Abstract
The hairpin ribozyme catalyses RNA cleavage by a mechanism utilizing its conformational flexibility during the docking of two independently folded internal loop domains A and B. Based on this mechanism, we designed hairpin ribozyme variants that can be induced or repressed by external effector oligonucleotides influencing the docking process. We incorporated a third domain C to assimilate alternate stable RNA motifs such as a pseudo-half-knot or an internal stem–loop structure. Small sequence changes in domain C allowed targeted switching of ribozyme activity: the same effector oligonucleotide can either serve as an inducer or repressor. The ribozymes were applied to trp leader mRNA, the RNA sequence tightly bound by l-tryptophan-activated trp-RNA-binding attenuation protein (TRAP). When domain C is complementary to this mRNA, ribozyme activity can be altered by annealing trp leader mRNA, then specifically reverted by its TRAP/tryptophan-mediated sequestration. This approach allows to precisely sense the activity status of a protein controlled by its metabolite molecule.
INTRODUCTION
Small catalytic RNAs are flexible molecules that can adopt alternative conformations during catalysis (1,2). Among them, the hairpin ribozyme, when annealed to its RNA substrate oligonucleotide, shows a unique secondary structure comprising two internal loops A and B, each flanked by two helices (3–5) (Figure 1). Crystallographic and biochemical studies (6,7) revealed that the mechanism of cleavage is characterized by an oscillating reaction pathway starting with a conformation where these two domains are coaxially stacked and aligned in a linear extended fashion. This conformer then folds into a docked structure with loops A and B interacting mainly via non-canonical base pairs, resulting in the ribozyme bending sharply at a site defined as the hinge region (8–11). Only this docked conformation permits site-specific cleavage of the substrate. Following cleavage, the docked complex unfolds back into the extended structure and the cleaved products dissociate.
Figure 1.
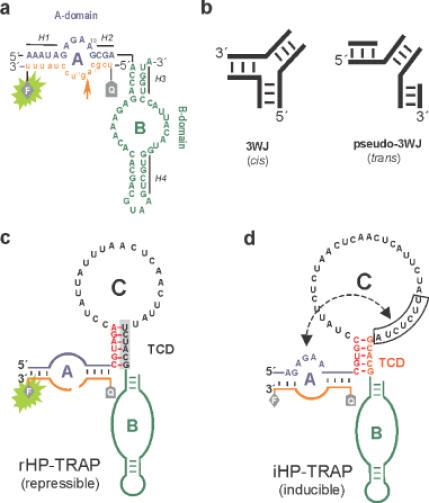
TRAP-responsive and mRNA-regulated hairpin ribozymes. (a) Hairpin ribozyme and fluorescent substrate (orange). Ribozyme and substrate sequences are in uppercase and lowercase letters, respectively. Substrate-binding arm A and the B domain are indicated in blue and green letters, respectively. The arrow indicates the substrate-cleavage site. Cleavage of substrate results in product dissociation and FRET decay (green light at 520 nm). F, fluorescent label (FAM); Q, fluorescence quencher (TAMRA). The flanking helical stems are abbreviated as H1 to H4. (b) Comparison of a classical three-way junction (3WJ) and the so-called pseudo-three-way junction (pseudo-3WJ). Insertion of a pseudo-3WJ motif at the hinge region allows the cleavage reaction to proceed in the trans-mode. (c) Design of the hairpin ribozyme rHP–TRAP, repressed by trp-mRNA. The colour scheme is the same as in (a). The sequence shown in black letters is complementary to trp-mRNA. Domain C is inserted at the hinge region between H2 and H3 [between positions A14 and A15 in (a)] and comprises the terminal connecting domain, TCD and the unpaired loop. The grey shadowed TCD sequence plus the unpaired loop of domain C hybridize to trp-mRNA, resulting in repression of rHP–TRAP catalysis. (d) Insertion of an additional seven nucleotides (boxed) into the trp-mRNA binding region of domain C produces an inducible trp-mRNA-responsive ribozyme (iHP–TRAP). The boxed region in domain C can hybridize to the ribozyme-part of domain A (the sequence shown in blue), preventing substrate (orange) binding to the ribozyme to create an inactive ribozyme construct.
The hinge region acts as a flexible linker mediating the docking process by stabilizing the active conformation and facilitating the tertiary contacts that define the active site within the interface between the internal loops (12,13). This fairly detailed understanding of the molecular mechanism involved in hairpin ribozyme catalysis highlights the hinge region as a promising target domain for controlling the ribozyme's structure and folding pathway by potentially binding effector oligonucleotides. Recently, several studies reported the activation of diverse inactive ribozyme variants by external effector oligonucleotides (14–20). These describe a number of allosteric hammerhead ribozyme variants responding to small-molecule effectors such as ATP (21), FMN (22–26), theophylline (24–26), doxycycline (27), pefloxacin (28), cAMP and cGMP (29) that were either designed or obtained by in vitro selection approaches.
Here, we describe a new method for engineering hairpin ribozyme variants that can be either induced or repressed by external oligonucleotide effectors. This activity is based on introducing a third regulatory domain, called domain C, at the H2–H3 junction. This new domain confers precise control over the formation of distinct structural motifs mediated by adaptive binding to defined effector mRNA sequences. As a prototype, we used trp-mRNA as the effector molecule, exploiting its involvement, together with the trp-mRNA-binding attenuation protein (TRAP) (30), in the regulatory pathway of tryptophan metabolism in Bacillus subtilis. TRAP regulates the expression of B.subtilis l-tryptophan biosynthetic genes in response to changes in the intracellular level of l-tryptophan (31). Comprising 11 identical protein subunits arranged in a single ring, TRAP binds a specific RNA sequence in the nascent trp-mRNA transcript leader region, but only when cooperatively activated by 11 bound l-tryptophan molecules (32–34). The RNA target contains between 9 and 11 G/UAG repeats which wrap around 11 KKR consensus motifs along the circumference of the l-tryptophan–TRAP complex (35,36). This means that after binding l-tryptophan, the TRAP protein interacts with the trp-mRNA transcript and prevents gene translation.
We set out to generate modified hairpin ribozymes that can be either activated or inhibited by trp-mRNA in a sequence-specific manner. We then used this system to monitor TRAP protein activation by its effector molecule l-tryptophan, and the subsequently induced trp-mRNA–TRAP complex formation. This approach allows the monitoring of binding events of metabolic regulatory networks by engineered ribozymes able to exert domain-specific conformational changes in response to their molecular environment.
MATERIALS AND METHODS
Synthesis of TRAP-responsive hairpin ribozymes
Synthetic DNA oligomers and fluorescence-labelled RNA substrates were purchased from MWG Biotech (Germany) or IBA (Göttingen, Germany), respectively. Ribozymes were synthesized by in vitro transcription of the following appropriate DNA templates: rHP–TRAP: 5′-AAA TAG AGA AGC GAC GTA GAC CTA TTA TTT AAC TCA ACT TAT TCT ACG ACC AGA GAA ACA CAC GAC GTA AGT CGT GGT ACA-3′; iHP–TRAP: 5′-AAA TAG AGA AGC GAC GUG CCU AUU CUC UAA CUC AAC UCA UUC UAT TCT CTA GCA CGA CCA GAG AAA CAC ACG ACG TAA GTC GTG GTA CA-3′; hairpin wild-type control HPwt: 5′-TCT AAT ACG ACT CAC TAT AGG GTC CTC TGA TGA GGC CGT TAG GCC GAA ACT CGT-3′; FRET-labelled RNA substrate, 5′-Tamra-UCGCAGUCCUAUUU-FAM-3′; mRNA DNA template: 5′-TAG AAT GAG TTG AGT TAG AGA ATA GGG TAG CAG AGA ATG AGT TTA GTT GAG CTG AGA-3′; 5′-primer: 5′-TCT AAT ACG ACT CAC TAT AGG GCG TAG AAT GAG TTG AGT TAG AG-3′; 3′-primer: 5′-TCT CAG CTC AAC TAA ACT CA-3; trp-mRNA sequence: 5′-GGG CGU AGA AUG AGU UGA GUU AGA GAA UAG GGU AGC AGA GAA UGA GUU UAG UUG AGC UGA GA-3′ (bold, T7 promoter sequence; italic, sequence not contained in the wild-type trp-mRNA). The ssDNA sequences were amplified by PCR using Dap polymerase (Eurogentec, Belgium). Transcription reactions (100 μl) containing 300 pmol dsDNA template were performed as described (37).
mRNA-dependent ribozyme assay
All reactions were carried out under multiple turnover conditions (MTO) with excess substrate (1 μM) over ribozyme (50 nM) in HP-buffer (50 mM Tris-HCl, pH 7.5, 20 mM MgCl2) at 37°C in 96-well plates (Corning Inc.). Ribozymes were pre-incubated with mRNA for 15 min and the cleavage reaction was initiated upon addition of the FRET labelled substrate. All reactions were further incubated for 20 min and the cleavage reaction was monitored in real-time by measuring FAM-dye fluorescence (Fluoroscan Ascent FL) as described previously(38).
TRAP-detecting ribozyme assay
Reactions were carried out under multiple turnover conditions (ribozyme/substrate = 1/20) in FBB reaction buffer [16 mM HEPES, 250 mM potassium glutamate (51), pH 8.0, 20 mM MgCl2]. TRAP activation was achieved in the presence of l-tryptophan for 11 min at room temperature. After addition of the mRNA the reaction mixture was further incubated for 16 min at 37°C to allow TRAP-mRNA complex formation. Subsequently, the hairpin ribozymes were added as indicated and the reaction mixture was incubated for an additional 6 min. Cleavage reaction was initiated upon addition of the FRET labelled substrate as described above.
RESULTS
Design of a repressible hairpin ribozyme (rHP–TRAP)
The regulatory mechanism we applied is based on a structural switch between bent and extended conformations caused by binding of a 62 nt fragment of trp-mRNA (Figure 2a). In this rational design strategy a new domain, called domain C, comprising 28 additional nucleotides complementary to the 3′-region of the effector trp-mRNA (Figure 1c and d, black letters), was inserted between H2 and H3 to act as an allosteric effector region specific for trp-mRNA. Domain C was linked via a pseudo-three-way junction at the H2 and H3 connection in the original hairpin ribozyme (Figure 1b). Six Watson–Crick base pairs were retained as a paired helix, called the ‘terminal connecting domain’ (TCD), to selectively favour the active bent conformation, as well as to avoid unspecific interference from domain C during catalysis by stabilizing stem–loop formation, and assure the required functional interaction between domains A and B. Repression was achieved by effector trp-mRNA annealing to domain C, thereby inducing a conformation that precludes stable docking of domains A and B.
Figure 2.
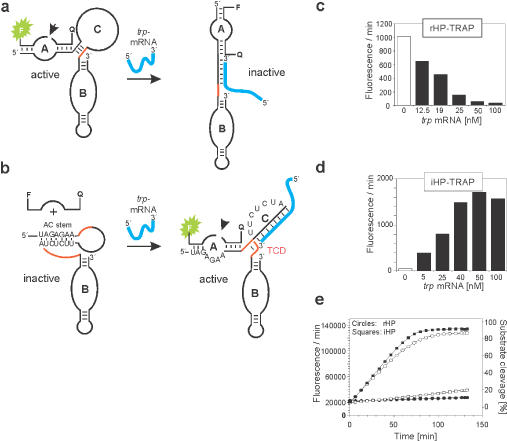
Trp-mRNA-dependent repression and induction of hairpin ribozymes. (a) Hybridization of trp-mRNA to rHP–TRAP inhibits catalysis by forcing A and B domains into an extended conformation. (b) The same trp-mRNA induces a conformational switch in iHP–TRAP which creates a pseudo-half-knot structure stabilized by the terminal helical sequence shown in red, thereby inducing cleavage activity. (c) Inactivation of rHP–TRAP and (d) induction of iHP–TRAP in the presence of increasing concentrations of trp-mRNA as indicated. Maximal rate enhancement occurrs at a ratio of 1 : 1 of trp-mRNA and iHP–TRAP, or rHP–TRAP, respectively. Reactions were carried out under MTO conditions with 50 nM iHP–TRAP/rHP–TRAP; 1 μM substrate. (e) Cleavage reactions were monitored using fluorescence signal measurement as previously described (38). Shown here is a time course of substrate-RNA cleavage catalysed by rHP–TRAP (circles) and iHP–TRAP (squares) in the presence of equimolar trp-mRNA (filled squares and circles) or the absence of any mRNA (open squares and circles). Reactions were carried out under multiple turnover conditions (MTO) to show maximum substrate turnover.
In the absence of trp-mRNA oligonucleotide, domain C forms a stem–loop structure that guides domains A and B into a side-by-side orientation by destabilizing the undocked conformation. In accordance with other studies, this hairpin ribozyme configuration is known to be kinetically more homogenous with respect to its cleavage rate (12). However, upon trp-mRNA oligonucleotide annealing, domain C should fully convert into a double-stranded RNA/RNA hybrid (Figure 2a), which is expected to abolish the docking event. Importantly, domain C is predisposed to act as a receptor for oligonucleotide effector molecules, a prerequisite for allosteric ribozyme regulation. Figure 1c shows the ribozyme construct rHP–TRAP investigated in our experiments.
To experimentally validate the engineered rHP–TRAP ribozyme, we monitored substrate cleavage reactions in real-time using fluorescence resonance energy transfer (FRET)-based detection in the presence and absence of trp-mRNA. The dye molecules FAM (donor) and TAMRA (acceptor) were attached to the 3′ and 5′ ends of the substrate strand as a FRET pair (39,40). An rHP–TRAP enzyme maximal turnover of 0.16 min−1 was observed under multiple turnover conditions in the absence of trp-mRNA (Figure 2e, and supplementary Table S). Repression of ribozyme activity in the presence of trp-mRNA was concentration dependent. At equal concentrations of rHP–TRAP and trp-mRNA effector (50 nM), ribozyme activity was reduced to nearly background levels (Figure 2c). Addition of our negative control tRNA from yeast had no effect on the catalytic activity of rHP–TRAP (data not shown). Annealing of the trp-mRNA effector oligonucleotide to one of the TCD paired strands (Figure 1c, shaded panel) was absolutely necessary to achieve inhibition: annealing to the unpaired loop region of domain C alone did not force the extended conformation and therefore did not inhibit rHP–TRAP activity (supplementary Figure 1S).
Design of an inducible hairpin ribozyme (iHP–TRAP)
These data verify the hinge region at the H2–H3 interface as an appropriate structural element suitable for allosteric ribozyme regulation. Taking this concept one step further, we set out to design an rHP–TRAP hairpin ribozyme counterpart with minimal basal cleavage activity but which can be induced by trp-mRNA. Such activity would increase the versatility of allosteric hairpin ribozyme regulators since it would allow any desired combination of on/off switching with the same effector oligonucleotide. We applied a strategy where the trp-mRNA oligonucleotide effector induces conformational changes that drive restoration of ribozyme activity from an originally inactive ribozyme construct. The regulation mechanism we used involved forming an alternate hairpin conformer that adopts a stable pseudo-half-knot structure upon activation. Pseudo-knots are naturally occurring RNA motifs where single-stranded regions of hairpin loops form intramolecular base pairs with each other, resulting in a characteristic stable conformation (41). However, if a complementary oligonucleotide hybridizes asymmetrically to the loop of a hairpin the topology of the resulting complex resembles one half of a pseudo-knot, or pseudo-half-knot (42).
This principle was used to generate a hairpin ribozyme that could be induced by trp-mRNA, called iHP–TRAP. We modified domain C in such a way that in the absence of trp-mRNA it's intramolecular interactions prevent substrate annealing and hence cleavage, i.e. abolishing ribozyme activity. Domain C was extended by seven consecutive nucleotides (5′-UUCUCUA-3′), complementary to the conserved sequences in the substrate binding arm (residues 4–10) that mediate the intramolecular interaction between domains A and B (Figure 1d). To promote preferential hybrid formation, this structural element, called the AC-stem, was introduced downstream of the trp-mRNA-binding region (Figure 2b). In addition, the terminal connecting domain TCD was changed into a five-nucleotide stem that is no longer complementary to the trp-mRNA sequence. Upon trp-mRNA hybridization, annealing of this stem should lead to a coaxially stacked, connective double-stranded helix resulting in a half-pseudo-knot that should contribute to cooperative stabilization of this structural RNA motif. In the absence of trp-mRNA, the TCD stem is predicted not to pair since it is thermodynamically less favoured than the alternative AC-stem at domain A. However, trp-mRNA association should prohibit AC-stem pairing and promote the formation of the pseudo-half-knot conformer, including the newly annealed TCD, to induce ribozyme activity. Figure 2b shows the predicted secondary structures of the free and the trp-mRNA-bound iHP–TRAP construct. This approach is somewhat related to the one investigated by Vaish et al. (43) who used a sequence in stem II complementary to the substrate binding arm of the hammerhead ribozyme to reduce ribozyme activity.
Experimental validation involved ribozyme assays in the absence and presence of increasing amounts of trp-mRNA (Figure 2d). The iHP–TRAP ribozyme shows virtually no basal cleavage activity in the absence of trp-mRNA. Induction of cleavage activity by trp-mRNA occurred in a strictly concentration-dependent manner with maximal enhancement of fluorescence intensity (1500-fold) obtained at equal concentrations of trp-mRNA and iHP–TRAP (50 nM). The effector sensitivity observed with the iHP–TRAP ribozyme is unique and distinguishes it from previously reported oligonucleotide-responsive ribozymes that require at least a 4-fold excess of their cognate effector molecule to achieve maximal activation (15). Even higher activation was achieved by increasing Mg2+ concentrations (Figure 3a). This further stimulation of ribozyme activity probably reflects the influence of domain C, which may ‘consume’ divalent cations to stabilize the mRNA hybrid formation. Interestingly, and consistent with previous studies, monovalent cations such as Na+ inhibited the Mg2+-promoted reaction, indicating that Na+ most likely competitively occupies Mg2+-binding sites in the ribozyme (data not shown) (44–49). Furthermore, even at low (4 mM) Mg2+-ion concentrations the cleavage activity of the iHP–TRAP ribozyme could be efficiently enhanced by addition of spermidine (2–8 mM) (Figure 3b). These data are in accordance with previous studies (45,47) and underline the relationship of the iHP–TRAP ribozyme to the wild-type hairpin ribozyme. The similar characteristics of our ribozyme constructs show that the cleavage mechanism itself is not altered by the introduction of domain C and that domain C acts as an allosteric effector site.
Figure 3.
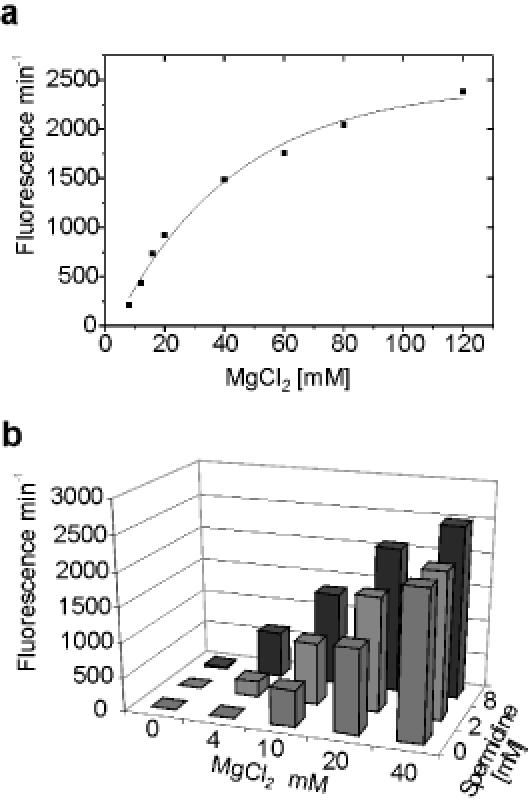
(a) Mg2+ dependence of the inducible construct iHP–TRAP in the presence of 50 nM trp-mRNA. (b) Spermidine dependence, with increasing Mg2+ concentrations, of the inducible construct iHP–TRAP in the presence of 50 nM trp-mRNA, in FBB and 1.0 mM l-tryptophan at 37°C. Notably, at a concentration of 8.0 mM spermidine iHP–TRAP shows activity in the presence of 50 nM mRNA despite the low concentration of Mg2+ (4.0 mM).
The remarkable sensitivity of the iHP–TRAP ribozyme permits very efficient and specific detection of trp-mRNA. We have shown elsewhere that this principle can be applied to a large variety of functional RNA-effectors, indicating that it provides a general strategy for rationally designing ribozymes capable of sensing tiny amounts of RNA in a highly sequence-specific fashion (50).
Sensing the TRAP activation state
Having demonstrated that our hairpin ribozyme variants can be efficiently regulated by annealing trp-mRNA, we further investigated whether the TRAP protein can reverse trp-mRNA-regulated hairpin ribozyme activity by sequestering trp-mRNA. Each TRAP protein subunit binds one l-tryptophan in its peripherically located cavity, which subsequently activates TRAP binding to its cognate trp-mRNA leader sequence. The trp-mRNA–TRAP-binding region comprises 53 nt that wrap around the TRAP molecule like a lasso, with a Kd of 0.12 nM (51). Figure 4a schematically depicts the TRAP-sensitive system where either ribozyme can be regulated in response to its molecular environment. All experiments were carried out in the filter binding buffer (FBB) to ensure efficient trp-mRNA/TRAP association as previously reported by Gollnick and co-workers (51) (also see supplementary Figure 2S for a filter binding assay with radioactively-labelled trp-mRNA). In fact, our effector-responsive ribozymes, like the wild-type hairpin ribozyme (52), possess comparative cleavage activities in different buffer conditions [Tris pH = 7.5 (see Figure 2c and d); Hepes pH = 8.0 (see Figure 4b and c)].
Figure 4.
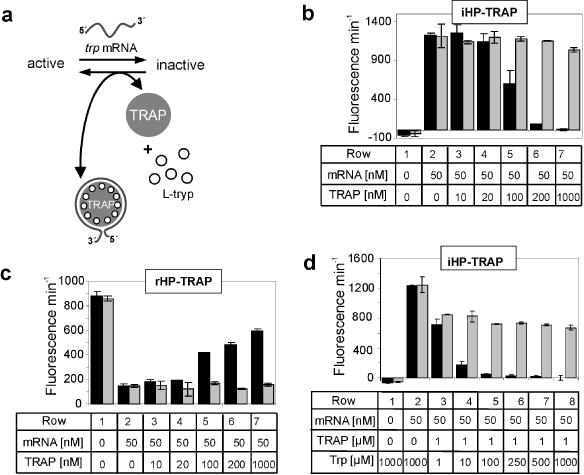
Sensing TRAP regulatory networks dependent on l-tryptophan. (a) In the presence of l-tryptophan (open circles) TRAP protein (grey shaded circle) is able to bind trp leader mRNA (grey). The presence or absence of l-tryptophan and TRAP causes the sequestration or release of trp-mRNA and thus shifts the equilibrium either in favour of inactive iHP–TRAP or active rHP–TRAP ribozyme conformations. (b) iHP–TRAP ribozyme activity regulated by increasing TRAP protein concentrations. Column 1, ribozyme alone; column 2, ribozyme/trp-mRNA (1 : 1); columns 3–7, ribozyme/trp-mRNA (1 : 1) with increasing concentrations of l-tryptophan-activated TRAP (10-1000 nM) (black bars). l-tryptophan concentration was 1.0 mM. The grey bars represent similar negative control experiments with 1.0 mM d-tryptophan. (c) Analogous to (b), but with the rHP–TRAP construct. Both ribozyme formats specifically respond to the interaction of l-tryptophan with the TRAP protein and the resulting formation of the TRAP/trp-mRNA complex. (d) l-tryptophan dependence of TRAP protein activation investigated using the iHP–TRAP ribozyme and a constant concentration of TRAP (1 μM). Assays were performed in the presence of l-tryptophan (black bars) or d-tryptophan (grey bars) at the indicated concentrations.
First, we tested whether iHP–TRAP can specifically respond to the presence of TRAP in its l-tryptophan-bound conformation. Figure 4b (black bars) demonstrates that l-tryptophan-activated TRAP sequesters the free trp-mRNA oligonucleotide, favouring the inactive state of the ribozyme. With rHP–TRAP we could demonstrate that ribozyme activity was restored to a level corresponding to >80% of its mRNA-unbound state when increasing concentrations of activated TRAP protein were used (Figure 4c, black bars). d-tryptophan did not mediate TRAP activation nor influence iHP–TRAP or rHP–TRAP activity (Figure 4b–d, grey bars). This precise and highly specific molecular recognition further characterizes this system as a network where reciprocal regulation guides ribozyme conformation, and hence catalysis. To examine the specificity of TRAP-dependent regulation we used BSA and p52, a p50-homologous member of the NF-κB transcription factor family as negative-control proteins. As expected, both proteins failed to interfere with trp-mRNA binding to iHP–TRAP (data not shown). Moreover, neither activated nor inactivated TRAP protein influenced wild-type, inducible or repressible hairpin ribozyme activities in the absence of trp-mRNA (data not shown). In this context, both hairpin ribozyme formats can apparently specifically monitor the trp-mRNA/TRAP interaction and actually sense the activity status of TRAP.
Measuring the level of l-tryptophan required for TRAP activation
In B.subtilis the amount of the metabolite l-tryptophan directs the expression level of genes involved in l-tryptophan metabolism (30,34,36). This stoichiometric sensor controlled via the activity of TRAP in vivo, switches on the expression of genes required for tryptophan synthesis if tryptophan levels in the cell fall below a certain threshold. However, no available data describe the relative intracellular l-tryptophan levels required to trigger TRAP activation and its ability to bind its target trp-mRNA. We therefore sought to measure the minimal l-tryptophan concentration required for TRAP activation in vitro. We employed the more sensitive iHP–TRAP to assess TRAP molecule activation with increasing concentrations of l-tryptophan ranging from 1 to 1000 μM (Figure 4d, black bars). In the absence of TRAP, 1.0 mM concentrations of l-tryptophan had no effect on trp-mRNA-induced iHP–TRAP ribozyme activity (Figure 4d, column 2). Interestingly, at 1.0 μM TRAP, a significant decrease in ribozyme activity was detected with concentrations as low as 10 μM l-tryptophan, reflecting a 10 fold excess of l-tryptophan over the TRAP undecamer (Figure 4, row 4). At 100 μM concentrations of l-tryptophan, TRAP activation seems to be saturated and iHP–TRAP activity reverts to background levels (Figure 4, column 5). Thus, under the in vitro conditions used here, even concentrations of l-tryptophan in the lower micromolar range are sufficient to prime TRAP to sense and tightly bind trp-mRNA. This activity absolutely depends on l-tryptophan, since d-tryptophan did not abrogate the ribozyme's activity even at concentrations as high as 1 mM (Figure 4d, grey bars).
DISCUSSION
The precise understanding of the hairpin ribozyme's cleavage mechanism and reaction pathway permits the targeted design of new hairpin ribozyme variants with particular characteristics. Here, we describe a new method to create a series of hairpin ribozymes that can be either induced or repressed by the same mRNA effector molecule. To achieve specific regulation we introduced a novel, allosteric effector RNA domain called domain C. By introducing sequence changes within this region, we could design either constitutively active or inactive ribozymes that can be inactivated or activated by trp-mRNA, respectively. Specifically, we demonstrate that binding of trp-mRNA effector molecules to the ribozymes induces conformational changes and refolding into active or inactive structures.
Our strategy is distinct from previous approaches that relied on annealing short oligonucleotides to reconstitute a disordered domain B since it involves the sequestration of an effector-binding element in an alternate conformer from where catalysis can proceed. In this context, Wang et al. (20) described a more general approach for effector-driven regulation of RNA-cleaving ribozyme catalysis. They used short oligonucleotides that enable the formation of a branched three-way junction annealing simultaneously with the substrate and ribozyme. Similar results were reported in a different study by Ohtsuka and co-workers (15). They applied in vitro selection and identified short oligomeric sequences capable of activating hairpin ribozyme cleavage activity. A different approach was investigated by Vauléon and Mueller: they mutated an essential nucleotide within loop B of the wild-type hairpin ribozyme, C25, into a G-residue to construct an inactive ribozyme variant (16). Cleavage activity of the ribozyme could essentially be reassembled by providing an oligonucleotide, containing the wild-type C25 residue, that reconstitutes loop B.
All the studies described above start from an inactive hairpin ribozyme construct where activation involves applying specific oligonucleotides that either interfere directly with the substrate or the ribozyme itself and hence reassemble cleavage activity. Our approach to constructing controllable hairpin ribozymes involves highly specific conformational changes mediated by effector binding. Specifically, our mRNA effector molecule neither interacts directly with the functional ribozyme nor with the substrate-binding domain. Catalysis is regulated exclusively through conformational changes within the new binding domain C and thus can be referred to as allosteric. In the case of iHP–TRAP, catalysis is also regulated directly through induced contacts between domain A and C. This sets our hairpin ribozyme constructs apart form previously reported adenine-dependent hairpin ribozymes that require adenine more as a catalytic co-factor rather than an allosteric effector molecule (53). Furthermore, and in contrast to a classical three-way junction where the substrate is cleaved in cis, our hairpin ribozyme constructs allow catalysis to proceed in a trans-cleavage mode. The present study provides evidence that mRNA molecules can exert an allosteric effect on hairpin ribozyme-catalysed cleavage mediated by distinct conformational changes. The kobs values observed with our hairpin ribozyme constructs are similar to the wild-type ribozyme and comparable to those reported for equivalent allosteric hammerhead ribozymes responsive to small molecules such as FMN (23).
Expediently, the ribozymes we constructed are specifically regulated by trp-mRNA sequences. Leader trp-mRNA, in turn, specifically interacts with the l-tryptophan-activated version of the TRAP protein, thereby sequestering trp-mRNA molecules. This complex regulation provides a challenging test for monitoring complex regulatory pathways. We used our inducible hairpin ribozyme variant to show that the ribozyme's cleavage activity not only correlates strongly and quantitatively with the presence of trp-mRNA, but also indirectly provides a tool to measure the TRAP protein activation state. Moreover, we could determinate the minimal amount of the metabolite l-tryptophan necessary for efficient TRAP protein activation in vitro. The data in Figure 4 indicate how sensitive TRAP responds to its target molecule l-tryptophan and provide a better understanding on how TRAP might behave in its intracellular environment. On the other hand, it seems reasonable that the intracellular concentrations of tryptophan required to activate TRAP might be considerably lower than the concentrations needed to activate the ribozymes in the assay. For example, if TRAP is an undecamer requiring 11 tryptophan molecules to activate it for binding the trp-mRNA effector, a 10-fold higher concentration of tryptophan over TRAP would be required to detect in the ribozyme assay. Furthermore, the fact that we used a version of the trp-mRNA oligonucleotide that was considerably shorter than the wild-type trp-mRNA sequence might affect the Kd of the RNA/TRAP interaction and, hence, the concentration of TRAP/tryptophan needed to activate the ribozymes in our assay.
In summary, the new hairpin ribozymes described here represent the first allosterically regulated hairpin ribozymes that simultaneously allow sensitive analysis of metabolic networks consisting of small molecule metabolites, mRNAs and proteins. We demonstrate that our mechanism-based design strategy led to highly regulated, conformationally controlled hairpin ribozymes and we provide evidence that these enzymes can also be used as efficient sensor molecules. Using this method, various ribozyme constructs could be designed to serve as new precision sensors for metabolic pathways. Similar studies with other mRNA effector molecules should reveal whether this approach is also more generally applicable. It is feasible that such allosteric hairpin ribozymes can be used as biosensor components, precise switches in nanotechnology or genetic control elements (54) that can respond to the presence of almost any effector molecule.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Paul Gollnick for donation of TRAP protein. This work was supported by the Deutsche Forschungsgemeinschaft and the Volkswagen-Stiftung (to M.F.).
REFERENCES
- 1.Doudna J.A. and Cech,T.R. (2002) The chemical repertoire of natural ribozymes. Nature, 418, 222–228. [DOI] [PubMed] [Google Scholar]
- 2.Butcher S.E. (2001) Structure and function of the small ribozymes. Curr. Opin. Struct. Biol., 11, 315–320. [DOI] [PubMed] [Google Scholar]
- 3.Ferre-D'amare A.R. and Rupert,P.B. (2002) The hairpin ribozyme: from crystal structure to function. Biochem. Soc. Trans., 30, 1105–1109. [DOI] [PubMed] [Google Scholar]
- 4.Hampel K.J., Pinard,R. and Burke,J.M. (2001) Catalytic and structural assays for the hairpin ribozyme. Methods Enzymol., 341, 566–580. [DOI] [PubMed] [Google Scholar]
- 5.Esteban J.A., Banerjee,A.R. and Burke,J.M. (1997) Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J. Biol. Chem., 272, 13629–13639. [DOI] [PubMed] [Google Scholar]
- 6.Rupert P.B. and Ferre-D'Amare,A.R. (2001) Crystal structure of a hairpin ribozyme–inhibitor complex with implications for catalysis. Nature, 410, 780–786. [DOI] [PubMed] [Google Scholar]
- 7.Earnshaw D.J., Masquida,B., Muller,S., Sigurdsson,S.T., Eckstein,F., Westhof,E. and Gait,M.J. (1997) Inter-domain cross-linking and molecular modelling of the hairpin ribozyme. J. Mol. Biol., 274, 197–212. [DOI] [PubMed] [Google Scholar]
- 8.Pinard R., Lambert,D., Heckman,J.E., Esteban,J.A., Gundlach,C.W.t., Hampel,K.J., Glick,G.D., Walter,N.G., Major,F. and Burke,J.M. (2001) The hairpin ribozyme substrate binding-domain: a highly constrained D-shaped conformation. J. Mol. Biol., 307, 51–65. [DOI] [PubMed] [Google Scholar]
- 9.Butcher S.E., Allain,F.H. and Feigon,J. (1999) Solution structure of the loop B domain from the hairpin ribozyme. Nat. Struct. Biol., 6, 212–216. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang X., Kim,H., Pereira,M.J., Babcock,H.P., Walter,N.G. and Chu,S. (2002) Correlating structural dynamics and function in single ribozyme molecules. Science, 296, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 11.Walter N.G., Burke,J.M. and Millar,D.P. (1999) Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol., 6, 544–549. [DOI] [PubMed] [Google Scholar]
- 12.Esteban J.A., Walter,N.G., Kotzorek,G., Heckman,J.E. and Burke,J.M. (1998) Structural basis for heterogeneous kinetics: reengineering the hairpin ribozyme. Proc. Natl Acad. Sci., USA, 95, 6091–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porschke D., Burke,J.M. and Walter,N.G. (1999) Global structure and flexibility of hairpin ribozymes with extended terminal helices. J. Mol. Biol., 289, 799–813. [DOI] [PubMed] [Google Scholar]
- 14.Sargueil B., Pecchia,D.B. and Burke,J.M. (1995) An improved version of the hairpin ribozyme functions as a ribonucleoprotein complex. Biochemistry, 34, 7739–7748. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu Y., Nobuoka,K., Karino-Abe,N., Matsuda,A. and Ohtsuka,E. (2002) In vitro selection of hairpin ribozymes activated with short oligonucleotides. Biochemistry, 41, 9090–9098. [DOI] [PubMed] [Google Scholar]
- 16.Vauleon S. and Muller,S. (2003) External regulation of hairpin ribozyme activity by an oligonucleotide effector. Chembiochem, 4, 220–224. [DOI] [PubMed] [Google Scholar]
- 17.Porta H. and Lizardi,P.M. (1995) An allosteric hammerhead ribozyme. Biotechnology, 13, 161–164. [DOI] [PubMed] [Google Scholar]
- 18.Kuwabara T., Warashina,M. and Taira,K. (2000) Allosterically controllable maxizymes cleave mRNA with high efficiency and specificity. Trends Biotechnol., 18, 462–468. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu Y., Yamashita,S., Kazama,N., Nobuoka,K. and Ohtsuka,E. (2000) Construction of new ribozymes requiring short regulator oligonucleotides as a cofactor. J. Mol. Biol., 299, 1231–1243. [DOI] [PubMed] [Google Scholar]
- 20.Wang D.Y., Lai,B.H., Feldman,A.R. and Sen,D. (2002) A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res., 30, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang J. and Breaker,R.R. (1997) Rational design of allosteric ribozymes. Chem. Biol., 4, 453–459. [DOI] [PubMed] [Google Scholar]
- 22.Araki M., Okuno,Y., Hara,Y. and Sugiura,Y. (1998) Allosteric regulation of a ribozyme activity through ligand-induced conformational change. Nucleic Acids Res., 26, 3379–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soukup G.A. and Breaker,R.R. (1999) Engineering precision RNA molecular switches. Proc. Natl Acad. Sci., USA, 96, 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soukup G.A. and Breaker,R.R. (1999) Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Struct. Fold. Des., 7, 783–791. [DOI] [PubMed] [Google Scholar]
- 25.Robertson M.P. and Ellington,A.D. (2000) Design and optimization of effector-activated ribozyme ligases. Nucleic Acids Res., 28, 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jose A.M., Soukup,G.A. and Breaker,R.R. (2001) Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res., 29, 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piganeau N., Jenne,A., Thuiller,V. and Famulok,M. (2001) An allosteric ribozyme regulated by doxycycline. Angew. Chem. Int. Ed., 39, 4369–4373. [DOI] [PubMed] [Google Scholar]
- 28.Piganeau N., Thuillier,V. and Famulok,M. (2001) In vitro selection of allosteric ribozymes: theory and experimental validation. J. Mol. Biol., 312, 1177–1190. [DOI] [PubMed] [Google Scholar]
- 29.Koizumi M., Soukup,G.A., Kerr,J.N. and Breaker,R.R. (1999) Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat. Struct. Biol., 6, 1062–1071. [DOI] [PubMed] [Google Scholar]
- 30.Yanofsky C. (2001) Advancing our knowledge in biochemistry, genetics, and microbiology through studies on tryptophan metabolism. Annu. Rev. Biochem., 70, 1–37. [DOI] [PubMed] [Google Scholar]
- 31.Elliott M.B., Gottlieb,P.A. and Gollnick,P. (2001) The mechanism of RNA binding to TRAP: initiation and cooperative interactions. RNA, 7, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babitzke P. and Yanofsky,C. (1995) Structural features of l-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis. J. Biol. Chem., 270, 12452–12456. [DOI] [PubMed] [Google Scholar]
- 33.Antson A.A., Dodson,E.J., Dodson,G., Greaves,R.B., Chen,X. and Gollnick,P. (1999) Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature, 401, 235–242. [DOI] [PubMed] [Google Scholar]
- 34.Gollnick P., Baumann,C., Yang,M., Otridge,J. and Antson,A. (1995) Interaction of the 11-subunit trp RNA-binding attenuation protein (TRAP) with its RNA target. Nucleic Acids Symp. Ser., 43–45. [PubMed] [Google Scholar]
- 35.Antson A.A., Brzozowski,A.M., Dodson,E.J., Dauter,Z., Wilson,K.S., Kurecki,T., Otridge,J. and Gollnick,P. (1994) 11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis. J. Mol. Biol., 244, 1–5. [DOI] [PubMed] [Google Scholar]
- 36.Babitzke P., Bear,D.G. and Yanofsky,C. (1995) TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc. Natl Acad. Sci., USA, 92, 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 38.Hartig J.S., Najafi-Shoushtari,S.H., Grune,I., Yan,A., Ellington,A.D. and Famulok,M. (2002) Protein-dependent ribozymes report molecular interactions in real time. Nature Biotechnol., 20, 717–722. [DOI] [PubMed] [Google Scholar]
- 39.Jenne A., Gmelin,W., Raffler,N. and Famulok,M. (1999) Real-time characterization of ribozymes by fluorecence resonance energy transfer (FRET). Angew. Chem. Int. Ed., 38, 1300–1303. [DOI] [PubMed] [Google Scholar]
- 40.Vitiello D., Pecchia,D.B. and Burke,J.M. (2000) Intracellular ribozyme-catalyzed trans-cleavage of RNA monitored by fluorescence resonance energy transfer. RNA, 6, 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tinoco I. Jr (1997) From RNA hairpins to kisses to pseudoknots. Nucleic Acids Symp. Ser., 49–51. [PubMed] [Google Scholar]
- 42.Ecker D.J., Vickers,T.A., Bruice,T.W., Freier,S.M., Jenison,R.D., Manoharan,M. and Zounes,M. (1992) Pseudo-half-knot formation with RNA. Science, 257, 958–961. [DOI] [PubMed] [Google Scholar]
- 43.Vaish N.K., Dong,F., Andrews,L., Schweppe,R.E., Ahn,N.G., Blatt,L. and Seiwert,S.D. (2002) Monitoring post-translational modification of proteins with allosteric ribozymes. Nature Biotechnol., 20, 810–815. [DOI] [PubMed] [Google Scholar]
- 44.Hampel A. and Cowan,J.A. (1997) A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol., 4, 513–517. [DOI] [PubMed] [Google Scholar]
- 45.Stolze K., Holmes,S.C., Earnshaw,D.J., Singh,M., Stetsenko,D., Williams,D. and Gait,M.J. (2001) Novel spermine-amino acid conjugates and basic tripeptides enhance cleavage of the hairpin ribozyme at low magnesium ion concentration. Bioorg. Med. Chem. Lett., 11, 3007–3010. [DOI] [PubMed] [Google Scholar]
- 46.Earnshaw D.J. and Gait,M.J. (1999) The harpin ribozyme: the roles of ions in RNA cleavage. Nucleic Acids Symp. Ser., 273–274. [DOI] [PubMed] [Google Scholar]
- 47.Earnshaw D.J. and Gait,M.J. (1998) Hairpin ribozyme cleavage catalyzed by aminoglycoside antibiotics and the polyamine spermine in the absence of metal ions. Nucleic Acids Res., 26, 5551–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nesbitt S., Hegg,L. and Fedor,M.J. (1997) An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol., 4, 619–630. [DOI] [PubMed] [Google Scholar]
- 49.Young K.J., Gill,F. and Grasby,J.A. (1997) Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res., 25, 3760–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartig J.S., Grune,I., Najafi-Shoushtari,S.H. and Famulok,M. (2004) Sequence-specific detection of microRNAs by signal-amplifying ribozymes. J. Am. Chem. Soc., 126, 722–723. [DOI] [PubMed] [Google Scholar]
- 51.Baumann C., Otridge,J. and Gollnick,P. (1996) Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem., 271, 12269–12274. [DOI] [PubMed] [Google Scholar]
- 52.Nesbitt S.M., Erlacher,H.A. and Fedor,M.J. (1999) The internal equilibrium of the hairpin ribozyme: temperature, ion and pH effects. J. Mol. Biol., 286, 1009–1024. [DOI] [PubMed] [Google Scholar]
- 53.Meli M., Vergne,J. and Maurel,M.C. (2003) In vitro selection of adenine-dependent hairpin ribozymes. J. Biol. Chem., 278, 9835–9842. [DOI] [PubMed] [Google Scholar]
- 54.Winkler W.C. and Breaker,R.R. (2003) Genetic control by metabolite-binding riboswitches. Chembiochem, 4, 1024–1032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


