Abstract
PcrA is an essential helicase in Gram-positive bacteria, but its precise role in cellular DNA metabolism is currently unknown. The Staphylococcus aureus PcrA helicase has both 5′→3′ and 3′→5′ helicase activities. In this work, we have studied the binding of S.aureus PcrA to a variety of DNA substrates that represent intermediates in DNA replication, repair, recombination and transcription. PcrA bound poorly or not at all to single-stranded DNA, double-stranded DNA with blunt ends, partially double-stranded DNA containing fork and bubble structures, and duplex DNA substrates containing either 5′ or 3′ single-stranded oligo dT tails. Interestingly, PcrA bound with high affinity to partially duplex DNA containing hairpin structures adjacent to a 6 nt long 5′ single-stranded region and one unpaired nucleotide (flap) at the 3′ end. However, PcrA did not detectably bind to partial duplexes with folded regions adjacent to a 6 nt long 3′ single-stranded tail (with or without a 1 nt flap at the 5′ end). PcrA showed moderate helicase activity with partially double-stranded DNAs containing 3′ or 5′ single-stranded oligo dT tails, the 3′→5′ helicase activity being more efficient than its 5′→3′ helicase activity. Interestingly, PcrA showed maximal helicase activity with substrates containing folded structures and 5′ single-stranded tails, suggesting that its 5′→3′ helicase activity is greatly stimulated in the presence of specific structures. However, the 3′→5′ helicase activity of PcrA did not appear to be affected by the presence of folded substrates containing 3′ single-stranded tails. Our data indicate that PcrA may recognize DNA substrates with specific structures in vivo and its 5′→3′ and 3′→5′ helicase activities may be involved in distinct cellular processes.
INTRODUCTION
DNA helicases play essential roles in cellular DNA metabolism, including DNA replication, repair, recombination and transcription [for reviews, see (1–4)]. All organisms contain multiple helicases, and it has been postulated that although some helicase functions may be redundant, other helicases may be required for specific cellular processes. The pcrA gene was originally identified as being required for the replication of rolling circle-replicating (RCR) plasmids in Staphylococcus aureus (5). Subsequently, pcrA was found to be an essential gene in S.aureus and Bacillus subtilis (6–8). Moreover, the pcrA gene has been identified in all Gram-positive bacteria whose genomes have so far been sequenced. PcrA belongs to the superfamily I of DNA helicases, which also includes the Rep and UvrD (helicase II) helicases of Escherichia coli, and contain seven conserved helicase motifs (1,9). In pairwise comparisons, the PcrA helicases of different Gram-positive bacteria share ∼60% identity (10). The PcrA helicases also share ∼40% identity with Rep and UvrD helicases of E.coli. Although disruption of the pcrA gene in Gram-positive bacteria is lethal, the loss of either rep or uvrD does not result in lethality (11). Genetic and biochemical studies have shown that PcrA is involved in plasmid RC replication and in UV repair of cellular DNA (5–8,12–15). PcrA can also complement rep uvrD double mutants of E.coli (7). Studies in B.subtilis with conditional pcrA mutants have shown that PcrA does not play a major role in chromosomal replication (7). Mutations in the recF, recO and recR genes have been isolated that allow viability of pcrA mutants, suggesting that PcrA may be involved in the resolution of toxic recombination intermediates and/or stalled replication forks (8). However, the precise role of PcrA that makes it essential for cell viability is yet to be identified.
PcrA helicase was first isolated from Bacillus stearothermophilus and its biochemical properties studied (16–22). This PcrA was shown to be a 3′→5′ helicase using substrates that lack the potential to form folded structures (18,22). The B.stearothermophilus PcrA helicase has been crystallized and its three-dimensional structure determined (16,17). Mutational studies of B.stearothermophilus PcrA have also identified regions of the protein that are important for its ATPase, helicase and translocation activities (18). The S.aureus and Bacillus anthracis PcrA proteins have also been purified and characterized. Interestingly, both these helicases have robust 3′→5′ as well as 5′→3′ helicase activities (15,23). In vitro studies have shown that these two helicases can promote unwinding of plasmid pT181 DNA that has been nicked at the origin by the plasmid initiator protein (15,24). Furthermore, these helicases were shown to promote the in vitro replication of plasmid pT181 DNA that replicates by a RC mechanism (15,24).
We have investigated further the biochemical activities of S.aureus PcrA to obtain clues about its possible roles in cellular DNA metabolism. In DNA binding studies, PcrA was found to interact poorly or not at all with single-stranded (ss) oligonucleotides, with double-stranded (ds) blunt-ended oligonucleotides (with or without fork structures) and with partial duplex substrates containing 5′ or 3′ oligo dT ss tails. However, PcrA bound with high affinity to substrates with folded structures that also contained a 5′ ss tail and a 1 nt flap at the 3′ end (resembling double flap structures generated during the synthesis and processing of Okazaki fragments during lagging strand DNA synthesis), but failed to interact with similar substrates that contained a 3′ ss region. In general, the helicase activity of PcrA was correlated with its DNA binding activity, i.e. it showed a much stronger helicase activity with substrates to which it bound with high affinity. The 3′→5′ helicase activity of PcrA was more efficient than its 5′→3′ activity with partial duplex substrates containing oligo dT tails. Interestingly, the 5′→3′ helicase activity of PcrA was much more efficient than its 3′→5′ activity with substrates containing hairpin structures and flaps. Our results show that PcrA has DNA binding specificity for structured substrates, and it is likely that its bidirectional helicase activity might be involved in distinct biochemical pathways involving different DNA substrates.
MATERIALS AND METHODS
Purification of the His-PcrA protein
The pcrA gene was amplified by PCR using the chromosomal DNA from S.aureus strain S6 as the template. The sequences of the primers, which contained BamHI linkers, were 5′-CCGGATCCAATGCGTTATTAAATCATATGAATACAGAGCAAAGTG-3′ (forward) and 5′-CCGGATCCCGATAAATCAGCCATCCCTTAATCCTCC-3′ (reverse). His-PcrA protein was overexpressed by induction with 1 mM IPTG at 37°C for 2 h and purified by using nickel affinity chromatography as described previously (15). The concentration of the His-PcrA preparation reached ∼0.5 mg/ml in the peak fractions, and the purity was tested by SDS–PAGE and staining with Coomassie brilliant blue.
DNA binding assays
Binding of the PcrA helicase to various DNA substrates was studied by electrophoretic mobility-shift assays (EMSA). Various DNA substrates were prepared by labeling one strand of the oligonucleotides with 32P at the 5′ end using T4 polynucleotide kinase (25), and annealing the cold complementary strand at a 3-fold molar excess. The reactions were performed in a buffer consisting of 20 mM Tris–HCl (pH 8.0), 100 mM KCl, 1 mM EDTA, 100 ng poly (dI–dC), 5 mM DTT, 10% ethylene glycol, indicated amounts of PcrA and ∼3 nM DNA substrate (15). The reactions were incubated at RT for 15 min and the DNA–protein complexes resolved by electrophoresis on 6% native polyacrylamide gels (25). The gels were dried and subjected to autoradiography.
DNA helicase assays
DNA substrates were prepared by labeling one strand of the oligonucleotides with 32P as described above. Helicase reactions were performed at 37°C for 20 min in a buffer containing 20 mM Tris–HCl (pH 7.5), 100 mM KCl, 3 mM MgCl2, 3 mM ATP, 10 mM DTT, 10% glycerol, ∼4 nM labeled DNA substrate and the indicated amounts of the PcrA helicase. The reactions were stopped by the addition of SDS-dye and the products were analyzed by 10% native PAGE (15). Gels were subsequently dried and exposed to Kodak X-ray films.
RESULTS
Purification of the S.aureus PcrA helicase
We have described previously the cloning and overexpression of the pcrA gene of S.aureus (15). The original cloning was based on PCR amplification of the pcrA gene using primers based on the published sequence of this gene (5). However, after the sequence of the whole S.aureus genome was published (26), we noticed that the size of the pcrA gene was slightly larger than that reported originally (26). The revised sequence of pcrA indicated that it encodes 730 amino acids (26), whereas the original PcrA that we purified and characterized consisted of 693 amino acids (15). We, therefore, re-cloned the pcrA gene using the revised sequence information. Automated DNA sequencing of the cloned pcrA gene showed that its sequence was identical to that reported by Kuroda et al. (26). The full-length PcrA helicase was then overexpressed and purified as a His6 fusion by using nickel affinity chromatography as reported previously (15). The protein was ∼90% pure. The impurities essentially consisted of breakdown products of His-PcrA. For simplicity, the His-PcrA protein is simply referred to as PcrA.
DNA binding activity of the PcrA helicase
We utilized a variety of synthetic oligonucleotides (Table 1) to study the DNA binding activity of the PcrA helicase. The substrates consisted of ssDNA, blunt-ended duplex DNA, 5′ and 3′ overhang duplexes, and forked and bubble duplexes (Figure 1). We also utilized partial duplexes in which the ss region can fold into secondary structures, with or without additional 5′ or 3′ ss tails and double flap-like structures (Figure 1). The substrates used resemble those expected to be present in various DNA metabolic pathways, such as replication, repair, recombination and transcription. Incubation of PcrA with an ss oligonucleotide or blunt-ended DNA (substrates A and E, respectively) did not generate significant levels of DNA–protein complexes (Figure 2). Similarly, PcrA showed very little or no binding to substrates D and F representing duplex DNA with forked and bubble structures, respectively (Figure 2). These results indicate that PcrA does not interact stably with ssDNAs or dsDNAs lacking any ss overhangs. Incubation of PcrA with dsDNAs containing 3′ or 5′ oligo dT overhangs (substrates B and C, respectively) also failed to generate detectable levels of DNA–protein complexes (Figure 2), demonstrating that the presence of a ss–ds junction is not sufficient for binding by PcrA. We also tested the DNA binding activity of the PcrA helicase utilizing partial duplex substrates in which the ss regions can form secondary structures (Figure 1). These probes represent the plasmid pT181 origin of replication, and their 5′ and 3′ ss regions correspond to the IRII element that can assume a hairpin structure (27–29). The presence of the hairpin structure in these probes was confirmed by their sensitivity to digestion by HpaII, which is expected to cleave the oligonucleotides only if the stem region is present as a duplex (data not shown). No significant binding of PcrA to duplex substrates L and P containing folded regions at their 5′ or 3′ ends was observed, respectively (Figure 2). However, PcrA bound to limited but detectable levels to a similar substrate J that contained a folded region at its 5′ region along with a 6 nt overhang at the 5′ end (Figure 2). Interestingly, PcrA bound with high affinity to substrate G containing a hairpin at its 5′ region, a 6 nt ss overhang at the 5′ end and a 1 nt flap (unpaired nucleotide) at the 3′ end of the complementary strand (Figure 2). This substrate (which resembles a double flap structure) is identical to J, except for the presence of a 1 nt flap at the 3′ end of the top strand (Figure 1). However, the presence of a single nucleotide flap at the 3′ end in the absence of a 5′ ss region was not sufficient for PcrA binding (substrate K, Figure 2). PcrA also bound efficiently to substrate H which has a structure similar to that of G but in which the stem sequence has been inverted (Figure 2 and Table 1), demonstrating that the structure rather than the sequence provides the primary recognition site for PcrA. The loop was not important for PcrA binding since its deletion did not appreciably affect binding to PcrA (substrate I, Figure 2). Note that substrate I generates two bands, the slower being a dimer due to pairing of the stem regions of two molecules (Table 1). This was confirmed by the generation of an NcoI site upon dimerization of the probe (data not shown). Interestingly, substrates with similar secondary structures but containing a 6 nt ss region at their 3′ ends were not bound by the PcrA helicase regardless of whether or not they also contained 1 nt flap at their 5′ ends (substrates M, N, O and P: Figure 2). The above experiments showed that PcrA has structure-specific DNA binding activity, and the presence of an unpaired 5′ ss region and a flap-like structure promotes its binding to the DNA.
Table 1. Oligonucleotides used in this study.
| Oligo | Sequence |
|---|---|
| 1 | d(TTTTTTTTTTTTTTTTTTTTTGGCGACGGCAGCGAGGC) |
| 2 | d(GCCTCGCTGCCGTCGCCA) |
| 3 | d(TGGCGACGGCAGCGAGGCTTTTTTTTTTTTTTTTTTTT) |
| 4 | d(ACGTATCGTAGATATACTATGTATTGGCAGAGTGGACAATT) |
| 5 | d(AATTGTCCACTCTGCCAATACATAGTATATCTACGATACGT) |
| 6 | d(GCGATAGTCTCTAGACAGGTACAGGATCGTTACTAATCGTCTATGACGTG) |
| 7 | d(GCCTCGCTGCCGTCGCCTTTTTTTTTTTTTTTTTTTTT) |
| 8 | d(CACGTCATAGACGATTACATTGCTAGGACATGCTGTCTAGAGACTATCGC) |
| 9 | d(GATCCAACCGGCTACTCTAATAGCCGGTTGGACGCACATACTGTGTGCATATG)a |
| 10 | d(CATATGCACACAGTATGTGCGTCCAG) |
| 11 | d(GATCCAATCGGCCATTAGAGTGGCCGATTGGACGCACATACTGTGTGCATATG) |
| 12 | d(GATCCAATCGGCCATGGCCGATTGGACGCACATACTGTGTGCATATG) |
| 13 | d(CATATGCACACAGTATGTGCGTCCA) |
| 14 | d(ACCGGCTACTCTAATAGCCGGTTGGACGCACATACTGTGTGCATATG) |
| 15 | d(ATATGCACACAGTATGTGCGTCC) |
| 16 | d(AACCGGCTACTCTAATAGCCGGTTGGACGCACATACTGTGTGCATAT) |
| 17 | d(TATGCACACAGTATGTGCGTCCAACCGGCTATTAGAGTAGCCGGTTTGATCC) |
| 18 | d(GTGGACGCACATACTGTGTGCATATGGATC) |
| 19 | d(CATATGCACACAGTATGTGCGTCCAG) |
| 20 | d(GATCCATATGCACACAGTATGTGCGTCCAACCGGCTATTAGAGTAGCCGGT) |
aThe underlined sequences can basepair and form folded structures. The HpaII cleavage sequence is present within this region and cleavage with this enzyme was used to confirm the presence of basepaired regions in these oligonucleotides.
Figure 1.
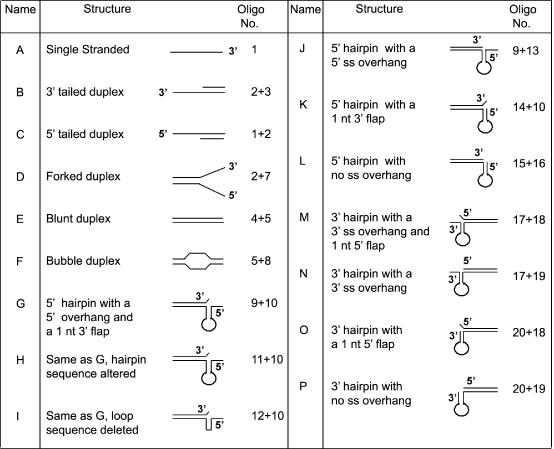
Structures of the various substrates used in this study.
Figure 2.
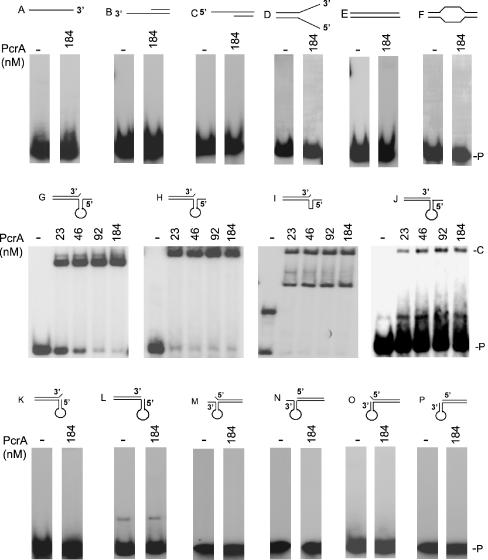
Structure-specific DNA binding activity of the PcrA helicase. EMSAs were carried out using 5′ 32P-labeled substrates (Table 1 and Figure 1) and the indicated amounts of PcrA. The DNA–protein complexes were resolved on polyacrylamide gels. P, probe; C, DNA–protein complex.
DNA helicase activity of PcrA
We also utilized the substrates used in DNA binding assays to test the specificity of the helicase activity of PcrA. PcrA was active in unwinding duplex substrates B and C containing either a 3′ or a 5′ oligo dT overhang (Figure 3), demonstrating that it has bipolar 3′→5′ and 5′→3′ helicase activities. Furthermore, its 3′→5′ helicase activity was more efficient than its 5′→3′ helicase activity. Also, PcrA unwound the 3′ and 5′ overhang substrates much more efficiently than the forked duplex (substrate D, Figure 3). PcrA did not significantly unwind blunt-ended substrates either lacking or containing ss bubble regions (substrates E and F, respectively; Figure 3). To rule out the possibility that a contaminating E.coli protein may be responsible for the unwinding activities of the PcrA preparation, we purified a His-fusion of the K33AQ250R double mutant of the S.aureus PcrA helicase. This mutant was found to be inactive in its ATPase and DNA helicase assays (data not shown).
Figure 3.
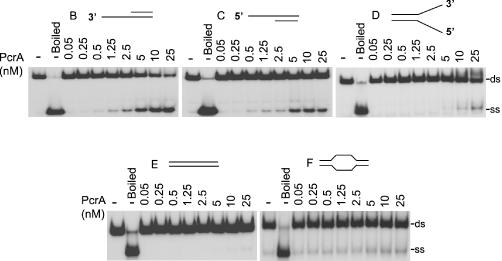
PcrA has bipolar 3′→5′ and 5′→3′ helicase activities. 32P-labeled substrates were incubated with the indicated amounts of PcrA, and the products resolved by native PAGE. The probes used are listed in Table 1 and their structures are depicted in Figure 1. ds, substrates containing partial or fully duplex DNA; ss, single-stranded DNA.
We also tested partial duplex substrates in which the 3′ and 5′ ss regions can fold into secondary structures. PcrA unwound substrate L containing a 5′ hairpin region without any ss overhang to a limited extent (Figure 4). PcrA showed a slightly higher helicase activity with substrate K, which is similar to L except that it also contains a 1 nt overhang at its 3′ end. This suggests that the 1 nt overhang at the 3′ end may facilitate the helicase activity of PcrA. Substrates G, H, I and J which all contain a 6 nt long 5′ ss overhang adjacent to the folded region were unwound efficiently by PcrA (Figure 4). The sequence of the stem, or the presence of the loop was not critical because PcrA also efficiently unwound substrates H and I (Figure 4). The above results showed that PcrA is highly active as a 5′→3′ helicase with substrates containing a 5′ region that can fold, and the presence of a 5′ ss tail significantly stimulates PcrA-driven unwinding. We also tested the helicase activity of PcrA utilizing duplex substrates containing a 3′ ss region that can fold into a hairpin structure. Substrate P which lacks a 3′ overhang, and substrate O that contains a 1 nt 5′ overhang, were unwound poorly by PcrA (Figure 5). The presence of a 6 nt overhang at the 3′ end in substrates M and N (with or without an unpaired 1 nt at the 5′ end, respectively), resulted in increased unwinding by PcrA (Figure 5). We conclude that, in general, the 5′→3′ helicase activity of PcrA is much more efficient than its 3′→5′ activity in the presence of folded substrates.
Figure 4.
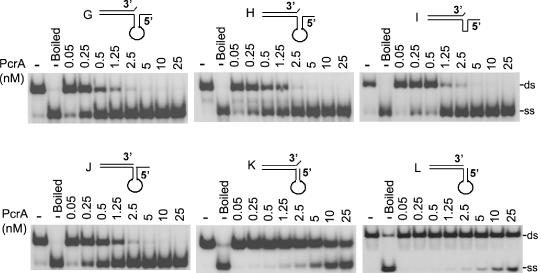
PcrA efficiently unwinds structured substrates with 5′ ss regions. Various 32P-labeled substrates containing a hairpin structure in their 5′ region were incubated with PcrA and subjected to EMSA.
Figure 5.
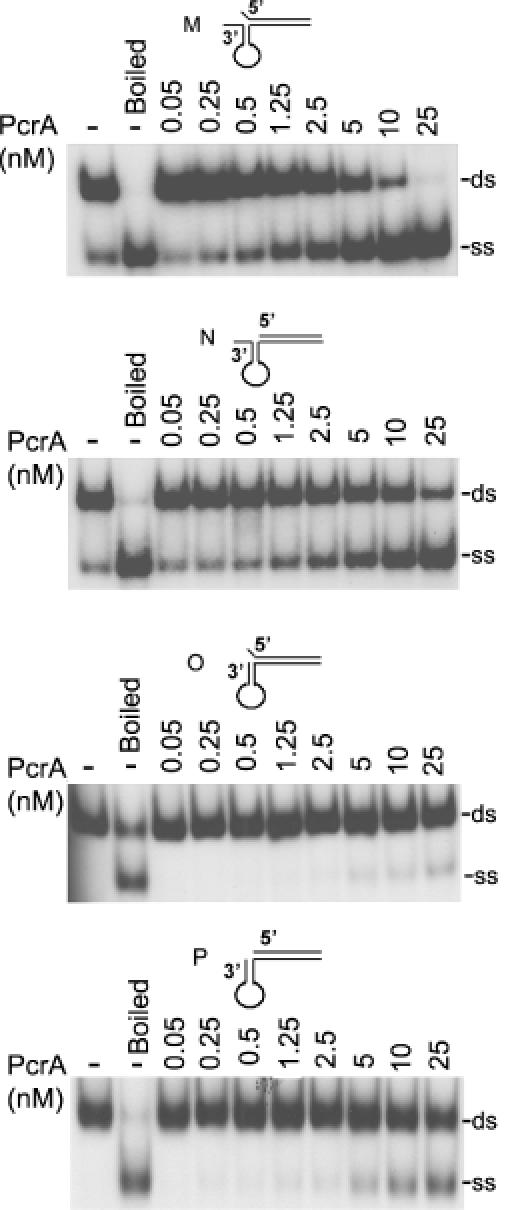
Unwinding of DNA substrates containing hairpin structures at their 3′ regions by PcrA. 32P-labeled substrates containing a hairpin structure in their 3′ region were incubated with PcrA and subjected to EMSA.
DISCUSSION
PcrA is an essential helicase found in Gram-positive bacteria that belongs to the SFI family of nucleic acid helicases (1,2). The PcrA helicases of Gram-positive bacteria share ∼40% identity with the UvrD and Rep helicases of E.coli. The UvrD helicase of E.coli is involved in nucleotide excision and mismatch repair pathways (1,30–32), whereas the Rep helicase is required for the RC replication of ssDNA phages (33). Also, rep mutants exhibit increased chromosomal breakage and decreased rate of chromosomal replication (34,35). The UvrD and Rep helicases of E.coli are not individually essential for cell survival, although a uvrD rep double mutant is not viable (11). The ability of PcrA to rescue uvrD rep double mutants of E.coli (7), suggests that PcrA can also perform an essential function. Studies in B.subtilis have suggested that PcrA does not appear to play a major role in chromosomal replication, but may be involved in the resolution of toxic recombination intermediates or stalled replication forks (7,8,36). It is also possible that PcrA may facilitate replication fork movement through secondary structures, such as hairpins. PcrA may also play a role in the maturation of Okazaki fragments during lagging strand synthesis by unwinding regions of secondary structures present in the displaced, adjacent Okazaki fragment.
To understand the possible roles of PcrA that make it an essential helicase, we investigated the DNA binding activity of PcrA utilizing a variety of substrates that are expected to be intermediates in various DNA transaction pathways, such as replication, repair, recombination and transcription. PcrA did not detectably bind to either ssDNA or to blunt-ended dsDNA with or without forked and bubble regions (Figure 2). Similarly, very little binding of PcrA to duplex substrates containing either 3′ or 5′ oligo dT tails was observed (Figure 2). We tested whether PcrA can specifically interact with DNA substrates containing hairpins in the presence or absence of 3′ or 5′ overhangs. We also utilized similar substrates that also contained a 1 nt 3′ tail (flap) which is generated during the processing of Okazaki fragments (37–39). Very little or no binding of PcrA was observed with substrates containing a hairpin structure at their 5′ end in the presence or absence of a 1 nt 3′ flap (Figure 3). However, the presence of a 6 nt long 5′ ss overhang resulted in robust binding of PcrA to such substrates (Figure 2). PcrA bound with the highest affinity to partial duplex substrates containing hairpin structures, a 5′ ss overhang and a 1 nt 3′ flap (Figure 2). The binding of PcrA to such substrates was not affected by sequence of the stem region or the presence of a loop, since substrates H and I were as efficient as substrate G for binding PcrA (Figure 2). Interestingly, PcrA failed to interact with similar substrates that contained hairpins and 6 nt long ss overhangs at their 3′ ends whether or not they also contained a 1 nt 5′ flap (Figure 2). The above results suggest that PcrA has structure-specific DNA binding activity, and the presence of a 5′ overhang is essential for its stable interaction with substrates containing folded regions. However, the presence of a 5′ overhang by itself is not sufficient for stable PcrA binding in the absence of a folded region in the DNA since PcrA did not stably bind to duplex substrates containing oligo dT tails.
Our studies showed that PcrA can unwind both 3′ and 5′ oligo dT tailed substrates, suggesting that it has bipolar 3′→5′ and 5′→3′ helicase activities (Figure 3). Furthermore, the 3′→5′ helicase activity of PcrA was stronger than its 5′→3′ helicase activity with such substrates. Interestingly, a forked duplex (D) containing a junction between duplex DNA and two single strands was a poorer substrate for the helicase activity of PcrA as compared with 3′ or 5′ tailed substrates that contain only one ss overhang (Figure 3). These results suggest that DNA unwinding may require direct binding of PcrA to a DNA branch with a free 5′ or 3′ end, and the presence of a 3′ or 5′ tail by itself (as is the case with the forked structure) is not sufficient for efficient unwinding. Since the structure of substrate D resembles a replication fork, our observations are consistent with the postulate that PcrA does not play a major role in chromosomal replication. Also, very little unwinding of blunt-ended substrates (with or without a bubble region) was observed with PcrA (Figure 3), suggesting that the presence of a free 5′ or 3′ end is required for its helicase activity. In general, PcrA showed maximal helicase activity with substrates to which it bound most efficiently in EMSA studies (Figures 2 and 4). Surprisingly, the 5′→3′ helicase activity of PcrA with substrates containing folded regions and single nucleolide flap such as G, H, I and J containing 5′ overhangs was much stronger than its helicase activity with the oligo dT-tailed substrate (Figures 3 and 4). Similarly, the 5′→3′ helicase activity of PcrA was much stronger than its 3′→5′ activity with substrates containing folded and flap regions (Figures 4 and 5). Quantification of the helicase activity of PcrA by counting the radioactivity in the gels shown in Figures 3–5 using the Amersham Typhoon 9400 Imager showed that 0.25-0.5 nM PcrA was sufficient for ∼50% unwinding of substrates G, H, I and J containing folded regions and ss overhangs at their 5′ ends. On the other hand, 10–50-fold higher levels of PcrA were required to obtain ∼50% unwinding of substrate B (3′ oligo dT-tail), and substrates M and N containing folded regions and ss overhangs at their 3′ ends. Other duplex substrates (C, D, E and F) and substrates with folded regions at their 3′ ends (O and P) failed to reach 50% unwinding even at the highest concentrations of PcrA used (25 nM). These results support the notion that recognition of specific DNA structures by PcrA may determine the polarity as well as the efficiency of its helicase activity. However, while PcrA interacted weekly with substrate J, it efficiently unwound this substrate (Figures 2 and 4). Similarly, while several substrates such as those containing oligo dT-tails or those with a hairpin structure at their 3′ regions along with single or double flaps (substrates M, N, O and P) were not detectably bound by PcrA (Figure 2), they were unwound to low to moderate levels by PcrA (Figures 3 and 5). These results suggest that transient/weak interaction between PcrA and DNA may be sufficient for DNA unwinding.
DNA helicases generally show a marked preference for substrates containing either a 3′ or a 5′ ss tail, and these activities are presumably related to their specific cellular functions (1–4,40). For example, the UvrD, Rep, PriA, RecB, WRN, BLM and Sgs1 proteins are 3′→5′ helicases whereas RecD and Dna2 proteins are 5′→3′ helicases (1,3,41–48). The PcrA helicase of B.stearothermophilus has been shown to have 3′→5′ helicase activity (18,22), although its 5′→3′ activity has not been evaluated using structured substrates. The S.aureus and B.anthracis PcrA helicases, on the other hand, have both 3′→5′ and 5′→3′ helicase activities [this study, and (15,23)]. Other helicases such as RecQ of E.coli are also known to have both 3′→5′ and 5′→3′ helicase activities (43,45). Also, helicases such as PriA, WRN, BLM and Sgs1 have been shown to contain structure-specific DNA binding and unwinding activities (41,42,44,46,49). Based on the DNA binding and bipolar helicase activities of PcrA reported here as well as previous studies, several possible roles of PcrA may be envisioned: (i) PcrA is required for the RC replication of pT181 and other RCR plasmids (12–15). It interacts with the RepC initiator protein of pT181 and unwinds DNA from the RepC-generated nick at the pT181 origin (15,24,50). The RepC nick site is located in the loop of a hairpin structure (27–29,51), and nicking of the DNA is expected to generate a 5′ ss region. It is likely that the 5′→3′ helicase activity of PcrA unwinds the DNA and promotes extension synthesis of the leading strand of the DNA. This is consistent with the role of PcrA in unwinding the DNA to facilitate progression of the replication fork during plasmid RC replication. (ii) PcrA has been suggested to play a role in the repair of UV lesions in the DNA based on the observations that it can restore UV resistance to a uvrD mutant of E.coli (7). Since UvrD is a 3′→5′ helicase (1), it is likely that the 3′→5′ helicase activity of PcrA is involved in UV repair. (iii) PcrA may be involved in facilitating replisome progression through regions of chromosomal DNA containing hairpin or other structures. This activity of PcrA, unlike the above two, may explain the requirement of PcrA for cell viability. A similar role for the RecQ family of helicases has also been proposed (43). (iv) Another possible role of PcrA may involve maturation of Okazaki fragments during lagging strand synthesis. During this process, DNA Polymerase III synthesizing a new Okazaki fragment may displace the previously synthesized downstream Okazaki fragment, generating a flap structure. If the displaced DNA has complementary sequences, it may fold generating a hairpin-like structure. PcrA may specifically bind and unwind such DNA, resulting in subsequent processing/degradation of the primer RNA and displaced DNA by RNaseH and other nucleases. A similar role in the processing of the Okazaki fragments is performed by the Dna2 protein of S. cerevisiae which has a 5′→3′ helicase as well as nuclease activities (37–39,52). (v) Since pcrA knockouts are viable in the presence of mutations in the recF, recO or recR genes (8), it is likely that PcrA is involved in resolution of toxic recombination structures or stalled replication forks that may impede replication fork progression. The helicase activity of PcrA may promote reverse branch migration at four-way Holliday junctions that may subsequently result in replication restart (53–55). Such an activity of PcrA may also promote replication re-initiation at sites of DNA damage, such as chemical lesions and nicks (53–55). However, other helicases such as PriA, RecBCD and RecQ may also be involved in these functions. Future investigations should reveal the mechanism of action of PcrA in essential cellular processes.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grant GM31685 (to S.A.K.).
REFERENCES
- 1.Hall M.C. and Matson,S.W. (1999) Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol., 34, 867–877. [DOI] [PubMed] [Google Scholar]
- 2.Bird L.E., Subramanya,S. and Wigley,D.B. (1998) Helicases: a unifying structural theme? Curr. Opin. Struct. Biol., 8, 14–18. [DOI] [PubMed] [Google Scholar]
- 3.Marians K.J. (2000) Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases. Structure, 8, R227–R235. [DOI] [PubMed] [Google Scholar]
- 4.Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 5.Iordanescu S. (1993) Identification and characterization of Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol. Gen. Genet., 241, 185–192. [DOI] [PubMed] [Google Scholar]
- 6.Iordanescu S. and Basheer,R. (1991) The Staphylococcus aureus mutation pcrA3 leads to the accumulation of pT181 replication initiation complexes. J. Mol. Biol., 221, 1183–1189. [DOI] [PubMed] [Google Scholar]
- 7.Petit M.-A., Dervyn,E., Rose,M., Entian,K.-D., McGovern,S., Ehrlich,S.D. and Bruand,C. (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol., 29, 261–273. [DOI] [PubMed] [Google Scholar]
- 8.Petit M.A. and Ehrlich,S.D. (2002) Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J., 21, 3137–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure–function relationship. Curr. Opin. Struct. Biol., 29, 419–429. [Google Scholar]
- 10.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 11.Taucher-Scholtz G., Abdel-Monem,M. and Hoffman-Berling,H. (1983) Functions of DNA helicases in Escherichia coli. In Cozzarelli,N.R. (ed.), Mechanism of DNA Replication and Recombination. Liss, New York, pp. 65–76. [Google Scholar]
- 12.del Solar G., Giraldo,R., Ruiz-Echevarria,M.J., Espinosa,M. and Diaz-Orejas,R. (1998) Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev., 62, 434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S.A. (1997) Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev., 61, 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan S.A. (2000) Plasmid rolling-circle replication: recent developments. Mol. Microbiol., 37, 477–484. [DOI] [PubMed] [Google Scholar]
- 15.Chang T.-L., Naqvi,A., Anand,S.P., Kramer,M.G., Munshi,R. and Khan,S.A. (2002) Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling-circle replication. J. Biol. Chem., 277, 45880–45886. [DOI] [PubMed] [Google Scholar]
- 16.Subramanya H.S., Bird,L.E., Branningan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box DNA helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- 17.Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,S. and Wigley,D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- 18.Soultanas P., Dillingham,M.S., Wiley,P., Webb,M.R. and Wigley,D.B. (2000) Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J., 19, 3799–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soultanas P. and Wigley,D.B. (2001) Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci., 26, 47–54. [DOI] [PubMed] [Google Scholar]
- 20.Soultanas P., Dillingham,M.S., Papadopoulos,F., Phillips,S.E.V., Thomas,C.D. and Wigley,D.B. (1999) Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res., 27, 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dillingham M.S., Wigley,D.B. and Webb,M.R. (2000) Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry, 39, 205–212. [DOI] [PubMed] [Google Scholar]
- 22.Bird L.E., Brannigan,J.A., Subramanya,H.S. and Wigley,D.B. (1998) Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res., 26, 2686–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naqvi A., Tinsley,E. and Khan,S.A. (2003) Purification and characterization of the PcrA helicase of Bacillus anthracis. J. Bacteriol., 185, 6633–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anand S.P., Mitra,P., Naqvi,A. and Khan,S.A. (2004) Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus. J. Bacteriol., 186, 2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Kuroda M., Ohta,T., Uchiyama,I., Baba,T., Yuzawa,H., Kubayashi,I., Cui,L., Oguchi,A., Aoki,K., Nagai,Y. et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet, 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 27.Jin R., Fernandez-Beros,M.-E. and Novick,R.P. (1997) Why is the initiation site of an AT-rich rolling circle plasmid at the tip of a GC-rich cruciform? EMBO J., 16, 4456–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin R. and Novick,R.P. (2001) Role of the double-strand origin cruciform in pT181 replication. Plasmid, 46, 95–105. [DOI] [PubMed] [Google Scholar]
- 29.Dempsey L.A., Birch,P. and Khan,S.A. (1992) Uncoupling of the DNA topoisomerase and replication activities of an initiator protein. Proc. Natl Acad. Sci. USA, 89, 3083–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 29, 43–81. [DOI] [PubMed] [Google Scholar]
- 31.Modrich P. (1994) Mismatch repair, genetic stability, and cancer. Science, 29, 1959–1960. [DOI] [PubMed] [Google Scholar]
- 32.Brosh R.M. Jr and Matson,S.W. (1997) A point mutation in Escherichia coli DNA helicase II renders the enzyme nonfunctional in two DNA repair pathways. Evidence for initiation of unwinding from a nick in vivo. J. Biol. Chem., 272, 572–579. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi S., Hours,C., Iwaya,M., Lane,H.E.D. and Denhardt,D.T. (1978) The Escherichia coli rep gene. In Denhardt,D.T., Dressler,D.H. and Ray,D.S. (eds), The Single-Stranded DNA phages. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 393–400. [Google Scholar]
- 34.Michel B., Ehrlich,S.D. and Uzest,M. (1997) DNA double-strand breaks caused by replication arrest. EMBO J., 29, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colasanti J. and Denhardt,D.T. (1987) Mechanism of replication of bacteriophage φX174. XXII. Site-specific mutagenesis of the A* gene reveals that A* protein is not essential for φX174 DNA replication. J. Mol. Biol., 29, 47–54. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto Y., Nakai,S., Moriya,S., Yoshikawa,H. and Ogasawara,N. (1995) The Bacillus subtilis dnaC gene encodes a protein homologous to the DnaB helicase of Escherichia coli. Microbiology, 29, 641–644. [DOI] [PubMed] [Google Scholar]
- 37.Kao H.I. and Bambara,R.A. (2003) The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol., 38, 433–452. [DOI] [PubMed] [Google Scholar]
- 38.Ayyagari R., Gomes,X.V., Gordenin,D.A. and Burgers,P.M. (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem., 278, 1618–1625. [DOI] [PubMed] [Google Scholar]
- 39.Bae S.-H. and Seo,Y.-S. (2000) Characterization of the enzymatic properties of the yeast Dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem., 275, 38022–38031. [DOI] [PubMed] [Google Scholar]
- 40.Delagoutte E. and von Hippel,P.H. (2003) Helicase mechanisms and coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Quart. Rev. Biophys., 36, 1–69. [DOI] [PubMed] [Google Scholar]
- 41.Mohaghegh P., Karow,J.K., Brosh,R.M.,Jr, Bohr,V.A. and Hickson,I.D. (2001) The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res., 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Kobbe C., Thoma,N.H., Czyzewski,B.K., Pavletich,N.P. and Bohr,V.A. (2003) Werner syndrome protein contains three structure-specific DNA binding domains. J. Biol. Chem., 278, 52997–53006. [DOI] [PubMed] [Google Scholar]
- 43.Hickson I.D. (2003) RecQ helicases: caretakers of the genome. Nature Rev. Cancer, 3, 169–178. [DOI] [PubMed] [Google Scholar]
- 44.Bennett R.J., Keck,J.L. and Wang,J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S.cerevisiae. J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- 45.Harmon F.G. and Kowalczykowski,S.C. (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev., 12, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marians K.J. (2000) PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci., 25, 185–189. [DOI] [PubMed] [Google Scholar]
- 47.Dillingham M.S., Spies,M. and Kowalczykowski,S.C. (2003) RecBCD enzyme is a bipolar DNA helicase. Nature, 423, 893–897. [DOI] [PubMed] [Google Scholar]
- 48.Taylor A.F. and Smith,G.R. (2003) RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature, 423, 889–893. [DOI] [PubMed] [Google Scholar]
- 49.Polard P., Marsin,S., McGovern,S., Velten,M., Wigley,D.B., Ehrlich,S.D. and Bruand,C. (2002) Restart of DNA replication in Gram-positive bacteria: functional characterisation of the Bacillus subtilis PriA initiator. Nucleic Acids Res., 30, 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iordanescu S. (1993) Plasmid pT181-linked suppressors of the Staphylococcus aureus pcrA3 chromosomal mutation. J. Bacteriol., 175, 3916–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koepsel R.R., Murray,R.W., Rosenblum,W.D. and Khan,S.A. (1985) The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc. Natl Acad. Sci. USA, 82, 6845–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weitao T., Budd,M., Hoopes,L.L. and Campbell,J.L. (2003) Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem., 278, 22513–22522. [DOI] [PubMed] [Google Scholar]
- 53.Morimatsu K. and Kowalczykowski,S.C. (2003) RecFOR proteins load RecA protein into gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell, 11, 1337–1347. [DOI] [PubMed] [Google Scholar]
- 54.Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 55.Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]


