcus neoformans, Filobasidiella neoformans . Zentralbl Bakteriol A. 1980;248:274–280. [PubMed] [Google Scholar]
- Shen WC, Davidson RC, Cox GM, Heitman J. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans . Eukaryot. Cell. 2002;1:366–377. doi: 10.1128/EC.1.3.366-377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Lengeler KB, Heitman J. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet. Biol. 2000;29:153–163. doi: 10.1006/fgbi.2000.1192. [DOI] [PubMed] [Google Scholar]
- Sionov E, Lee H, Chang YC, Kwon-Chung KJ. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. The Evolution of Sex. New York, NY: Cambridge University Press; 1978. [Google Scholar]
- Song W, Dominska M, Greenwell PW, Petes TD. Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA. 2014;111:E2210–8. doi: 10.1073/pnas.1406847111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig T, Regenfelder E, Reichmann M, Schauwecker F, Bohlmann R, Urban M, Bolker M, Kamper J, Kahmann R. Control of mating and development in Ustilago maydis . Anton. Van Leeuw. 1994;65:191–197. doi: 10.1007/BF00871946. [DOI] [PubMed] [Google Scholar]
- Sun S, Hsueh YP, Heitman J. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 2012;8:e1002810. doi: 10.1371/journal.pgen.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Xu J. Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans . Genetics. 2007;177:1475–1486. doi: 10.1534/genetics.107.078923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Xu J. Chromosomal rearrangements between serotype A and D strains in Cryptococcus neoformans . PLoS One. 2009;4:e5524. doi: 10.1371/journal.pone.0005524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- Torres-Guererro H, Edman JC. Melanin-deficient mutants of Cryptococcus neoformans . J. Med. Vet. Mycol. 1994;32:303–313. [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Tscharke RL, Lazera M, Chang YC, Wickes BL, Kwon-Chung KJ. Haploid fruiting in Cryptococcus neoformans is not mating type α-specific. Fungal Genet. Biol. 2003;39:230–237. doi: 10.1016/s1087-1845(03)00046-x. [DOI] [PubMed] [Google Scholar]
- Voelz K, Ma H, Phadke S, Byrnes EJ, Zhu P, Mueller O, Farrer RA, Henk DA, Lewit Y, Hsueh Y-P, Fisher MC, Idnurm A, Heitman J, May RC. Transmission of hypervirulence traits via sexual reproduction within and between lineages of the human fungal pathogen Cryptococcus gattii . PLoS Genet. 2013;9:e1003771. doi: 10.1371/journal.pgen.1003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan AA, Khankhet J, Xu J. Evidence for mitotic recombination within the basidia of a hybrid cross of Cryptococcus neoformans . PLoS One. 2013;8:e62790. doi: 10.1371/journal.pone.0062790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton G, White S, Teng G, Martienssen R, Allshire R. RNA interference is required for normal centromere function in fission yeast. Chromosome Research. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans . Mol. Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Lin X. Mechanisms of unisexual mating in Cryptococcus neoformans . Fungal Genet. Biol. 2011;48:651–660. doi: 10.1016/j.fgb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Wang L, Tian X, Gyawali R, Lin X. Fungal adhesion protein guides community behaviors and autoinduction in a paracrine manner. Proc. Natl. Acad. Sci. USA. 2013a;110:11571–11576. doi: 10.1073/pnas.1308173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tian X, Gyawali R, Upadhyay S, Foyle D, Wang G, Cai JJ, Lin X. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathog. 2014;10:e1004185. doi: 10.1371/journal.ppat.1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhai B, Lin X. The link between morphotype transition and virulence in Cryptococcus neoformans . PLoS Pathog. 2012a;8:e1002765. doi: 10.1371/journal.ppat.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Darwiche S, Heitman J. Sex-induced silencing operates during opposite-sex and unisexual reproduction in Cryptococcus neoformans . Genetics. 2013b;193:1163–1174. doi: 10.1534/genetics.113.149443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hsueh YP, Li W, Floyd A, Skalsky R, Heitman J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010;24:2566–2582. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang P, Sun S, Darwiche S, Idnurm A, Heitman J. Transgene induced co-suppression during vegetative growth in Cryptococcus neoformans . PLoS Genet. 2012b;8:e1002885. doi: 10.1371/journal.pgen.1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, Edman JC. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc. Natl. Acad. Sci. USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC. Sex and Evolution. Princeton, NJ: Princeton University Press; 1975. [Google Scholar]
- Xue C, Tada Y, Dong X, Heitman J. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe. 2007;1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Yeh YL, Lin YS, Su BJ, Shen WC. A screening for suppressor mutants reveals components involved in the blue light-inhibited sexual filamentation in Cryptococcus neoformans . Fungal Genet. Biol. 2009;46:42–54. doi: 10.1016/j.fgb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, Heitman J. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans . Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SH, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA. 1999;96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Nielsen K. Titan cells in Cryptococcus neoformans: cells with a giant impact. Curr. Opin. Microbiol. 2013;16:409–413. doi: 10.1016/j.mib.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zörgö E, Chwialkowska K, Gjuvsland AB, Garré E, Sunnerhagen P, Liti G, Blomberg A, Omholt SW, Warringer J. Ancient evolutionary tradeoffs between yeast ploidy states. PLoS Genet. 2013;9:e1003388. doi: 10.1371/journal.pgen.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]