Abstract
DExD/H proteins catalyze structural rearrangements in RNA by coupling ATP hydrolysis to the destabilization of RNA helices or RNP complexes. The Escherichia coli DExD/H protein DbpA specifically recognizes a region within the catalytic core of 23S rRNA. To better characterize the interaction of DbpA with this region and to identify changes in the complex between different nucleotide-bound states of the enzyme, RNase T1, RNase T2, kethoxal and DMS footprinting of DbpA on a 172 nt fragment of 23S rRNA were performed. A number of protections identified in helices 90 and 92 were consistent with biochemical experiments measuring the RNA binding and ATPase activity of DbpA with truncated RNAs. When DbpA was bound with AMPPNP, but not ADP, several additional footprints were detected in helix 93 and the single-stranded region 5′ of helix 90, suggesting binding of the helicase domains of DbpA at these sites. These results propose that DbpA can act at multiple sites and hint at the targets of its biological activity on rRNA.
INTRODUCTION
DExD/H proteins participate in practically all aspects of RNA metabolism, including transcription, splicing, ribosomal biogenesis, translation initiation and termination, nonsense-mediated decay and RNA interference (1–4). Typically termed helicases, DExD/H proteins use NTP hydrolysis to disrupt or rearrange RNA–RNA (5–7) and, possibly, RNA–protein interactions (8–10). DExD/H proteins are defined by seven conserved sequence motifs and share a common structural core, suggesting that these enzymes use similar mechanisms in binding and hydrolysis of NTP, as well as in unwinding and translocating along nucleic acids (11).
Most of the RNA helicases studied to date lack RNA specificity in vitro, presumably achieving their specific targets in vivo by interactions with auxiliary proteins (3,12). However, the Escherichia coli DEAD protein DbpA specifically recognizes 23S rRNA in vitro (13). Truncation experiments identified a 153 nt fragment of domain V of 23S rRNA, including parts of the central wheel and the A-loop, that has identical RNA-dependent ATPase activity as full-length 23S rRNA (14). However, shorter fragments showed reduced binding affinity in the ATPase assay, suggesting that a substantial portion of the 153mer is required for a complete interaction with DbpA (15). Mutagenesis studies identified hairpin 92 (the A-loop of 23S rRNA) (Fig. 1) as an important sequence-specific DbpA recognition element (15). Additional electrophoretic mobility shift binding experiments and helicase activity studies have confirmed the specificity of this RNA–protein interaction (16–18).
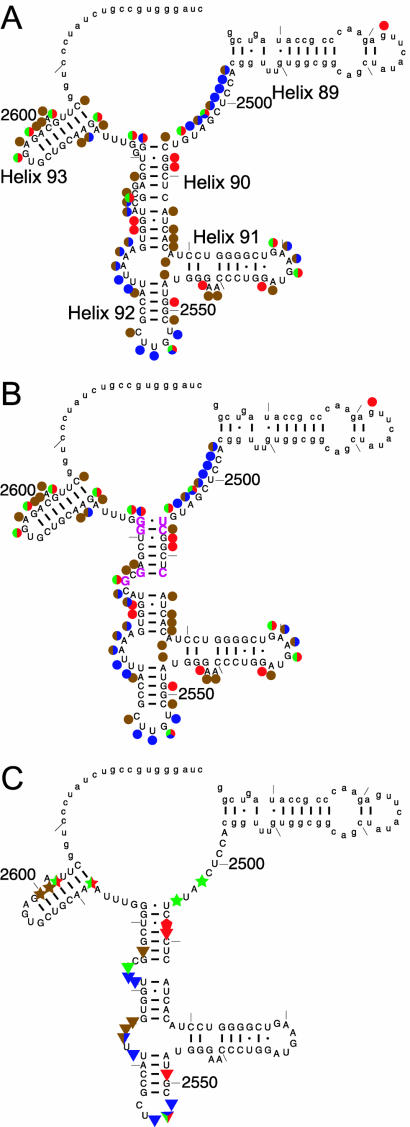
Figure 1.
(A) Cleavage and modification pattern of the 172mer by RNase T1 (green), RNase T2 (blue), kethoxal (red) and DMS (brown) shown on the phylogenetic secondary structure. (B) The cleavage/modification pattern shown on our proposed secondary structure. Rearranged residues are shown in magenta. (C) Footprinting of DbpA on the 172mer: apo-DbpA protection (triangles), AMPPNP-dependent DbpA protection (stars) and hypersensitivity (pentagons). Lower case denotes regions for which no data were collected.
Although the function of DbpA has not been established genetically, its in vitro specificity for 23S rRNA suggests a role in the ribosomal life cycle. Since the region of DbpA binding on rRNA composes part of the peptidyltransferase center (PTC) and is expected to be occluded by tRNAs during translation, DbpA is more likely to participate in ribosomal assembly than translation. The RNA specificity of the protein suggests that it would act near the PTC. Two other E.coli DEAD-box proteins (19,20) and 17 yeast DExD/H proteins (3) have been implicated in ribosome biogenesis.
DbpA has a 75 amino acid C-terminal extension outside the DEAD-box motifs that defines a group of bacterial homologs (21). Attachment of this extension to the DEAD-box domains of a non-specific RNA helicase, SrmB, yields a chimeric protein with specificity for hairpin 92, indicating that the C-terminal domain is the specific recognition element in the protein (22). In this study, we identify the RNA recognition elements of this complex interaction in further detail by chemical and enzymatic footprinting of DbpA. An extensive area of the RNA is protected by the helicase. In addition, several AMPPNP-dependent changes in the footprint reflect conformational rearrangements of the complex during the cycle of ATP binding and hydrolysis.
MATERIALS AND METHODS
Cloning and purification of His-tagged DbpA
A sequence coding for Met-His6 was appended to the 5′-end of the DbpA gene by PCR and cloned into the pET-3a vector between the NdeI and BamHI sites. A 3 l culture of transformed BL21(DE3) cells was grown to an OD600 of 0.4 at 37°C, cooled to room temperature for 15 min and induced with 0.5 mM IPTG for 30 min at room temperature. The cells were spun down and stored frozen. Cells were lysed by sonication in 50 ml of 50 mM Tris pH 7.5, 250 mM NaCl, 1 mM benzamidine, 10 mM MgCl2 and 1 mM PMSF. Cell debris and ribosomes were removed by centrifugation for 15 min at 13 000 r.p.m. in a Sorvall SS-34 rotor and 1 h at 40 000 r.p.m. in a Ti45 rotor. The supernatant was purified over a nickel column in buffer A (500 mM NaCl, 20 mM MOPS pH 6.8, 1 mM DTT, 10% glycerol, 0.02% NaN3) + 10 mM imidazole. His6-DbpA was eluted in buffer A + 0.5 M imidazole and purified further over a Pharmacia Superdex75 sizing column in buffer A. The protein was dialyzed into 500 mM NaCl, 20 mM MOPS pH 6.8, 1 mM DTT and 55% glycerol. Binding, ATPase and helicase activities of His6-DbpA were similar to the untagged protein. Where tested, DbpA and His-DbpA showed no differences in the footprinting assays (data not shown).
Preparation of RNA
23S rRNA and the 172mer substrate were prepared by run-off transcription with T7 RNA polymerase as described previously (14). The 172mer sequence was cloned into pUC19 between the EcoRI and BamHI sites. Linearization with BamHI and transcription yields nucleotides 2454–2625 (E.coli 23S rRNA numbering), plus an ATC-3′ end from the BamHI site.
Footprinting and reverse transcriptase readout
Footprinting reactions were carried out on 50 nM RNA in a buffer containing 50 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, 100 µM DTT and 0.2 mg/ml poly(A) in a total volume of 20 µl. Where noted, 1 µM His-DbpA, 1 mM ADP·Mg, 1 mM AMPPNP·Mg or 1 mM ATP·Mg were added. RNase T1 (Sigma) was used at 0.0002 U/µl, RNase T2 (Calbiochem) was used at 0.003 U/µl. Kethoxal was used at 25 mM and DMS at 2.5% (v/v). Reactions were incubated for 10 min at room temperature. RNase T1 and T2 reactions were phenol-chloroform extracted once and chloroform extracted once. DMS reactions were stopped with 5 µl of stop solution (1 M 2-mercaptoethanol, 0.75 M Tris–HCl pH 7.0). Kethoxal reaction products were stabilized by adding 1 µl of 1 M potassium borate (pH 7.0). All reactions were ethanol precipitated, washed twice in 70% ethanol and dried. The cleavage reaction RNA pellet was reconstituted in 6.6 µl of H2O, 2 µl of 500 mM Tris–HCl pH 8.3, 2 µl of 500 mM NaCl and separated into two 5.3 µl aliquots. For kethoxal reactions, the reconstitution included 0.3 µl of 1 M potassium borate (pH 7.0). Aliquots (1 µl) of 500 nM 5′-32P-GATCCC ACGGCAGATAG-3′ (complementary to the 3′-end of the 172mer and nucleotides 2612–2625 of 23S rRNA) or 5′-32P-TAAACCCAGCTCGCG-3′ (complementary to nucleotides 2573–2587) were added to the tubes. The primers were annealed by heating to 95°C for 1 min and 65°C for 3 min, adding 8 mM MgCl2 and letting it cool to room temperature. To these were added 50 µM DTT, 0.5 mM dNTPs and 1 U AMV reverse transcriptase (Amersham) to a total volume of 10 µl, followed by incubation at 42°C for 1 h and running on 12% denaturing polyacrylamide gels.
RESULTS AND DISCUSSION
The ATPase activity of DbpA is fully stimulated by a 153mer fragment of 23S rRNA containing the A-loop and parts of the central wheel of domain V, regions that are at the peptidyltransferase center of the ribosome (14). In this study, structure mapping and footprinting of DbpA were performed using two enzymatic probes, RNase T1 and T2, and two chemical probes, kethoxal and DMS. A slightly larger, 172 nt RNA was used to accommodate the reverse transcriptase (RT) primers. RNase T1 and kethoxal treatment targets single-stranded guanosines, DMS modification (with RT readout) reports on single-stranded adenosines and cytidines and RNase T2 cleaves single-stranded regions non-specifically.
Since the probes act preferentially on single-stranded and unstructured regions of RNA, the cleavage/modification pattern in the absence of DbpA allows us to assess whether the 172mer adopts a secondary structure similar to that predicted by phylogeny (23). While the observed pattern generally supports the phylogenetic secondary structure (Fig. 1A), two local rearrangements in the upper part of helix 90 are proposed in order to be consistent with the data (Fig. 1B). First, since G2574 is accessible to both RNase T1 and kethoxal and G2576 is not, it is likely that G2576, rather than G2574, pairs with C2512. Second, G2581, which is bulged in the phylogenetic structure, is not accessible in the 172mer, suggesting that it pairs with C2507. G2582 is likely to form a base pair with U2506, which is inaccessible to any of the probes in the 172mer. The resulting proposed secondary structure of helix 90 contains only a single bulge (Fig. 1B). The multiple bulges in the phylogenetic structure can be rationalized by the tertiary packing interactions of helix 90 with L3 and other parts of domain V observed in the folded ribosome that do not form in the 172mer (24). Several additional weak modifications at other positions in helix 90 suggest partial opening or ‘breathing’ of the helix under our experimental conditions. Finally, the moderate accessibility of the 2589–2603 region (helix 93) is inconsistent with a stable helix, suggesting breathing or an alternative structure.
To define the nucleotides that are protected by DbpA binding, treatment with the enzymatic and chemical probes was carried out in the presence of saturating concentrations of protein. The resulting footprinting patterns are shown in Figure 2 and summarized on the secondary structure of the 172mer in Figure 1C. Overall, apo-DbpA protects 13 of the 62 reactive sites in the 172mer. Several of the residues in hairpin 92 that were previously identified as important elements for the binding, ATPase and helicase activities of DbpA (15,17,18) are protected from RNase T1, T2 and kethoxal in the presence of DbpA (Fig. 2A–C, lanes 2 and 3). The protein also protects several sites in helix 90 and the single-stranded region between helices 90 and 92, consistent with the finding that RNA fragments containing this helix bind DbpA with higher affinity (17). No protection by apo-DbpA is observed in helices 91 and 93 and the region between helices 89 and 90. Overall, the protein appears to bind a relatively large, defined site on this rRNA fragment.
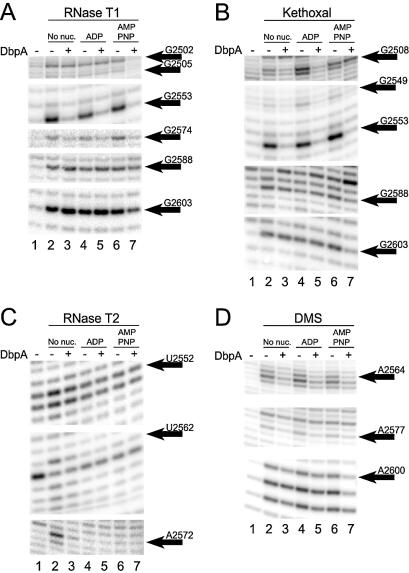
Figure 2.
Footprinting of DbpA on the 172mer by (A) RNase T1, (B) kethoxal, (C) RNase T2 and (D) DMS in the absence of nucleotide (lanes 2 and 3), with 1 mM ADP·Mg2+ (lanes 4 and 5) and with 1 mM AMPPNP·Mg2+ (lanes 6 and 7). Lane 1 is a no cleavage control.
The high resolution structures of 50S ribosomes show that the 172mer in the context of the ribosome adopts a fold that places nucleotides protected by DbpA into proximity (Fig. 3) (24,25). Helices 90 and 91 stack coaxially and helix 92 flips up and runs parallel to them, making a base triple between G2553 in helix 92 and the C2507–G2582 pair at the top of helix 90. Helix 89 also lies alongside and parallel to helices 90 and 91, forming a base pair between C2475 and G2529. While the structure of the 172mer in solution is not known, the large extent of the DbpA footprint and its compact mapping onto the ribosomal fold (Fig. 3, in blue) suggest that the 172mer may exist as a similar structure when bound to DbpA.
Figure 3.
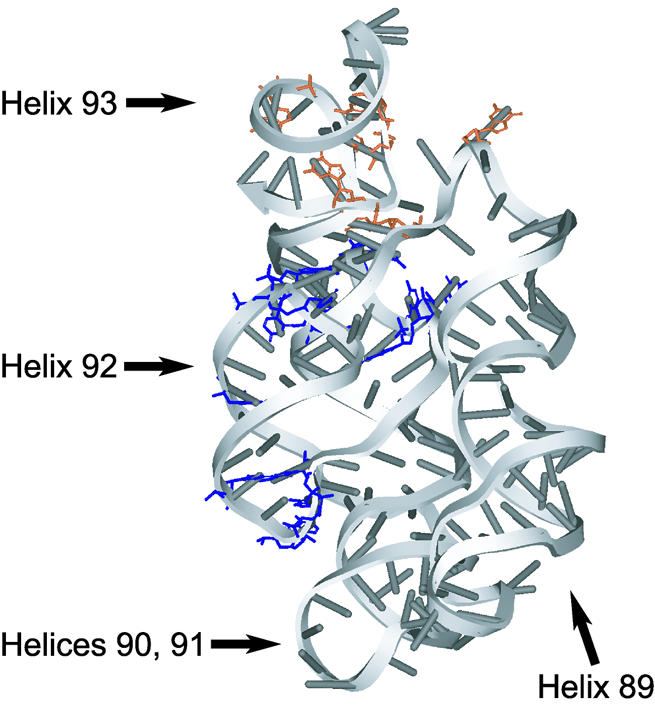
Three-dimensional structure of the RNA region encompassing helices 89–93 in the 50S ribosome. Positions protected by apo-DbpA are shown in blue. AMPNP-dependent DbpA protections are shown in orange.
As enzymes that translocate along single-stranded and unwind double-stranded nucleic acids, helicases couple NTP hydrolysis to conformational changes that lead to mechanical movement. Conformational changes in the protein between the apo- and various nucleotide-bound states have been observed by partial protease digestion assays with DbpA (26), eIF4A (27), Rep and UvrD (28) and RecB (29). It was thus of interest to detect whether conformational changes in the RNA could be observed at different stages of the catalytic cycle of DbpA. To this end, treatment with the chemical and enzymatic probes was performed in the presence of saturating ADP and AMPPNP, a non-hydrolyzable ATP analog (Fig. 2, lanes 4–7, summarized in Figs 1C and 3). RNase T2 cleavage was inhibited by the nucleotides (30) (Fig. 2C, lanes 4–7 versus lane 1), prohibiting interpretation of these data. Based on data from the other probes, the addition of ADP or AMPPNP in the absence of DbpA generally does not affect cleavage (Fig. 2A, B and D, lanes 2, 4 and 6). However, positions G2508, G2509 and G2549 reproducibly appear hypersensitive to kethoxal with 1 mM ADP (Fig. 2B, lane 2 versus 4). While the nature of this effect is unclear, RNA sequences that bind ATP have been identified (31). In the presence of DbpA, the protection pattern was not affected by the addition of 1 mM ADP (Fig. 2A, B and D, lane 3 versus 5). In contrast, when 1 mM AMPPNP was added, several changes in the footprint of DbpA were observed (Fig. 2A, B and D, lane 3 versus 7). The kethoxal protection at G2508 disappeared, while new RNase T1 protections at G2502 and G2505, and T1, DMS and kethoxal protections at G2588, A2600, C2601 and G2603 emerged. The affected regions were distinct from, but adjacent to, the apo-DbpA footprint. In the presence of 1 mM ATP, the G2502 and G2505 positions were not protected, while G2588 and G2603 were (data not shown).
The presented footprinting results elaborate on our current model of interaction of DbpA with RNAs such as the 153mer or the 172mer. In this model, the apo-DbpA protection of hairpin 92 and helix 90 represents specific binding of the C-terminal domain of DbpA. The additional AMPPNP-dependent footprints 3′ and 5′ of helix 90 are the result of helicase domain binding to those regions. The fact that positions 2502 and 2505 are protected from RNase T1 cleavage of the ribose backbone, but not from kethoxal modification of the guanine base, is consistent with binding of the DExD/H helicase domains, which are known to interact with the backbone (32). The two groups of AMPPNP-dependent contacts may arise from a single DbpA–RNA complex or from a heterogeneous population, where the helicase domains bind either 3′ or 5′ of helix 90. Finally, the difference between the AMPPNP- and ATP-dependent footprints suggests that the time-averaged conformational state of the ATP hydrolyzing system that is reported by the assay is different from the ADP- or AMPPNP-bound static state.
DbpA is targeted to hairpin 92 by its C-terminal domain and is capable of unwinding several different helices near hairpin 92 in small RNA substrates (18) or in larger 153mer-like RNAs (C.Diges and O.C.Uhlenbeck, unpublished results). Since the region of helicase domain binding identifies where the helicase acts, the presence of AMPPNP-dependent footprints in or near helices 89 and 93 suggests that they are the preferred sites of helicase activity in the 172mer. Furthermore, it raises the possibility that DbpA function is targeted to this region on its in vivo substrate, presumably a pre-50S ribosomal intermediate. As an alternative possibility, the helicase domains of DbpA may act at another site or sites that are spatially close to the A-loop. The secondary and tertiary structure of the PTC in the mature ribosome may represent either the substrate or product of DbpA activity.
RNase T1 footprinting of apo-DbpA on intact 23S rRNA (either native or transcribed) indicates that DbpA is still targeted to hairpin 92 in the context of this large RNA (data not shown). A preliminary search through the rest of 23S rRNA yielded no additional protection sites of apo-DbpA (K.Polach and O.C.Uhlenbeck, unpublished results). However, the AMPPNP-dependent footprint positions are inaccessible to RNase T1 cleavage in 23S rRNA, leaving open the question of helicase domain binding in a larger RNA.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Karl Kossen, Kevin Polach and Camille Diges for helpful discussions and support. This work was funded in part by a National Science Foundation graduate fellowship to F.V.K.
REFERENCES
- 1.de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- 2.Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- 3.Tanner N.K. and Linder,P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell, 8, 251–262. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 5.Pause A. and Sonenberg,N. (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J., 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers G.W. Jr, Richter,N.J. and Merrick,W.C. (1999) Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem., 274, 12236–12244. [DOI] [PubMed] [Google Scholar]
- 7.Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2000) The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature, 403, 447–451. [DOI] [PubMed] [Google Scholar]
- 8.Chen J.Y., Stands,L., Staley,J.P., Jackups,R.R.,Jr, Latus,L.J. and Chang,T.H. (2001) Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell, 7, 227–232. [DOI] [PubMed] [Google Scholar]
- 9.Kistler A.L. and Guthrie,C. (2001) Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev., 15, 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jankowsky E., Gross,C.H., Shuman,S. and Pyle,A.M. (2001) Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science, 291, 121–125. [DOI] [PubMed] [Google Scholar]
- 11.Caruthers J.M. and McKay,D.B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol., 12, 123–133. [DOI] [PubMed] [Google Scholar]
- 12.Silverman E., Edwalds-Gilbert,G. and Lin,R.J. (2003) DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene, 312, 1–16. [DOI] [PubMed] [Google Scholar]
- 13.Fuller-Pace F.V., Nicol,S.M., Reid,A.D. and Lane,D.P. (1993) DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J., 12, 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsu C.A. and Uhlenbeck,O.C. (1998) Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry, 37, 16989–16996. [DOI] [PubMed] [Google Scholar]
- 15.Tsu C.A., Kossen,K. and Uhlenbeck,O.C. (2001) The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA, 7, 702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pugh G.E., Nicol,S.M. and Fuller-Pace,F.V. (1999) Interaction of the Escherichia coli DEAD box protein DbpA with 23S ribosomal RNA. J. Mol. Biol., 292, 771–778. [DOI] [PubMed] [Google Scholar]
- 17.Polach K.J. and Uhlenbeck,O.C. (2002) Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry, 41, 3693–3702. [DOI] [PubMed] [Google Scholar]
- 18.Diges C.M. and Uhlenbeck,O.C. (2001) Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J., 20, 5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moll I., Grill,S., Grundling,A. and Blasi,U. (2002) Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol. Microbiol., 44, 1387–1396. [DOI] [PubMed] [Google Scholar]
- 20.Charollais J., Pflieger,D., Vinh,J., Dreyfus,M. and Iost,I. (2003) The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol., 48, 1253–1265. [DOI] [PubMed] [Google Scholar]
- 21.Kossen K. and Uhlenbeck,O.C. (1999) Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res., 27, 3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kossen K., Karginov,F.V. and Uhlenbeck,O.C. (2002) The carboxy-terminal domain of the DExD/H protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol., 324, 625–636. [DOI] [PubMed] [Google Scholar]
- 23.Cannone J.J., Subramanian,S., Schnare,M.N., Collett,J.R., D’Souza,L.M., Du,Y., Feng,B., Lin,N., Madabusi,L.V., Muller,K.M. et al. (2002) The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron and other RNAs. BMC Bioinformatics, 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 25.Harms J., Schluenzen,F., Zarivach,R., Bashan,A., Gat,S., Agmon,I., Bartels,H., Franceschi,F. and Yonath,A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell, 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 26.Henn A., Shi,S.P., Zarivach,R., Ben-Zeev,E. and Sagi,I. (2002) The RNA helicase DbpA exhibits a markedly different conformation in the ADP-bound state when compared with the ATP- or RNA-bound states. J. Biol. Chem., 277, 46559–46565. [DOI] [PubMed] [Google Scholar]
- 27.Lorsch J.R. and Herschlag,D. (1998) The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry, 37, 2194–2206. [DOI] [PubMed] [Google Scholar]
- 28.Chao K. and Lohman,T.M. (1990) DNA and nucleotide-induced conformational changes in the Escherichia coli Rep and Helicase II (UvrD) proteins. J. Biol. Chem., 265, 1067–1076. [PubMed] [Google Scholar]
- 29.Phillips R.J., Hickleton,D.C., Boehmer,P.E. and Emmerson,P.T. (1997) The RecB protein of Escherichia coli translocates along single-stranded DNA in the 3′ to 5′ direction: a proposed ratchet mechanism. Mol. Gen. Genet., 254, 319–329. [DOI] [PubMed] [Google Scholar]
- 30.Irie M. and Ohgi,K. (2001) Ribonuclease T2. Methods Enzymol., 341, 42–55. [DOI] [PubMed] [Google Scholar]
- 31.Sassanfar M. and Szostak,J.W. (1993) An RNA motif that binds ATP. Nature, 364, 550–553. [DOI] [PubMed] [Google Scholar]
- 32.Kim J.L., Morgenstern,K.A., Griffith,J.P., Dwyer,M.D., Thomson,J.A., Murcko,M.A., Lin,C. and Caron,P.R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure, 6, 89–100. [DOI] [PubMed] [Google Scholar]