Abstract
The Mediator multiprotein complex (‘Mediator’) is an important transcriptional coregulator that is evolutionarily conserved throughout eukaryotes. Although some Mediator subunits are essential for the transcription of all protein-coding genes, others influence the expression of only subsets of genes and participate selectively in cellular signaling pathways. Here, we review the current knowledge of Mediator subunit function in the nematode Caenorhabditis elegans, a metazoan in which established and emerging genetic technologies facilitate the study of developmental and physiological regulation in vivo. In this nematode, unbiased genetic screens have revealed critical roles for Mediator components in core developmental pathways such as epidermal growth factor (EGF) and Wnt/β-catenin signaling. More recently, important roles for C. elegans Mediator subunits have emerged in the regulation of lipid metabolism and of systemic stress responses, engaging conserved transcription factors such as nuclear hormone receptors (NHRs). We emphasize instances where similar functions for individual Mediator subunits exist in mammals, highlighting parallels between Mediator subunit action in nematode development and in human cancer biology. We also discuss a parallel between the association of the Mediator subunit MED12 with several human disorders and the role of its C. elegans ortholog mdt-12 as a regulatory hub that interacts with numerous signaling pathways.
INTRODUCTION
In eukaryotes, gene transcription is regulated by transcription factors that recognize and bind specific DNA sequences in promoters, enhancers or silencers (1). Through these physical interactions, transcription factors activate or repress nearby or distant genes. Although the ability to interact functionally with specific DNA elements is a key determinant in the selective regulation of gene expression, transcription factors do not regulate genes in isolation. Instead, they form regulatory complexes with transcriptional coregulators (coactivators and corepressors). Coregulators are essential accessory proteins that link transcription factors to the core transcriptional machinery such as RNA polymerase II (Pol II), and that modulate the structure of chromatin (2–4). The combinations of individual coregulators that act at a certain promoter ultimately determine whether a corresponding gene is induced or repressed in a particular cell type or physiological condition. Among the many coregulators that are potentially available to transcription factors, the multiprotein Mediator complex (henceforth ‘Mediator’) plays a particularly interesting and central role (5–7). Originally discovered and purified in yeast as a factor that promotes activator-dependent gene transcription (8–11), Mediator mechanistically influences transcription, Pol II activity and chromatin structure and function in numerous ways; these functions have been expertly reviewed elsewhere (7,12) and are not covered herein.
Mediator is evolutionarily conserved from yeast to humans, although many of its subunits display limited sequence similarity between species; some homology assignments (e.g. MED2/MED29, MED3/MED27 and MED5/MED24) are thus tenuous and await experimental validation (13). Overall Mediator composition is also variable, typically including 25–30 subunits, dependent on species. Nevertheless, the key overall features of Mediator structure and function are evolutionarily conserved (13,14). Mediator from yeast and human cells exhibits a similar overall architecture comprising four modules that perform somewhat separable functions: the head and middle modules contact Pol II, the tail module serves as a docking site for Mediator-binding transcription factors, and the dissociable kinase module regulates the activity of Mediator and of Mediator-binding transcription factors (14,15). Head or middle module subunits are often broadly required for Mediator function and transcription, although certain subunits play distinct roles in recruitment of preinitiation complex components; in contrast, some tail and kinase module subunits apparently are not broadly required for transcription and instead are essential in specialized developmental and physiological gene programs (5,16,17). We note that, although emerging evidence hints at potential functions for some Mediator subunits independently of the complex and outside the nucleus (18,19), the specialized roles reviewed here are thought to originate not from subunit dissociation but due to individual activities while part of the complex. Specificity likely arises from selective physical interactions with transcription factors that contact Mediator and couple it to a vast number of signaling pathways (20). Such binding events may produce a specific output by inducing particular conformational changes within Mediator, a known effect of transcription factor binding to Mediator (7,12).
In line with this broad range of biological activities, numerous studies now implicate mutations in Mediator subunit genes in diverse human diseases, including cancers, neurological diseases and developmental disorders (21,22). The broad spectrum of disease phenotypes and the fact that the mutations occur in distinct Mediator subunit genes supports the notion that at least some subunits influence gene programs in a specific rather than a general manner. However, in many cases the mechanisms underlying the pathogenicity of these mutations remain poorly understood. This underscores the need to study individual Mediator subunits and their in vivo functions.
Genetically tractable model organisms offer a powerful approach to investigate the roles that individual Mediator subunits play in animal development and adult physiology (23). As many aspects of Mediator-dependent gene regulation are evolutionarily conserved, such studies can provide insight into the regulatory functions of Mediator and how Mediator subunit mutations might impinge on these activities. In this review, we summarize the current knowledge of Mediator function in the nematode Caenorhabditis elegans, a metazoan that provides excellent opportunities to study developmental regulation as well as adult physiology.
TOOLS AVAILABLE AND GENERAL INSIGHTS INTO MEDIATOR FUNCTION IN C. ELEGANS
Although it has not yet been purified biochemically, sequence comparisons predict that C. elegans Mediator consists of 29 subunits (Table 1; for nomenclature conventions, see (24)). The availability of a fully annotated genome, genome-wide RNA interference (RNAi) libraries, large-scale collections of promoter reporters, and thousands of mutant strains, including 2000 completely sequenced strains jointly harbouring nearly a million mutations, collectively provide some insight into Mediator subunit function in this metazoan (25–28). For example, the expression patterns of one third of the 29 C. elegans Mediator subunits have been examined to date by in vivo promoter::gfp fusions, and most are broadly expressed (e.g. (25,29,30)). These findings align well with the view that Mediator is likely required for the bulk of Pol II transcription, and suggest that tissue-restricted expression is unlikely to be a key driver of Mediator subunit selectivity.
Table 1. List of C. elegans Mediator subunits.
| C. elegans subunit | Mammalian ortholog | Alternative C. elegans names | Sequence number | Module (14,15) | Known functions |
|---|---|---|---|---|---|
| MDT-1.1 | MED1 | SOP-3 | Y71F9B.10 | Middle | Canonical Wnt signaling (68); cell cycle quiescence (72) |
| MDT-1.2 | MED1L | T23C6.1 | Middle | ||
| MDT-4 | MED4 | ZK546.13 | Middle | ||
| MDT-6 | MED6 | LET-425 | Y57E12AL.5 | Head | EGFR signaling; canonical Wnt signaling (43); embryonic viability (75) |
| MDT-7 | MED7 | LET-49 | Y54E5B.3 | Middle | Embryonic viability (75) |
| MDT-8 | MED8 | Y62F5A.1 | Head | ||
| MDT-9 | MED9 | Y62E10A.11 | Middle | ||
| MDT-10 | MED10 | T09A5.6 | Middle | Embryonic viability (75) | |
| MDT-11 | MED11 | R144.9 | Head | ||
| MDT-12 | MED12 | DPY-22, SOP-1, PSA-6 | F47A4.2 | Kinase | EGFR signaling (50); canonical and divergent canonical Wnt signaling (31,43,67); cell cycle quiescence (72); embryonic viability (31); axon guidance (30) |
| MDT-13 | MED13 | LET-19, PSA-7, PQN-49 | K08F8.6 | Kinase | Canonical and divergent canonical Wnt signaling (31,43); cell cycle quiescence (72); embryonic viability (31); axon guidance (30) |
| MDT-14 | MED14 | RGR-1 | C38C10.5 | Tail | Embryonic viability (76) |
| MDT-15 | MED15 | R12B2.5 | Tail | Lipid metabolism: FA desaturation and β-oxidation (29,81,83); xenobiotic detoxification: fluoranthene, β-naphthoflavene, phenazines (91,100); heavy metal detoxification (91); oxidative stress responses: arsenite, t-BOOH (82); innate immunity: p38 MAPK signaling (100) | |
| MED16 | Absent? | Tail | |||
| MDT-17 | MED17 | Y113G7B.18 | Head | ||
| MDT-18 | MED18 | C55B7.9 | Head | ||
| MDT-19 | MED19 | Y71H2B.6 | Middle | ||
| MDT-20 | MED20 | Y104H12D.1 | Head | ||
| MDT-21 | MED21 | C24H11.9 | Middle | ||
| MDT-22 | MED22 | ZK970.3 | Head | ||
| MDT-23 | MED23 | SUR-2 | F39B2.4 | Tail | EGFR signaling (41,44); cell cycle quiescence (72); muscle development (78); axon guidance (30); innate immunity: ERK/MAPK signaling (112) |
| MDT-24 | MED24 (MED5 in Saccharomyces cerevisiae) | LIN-25 | F56H9.5 | Tail | EGFR signaling (42); cell cycle quiescence (72); Innate immunity (112) |
| MED25 | Absent? | Tail | |||
| MDT-26 | MED26 | C25H3.6 | Middle | ||
| MDT-27 | MED27 (MED3 in S. cerevisiae) | T18H9.6 | Density between head and tail | ||
| MDT-28 | MED28 | W01A8.1 | Density between head and tail | ||
| MDT-29 | MED29 (MED2 in S. cerevisiae) | K08E3.8 | Density between head and tail | ||
| MDT-30 | MED30 | PQN-38 | F44B9.7 | Density between head and tail | |
| MDT-31 | MED31 | F32H2.2 | Middle | ||
| CDK-8 | CDK8 | F39H11.3 | Kinase | Cell cycle quiescence (72); axon guidance (30) | |
| CIC-1 | CycC | H14E04.5 | Kinase | Axon guidance (30) |
The table lists the 29 putative C. elegans Mediator subunits, their mammalian orthologs, alternative names, sequence number, hypothetical location within Mediator (inferred from (14)), and functions. Orthologs for MDT-16 and MDT-25 have not yet been identified and may be absent in C. elegans. The table is based on homologies described by Bourbon (13).
Functional studies using genome-wide RNAi libraries and/or strains carrying mutations in Mediator subunit genes suggest that many Mediator components are essential for viability or fertility (28). These requirements are underscored by the fact that three Mediator subunit genes were initially discovered as essential (lethal, let) genes: mdt-13 as let-19, mdt-6 as let-425 and mdt-7 as let-49 (31–33) (Table 1). Although these studies likely underestimate the number of essential Mediator subunits (due to RNAi causing a reduction rather than a loss of gene function and due to the fact that certain tissues are refractory to RNAi), it is also worth noting that some Mediator subunit null mutants do not display lethality or sterility (e.g. cdk-8) (30). Thus, rather than being universally required for gene transcription, at least some Mediator subunits appear to participate in specific signaling events. Below, we outline the current understanding of how individual Mediator subunits regulate specific developmental and physiological pathways in C. elegans.
MEDIATOR REGULATES DEVELOPMENT VIA ACTION UPON DIVERSE PATHWAYS
The power of genetic analysis in C. elegans and in particular of large-scale forward and reverse genetic screens has brought to light the key roles that some Mediator subunits play during development. Here, we describe the developmental phenotypes associated with mutations in certain Mediator subunits, and the mechanisms by which these subunits regulate cell fate by modulating the epidermal growth factor receptor (EGFR)–Ras–mitogen activated protein kinase (MAPK) signaling pathway, the Wnt signaling pathway or the cell cycle.
Mediator subunits are required to activate and repress EGFR–Ras–MAPK signaling
The EGFR–Ras–MAPK pathway is a conserved signaling cascade that regulates cell proliferation and differentiation in eukaryotes (34,35). Mutations in the EGFR signaling pathway cause several human developmental syndromes and underlie numerous human cancers (36,37). Several Mediator subunits are required for activation or repression of EGFR pathway activity. Many of these regulatory roles were first discovered in studies on C. elegans development, and were followed later by the description of similar regulatory mechanisms in human cancers.
The EGFR signaling pathway is broadly required for C. elegans development, but the hermaphrodite vulva is the best-studied EGFR signaling-dependent organogenesis paradigm (38–40). In the nascent vulva, EGFR and Notch signaling direct three of six equivalent vulva precursor cells (VPCs) to adopt a vulval cell fate, while the remaining VPCs adopt a hypodermal fate. VPC fates are exquisitely sensitive to perturbations in the EGFR–Ras–MAPK pathway: increased EGFR signaling activity promotes additional VPCs to adopt the vulval cell fate, leading to a multivulva phenotype with ectopic, non-functional pseudovulvae; vice versa, reduced EGFR signaling activity inhibits the vulval cell fate and leads to a vulvaless phenotype (38). These phenotypes have been used to identify positive and negative regulators of EGFR signaling, including several Mediator subunits.
Forward genetic screens for positive regulators of EGFR signaling (41,42) uncovered requirements for mdt-23/sur-2 and for the divergent MED24 ortholog lin-25 (which we hereby propose to rename mdt-24, in line with the analysis by Bourbon (13)). Both mdt-23 and mdt-24 are essential for vulva development in C. elegans, as mutants display a completely penetrant vulvaless phenotype. Both subunits are required downstream of Ras, as loss of mdt-23 or mdt-24 suppresses the multivulva phenotype of let-60/Ras gain-of-function mutants (41,42). Similarly, the Mediator head module subunit mdt-6 may also be essential downstream of Ras, as mdt-6 depletion partially suppresses the multivulva phenotype in let-60/Ras gain-of-function mutants (43).
Mechanistically, mdt-23 is required downstream of the core EGFR signaling cascade (44,45). Specifically, mdt-23 is essential to coactivate a positive regulatory element in the lag-2 gene, a key EGFR target. In contrast to mdt-23, the LIN-1/Ets transcription factor represses lag-2 through a distinct regulatory element. Together, mdt-23 and lin-1 are required to restrict lag-2 expression to the appropriate VPCs. However, the transcription factor(s) that partners with MDT-23 to promote lag-2 transcription has not yet been identified (44). Interestingly, in murine cells, MED23 binds the Ets-family transcription factor Elk-1 and is required to coactivate serum response genes (46). However, at the C. elegans lag-2 promoter, the Elk-1 ortholog LIN-1/Ets acts exclusively as a repressor (44), suggesting that MDT-23 may not cooperate with LIN-1/Ets to activate this EGFR target.
mdt-23 and mdt-24 are required for additional developmental events regulated by EGFR–Ras–MAPK (47), suggesting that these Mediator subunits are generally essential within this pathway. However, mdt-23 and mdt-24 do not appear to be required in certain developmental events (e.g. fluid homeostasis, oogenesis) wherein LET-60/Ras is activated by an alternate receptor tyrosine kinase, such as fibroblast growth factor receptor or others (47–49). The mechanism for this specificity remains unclear, but likely depends on interactions between Mediator subunits and specific downstream transcription factors.
In contrast to mdt-6, -23 and -24, a requirement for the Mediator kinase module subunit mdt-12 was identified in a forward genetic screen for negative regulators of EGFR signaling (50). Reduction-of-function mutations in mdt-12 cause a low penetrance multivulva phenotype in wildtype worms and strongly enhance multivulva penetrance in worms heterozygous for gain-of-function mutations in let-60/Ras or the EGFR ortholog let-23. mdt-12 is required downstream of EGFR, as reduction-of-function of mdt-12 partially suppresses the vulvaless phenotype of a let-23/EGFR reduction-of-function mutant (50). Beyond this, the position of mdt-12 in the EGFR–Ras–MAPK pathway is unclear, although an MDT-12::GFP fusion protein is expressed in VPCs (50), suggesting a function in these cells, perhaps in conjunction with LIN-1/Ets. Alternatively, mdt-12 may prevent ectopic EGF ligand expression in the surrounding hypodermis (skin), as do several other transcriptional coregulators (51).
mdt-23 and mdt-24 promote, whereas mdt-12 inhibits EGFR-Ras-MAPK signaling. Thus, it would be informative to conduct genetic epistasis analyses to determine whether these Mediator subunits regulate each other's activity in this pathway. Such epistatic relationships have been observed in yeast between the kinase module subunit CDK8 and the tail module subunits MED2, MED3, and MED15 (52,53); it will be of interest to determine whether a similar relationship exists in metazoans. Furthermore, other Mediator subunits might also regulate EGFR signaling, and the C. elegans vulva provides an excellent system to explore this possibility. In particular, since the kinase module subunit mdt-12 has already been implicated in vulva development, the other kinase module subunits, cdk-8, cic-1/Cyclin C and mdt-13 may also negatively regulate this process.
In the last few years, strong parallels have emerged between the requirements for mdt-23 and mdt-12 in EGFR-regulated development in C. elegans and the roles of MED23 and MED12 in EGFR-driven tumorigenesis in humans. MED23 is essential for the proliferation and tumorigenicity of human lung cancer cells with hyperactive Ras mutations, but not in cells lacking Ras mutations (54). Thus, just as observed decades earlier in C. elegans (41,50), loss of human MED23 suppresses the effects of activated Ras, suggesting a potential role in promoting transcriptional activation downstream of Ras. Moreover, in human melanoma cells, loss of MED12 promotes cellular resistance to chemotherapeutic agents that inhibit the MAPK signaling pathway component BRAF (55,56). This suggests that, like C. elegans mdt-12 (50), MED12 is required to inhibit MAPK signaling in melanoma cells by acting downstream of BRAF, perhaps by promoting transcriptional repression of downstream target genes. Lastly, the two Mediator subunits MED12 and CDK8 are mutated or amplified in human cancers (57), but their impact upon EGFR signaling in these contexts has not been examined. Overall, the parallels between C. elegans development and human cancer indicate that further study of C. elegans Mediator subunits in the context of EGFR signaling may yield clinically applicable findings for cancers bearing activating mutations in this signaling cascade.
Mediator action in the canonical Wnt signaling pathway
The Wnt signaling pathway is an important regulator of cell fate and of proliferation and self-renewal of stem and progenitor cells (58). Reduction of Wnt signaling causes degenerative diseases such as syndromic osteoporosis, whereas increased Wnt signaling causes multiple human cancers including colon cancer and melanoma (59). Several Mediator subunits are required for canonical Wnt signaling in C. elegans development, and some also modulate Wnt signaling in Drosophila and in human colon cancer (60–62).
The components of the canonical Wnt signaling pathway are evolutionarily conserved. In the absence of Wnt, the transcriptional coactivator BAR-1/β-catenin is targeted for proteasomal degradation (63). Binding of a Wnt ligand to a Frizzled receptor relocates the BAR-1/β-catenin degradation complex to the plasma membrane, allowing unphosphorylated BAR-1/β-catenin to accumulate, enter the nucleus, and coactivate transcription by the TCF/LEF family transcription factor POP-1 (63). This canonical Wnt signaling pathway regulates various aspects of C. elegans development, including VPC fusion, generation of seam cell-derived structures (e.g. male tail rays), neuroblast migration, and somatic gonad development. Additionally, a divergent canonical Wnt signaling pathway, involving the β-catenin orthologs WRM-1 and SYS-1, regulates asymmetric cell division and endoderm induction (63).
In the C. elegans vulva, mdt-12 and mdt-13 are required for cell fusion events controlled by BAR-1/β-catenin (31). In the VPCs, BAR-1/β-catenin prevents fusion of the VPCs with the surrounding hypodermal tissue by inducing the Homeobox (Hox) transcription factor lin-39 (64). Loss of mdt-12 or mdt-13 inhibits VPC fusion and suppresses the ectopic cell fusion phenotype of bar-1/β-catenin mutants, but not of lin-39 mutants (31). Thus, mdt-12 and mdt-13 apparently act parallel to or downstream of bar-1/β-catenin, but upstream of lin-39 to promote cell fusion.
Forward genetic screens additionally identified requirements for several Mediator subunit genes in Wnt-dependent cell fate patterning of C. elegans male tail rays. This process is governed by Hox factors pal-1/Caudal, mab-5/Antennapedia and egl-5/Abdominal-B (65,66). The kinase module subunits mdt-12 and mdt-13, and the middle module subunit mdt-1.1 are required to prevent aberrant activation of this transcription factor cascade by BAR-1/β-catenin (67,68). Conversely, the head module subunit mdt-6 is essential for the aberrant BAR-1/β-catenin-driven pal-1 activation in mdt-12 mutants (43). Thus, as seen in yeast (52,53) and as suggested for Mediator subunit action in EGFR-driven development, functional opposition between Mediator subunits apparently occurs in the Wnt signaling pathway. Interestingly, mdt-23 is not required for BAR-1/β-catenin-dependent transcriptional regulation of pal-1 (68), demonstrating that only some Mediator subunits are required for Wnt signaling.
Mediator subunits are also essential for asymmetric cell division mediated by divergent canonical Wnt signaling in C. elegans. Asymmetric division of the T blast cell, whose daughters can adopt neural or hypodermal fates, requires lin-44/Wnt and lin-17/Frizzled to establish polarity of the asymmetrically expressed transcription factors POP-1/TCF and TLP-1/Sp1 (69,70). In the T cell daughters, the Mediator kinase module subunits mdt-12 and mdt-13 are required downstream of lin-17/Frizzled to establish the asymmetric expression of TLP-1/Sp1 but not of POP-1/TCF (31,43). Unexpectedly, the MDT-12 and MDT-13 proteins themselves are distributed symmetrically in the T-cell daughters (31), suggesting that their activity may itself be regulated by the Wnt pathway.
Although mdt-12 and mdt-13 are needed in at least three developmental processes governed by Wnt signaling, they do not indiscriminately influence all Wnt-regulated developmental events. QL neuroblast migration, somatic gonad development and endoderm induction, all regulated by Wnt signaling, are refractory to loss of mdt-12 or mdt-13 (31). Thus, various tissues may differentially require mdt-12 and mdt-13 to implement Wnt-dependent developmental events.
The requirement for C. elegans Mediator kinase module subunits in Wnt/β-catenin signaling is intriguing, as human CDK8 influences Wnt signaling in colon cancers. Specifically, CDK8 expression is upregulated in a substantial fraction of human colon cancer specimens, often due to CDK8 copy number gain (57). Colon cancer cell lines with CDK8 amplification and high β-catenin activity require CDK8 for cell proliferation and tumorigenicity (61). Additionally, CDK8 binds and inhibits E2F1, a negative regulator of β-catenin activity (60). Thus, CDK8 directly and indirectly promotes oncogenic action of β-catenin in human colon cancer. These data contrast with the inhibitory role of C. elegans mdt-12 and mdt-13 in Wnt/β-catenin signaling in male tail patterning (67) and VPC fusion (31). This may suggest evolutionary divergence of the kinase module's role in Wnt signaling, or divergence of the roles of individual kinase module subunits in Wnt regulation. Further work is needed to answer these questions, particularly to elucidate whether MDT-12 and MDT-13 regulate BAR-1/β-catenin directly or indirectly in the context of cell differentiation and fusion.
Developmental regulation by Mediator control of cell cycle progression
Cell cycle regulation is essential for animal development, and certain Mediator subunits influence cell fate by regulating this process. During vulva development, six VPCs enter a period of cell cycle quiescence, arresting in an extended G1 phase that lasts from the first larval stage L1 until the time of their cell fate determination (vulval versus hypodermal fate) in the third larval stage L3 (71). An elegant genetic screen for regulators of VPC quiescence revealed that multiple Mediator subunits are required for cell cycle entry or exit (72). Loss of the middle module subunit mdt-1.1, the tail module subunits mdt-23 or mdt-24, or the kinase module subunits cdk-8, mdt-12 or mdt-13 prevents establishment or maintenance of VPC quiescence. Depletion of 13 additional Mediator subunits caused no observable effect on cell cycle quiescence, suggesting that only some Mediator subunits are required for this process (72). Mechanistically, mdt-13 was not required to express the cyclin-dependent kinase inhibitor cki-1, a key effector of VPC quiescence, implying that Mediator regulates the transcription of other, unknown genes involved in cell cycle quiescence (72). The six Mediator subunits identified in this screen are required in multiple cell signaling pathways (see above), suggesting that Mediator may integrate information from various signaling cascades to coordinate cellular quiescence in development.
The requirement for Mediator kinase module subunits in cell cycle regulation in C. elegans parallels a function of human MED13L. The cellular senescence or cell cycle arrest induced by a constitutively active Retinoblastoma (RB) mutant depends on MED13L (73). In this context, MED13L corepresses RB/E2F, as loss of MED13L causes derepression of the RB/E2F target Cyclin A even when constitutively active RB is present (73). This cooperation between a kinase module subunit and RB is similar to the role of Drosophila CDK8 in parallel to RB as a corepressor for E2F1 (60). Together, these data suggest that the Mediator kinase module and RB may target common transcription factors to regulate the cell cycle. It may be interesting to explore this hypothesis further using the C. elegans VPC quiescence model, to determine whether the kinase module subunits, and other Mediator subunits required in cell cycle quiescence (72), cooperate with RB in this capacity.
Other developmental phenotypes of Mediator mutants
Besides the roles described above, Mediator subunits are essential for additional aspects of C. elegans development. The underlying molecular pathways are less well defined in these instances, yet these studies elegantly reveal differential Mediator subunit requirements during embryonic and post-embryonic development.
C. elegans provides an ideal system to study the requirements for Mediator subunits in gene-specific versus general embryonic transcription, as maternal gene products allow embryos with null mutations in essential genes to survive to at least the 100-cell stage despite drastically perturbed transcription (74). In this system, depletion of the head module subunit mdt-6, the middle module subunits mdt-7 or mdt-10, or the kinase module subunit mdt-13 causes embryonic arrest at the ∼300-cell stage, before cell differentiation and body morphogenesis begin (32,75). Examination of transcriptional reporters demonstrated that the depletion of these Mediator subunits prevents the expression of some stage- or lineage-specific genes, but not of ubiquitously expressed genes (32,75). In contrast, depleting mdt-14 causes embryonic arrest at the 100-cell stage due to a broad loss of transcription as evidenced by loss of Pol II C-terminal domain phosphorylation (76). Thus, whereas some Mediator subunits are required for specific gene programs in early embryogenesis, mdt-14 is broadly required for transcription. The latter is in line with the role of MED14 as a structural backbone of Mediator, connecting the head, middle and tail modules (14) (Figure 1).
Figure 1.
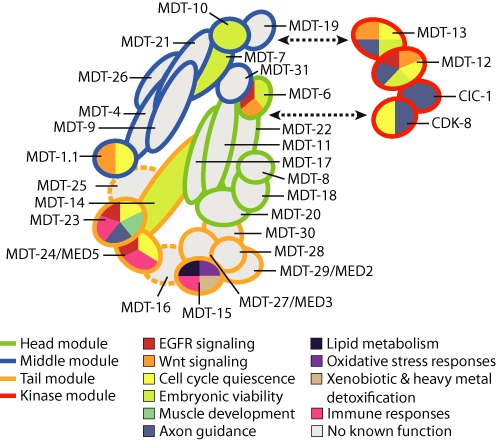
Overview of the biological activities of Mediator subunits in C. elegans. The figure summarizes the roles of C. elegans Mediator subunits in developmental and physiological pathways. The model is a hypothetical adaptation of C. elegans Mediator, based on the architecture of human Mediator (14). Subunits labeled with a dashed outline lack apparent C. elegans orthologs. In cases where the homology between yeast and metazoan Mediator subunits is tenuous (13), both C. elegans and yeast names are listed (MDT-24/MED5, MDT-27/MED3 and MDT-29/MED2); note, that the positioning of these three subunits is particularly speculative in our model. Putative functions identified in large-scale genetic screens only (i.e. without further experimental validation) are not listed.
Interestingly, Mediator subunit knockout mice similarly display differing phenotypes in early embryogenesis. Some subunits are required for early events such as compaction, gastrulation or blastocyst differentiation (CDK8, MED12 and MED21), whereas others are required for later events such as placental, circulatory system, nervous system or limb bud development (MED1, MED23, MED24 and MED31, respectively; reviewed in (77)). In the future, it would be interesting to conduct a systematic study to determine the embryonic requirements for all C. elegans Mediator subunits, and whether these requirements involve gene-specific or global transcriptional regulation.
Several processes during C. elegans larval development also depend on Mediator subunits. The tail module subunit mdt-23 was identified in a screen for genes required for muscle development, as mdt-23 depletion causes aggregation of muscle myosin myo-3 in myofilaments (78). Furthermore, mdt-23 and the kinase module subunits cdk-8, cic-1/Cyclin C, mdt-12 and mdt-13 are required for correct navigation of ventral nerve cord axons (30,79). Genetic epistasis analysis suggested that, in this context, the kinase module may suppress the sax-3/ROBO pathway by molecular mechanisms that remain to be elucidated (30).
MEDIATOR REGULATES DIVERSE ASPECTS OF ADULT PHYSIOLOGY
A growing number of studies have identified roles for Mediator in regulating adult physiology in C. elegans. Below, we review Mediator-dependent regulation of three processes: lipid metabolism, stress responses and innate immune responses.
Regulation of lipid metabolism by the Mediator subunit MDT-15
The tail module subunit gene mdt-15 is essential for the regulation of lipid metabolism in C. elegans. MDT-15 physically interacts with nuclear hormone receptors (NHRs) homologous to mammalian hepatocyte nuclear factor 4α (HNF4α), such as NHR-49 and NHR-64, and also binds SBP-1, the worm homolog of sterol regulatory element binding protein (SREBP) (29,80,81). Like sbp-1 and nhr-49, mdt-15 is required for the expression of the fatty acid (FA) desaturase genes fat-5, -6 and -7, whose products generate mono- and polyunsaturated FAs (MUFAs and PUFAs). These FAs are required for many aspects of C. elegans physiology; accordingly, worms depleted of mdt-15 exhibit pleiotropic phenotypes that can be partially rescued with dietary PUFAs (29,82,83). The partial nature of this rescue suggests that, although a substantial fraction of its role in animal physiology entails maintaining normal FA desaturation, mdt-15 must also be essential for additional processes (see below).
mdt-15-dependent FA production is particularly important for membrane lipid homeostasis. For instance, worms depleted of mdt-15 activate the endoplasmic reticulum (ER) stress response due to imbalances in ER membrane lipid composition (83). Moreover, an mdt-15 gain-of-function mutation suppresses the cold-sensitive phenotype of a C. elegans adiponectin receptor mutant (84). Cold sensitivity is intrinsically linked to membrane fluidity, which in turn is affected by membrane lipid composition, again implicating mdt-15 in membrane lipid homeostasis. Additional mutations that increase unsaturated FA production and suppress this cold-sensitive phenotype were identified in nhr-49 (84). Intriguingly, the mutations in mdt-15 and nhr-49 lie within or near domains that mediate the physical interaction between the two proteins (84), suggesting that these mutations may modulate the MDT-15:NHR-49 interaction to alter downstream gene expression and membrane lipid composition. Lastly, mdt-15 was also identified in genetic screens for genes involved in yolk receptor endocytosis or endosome trafficking (85,86). Although it is unclear why mdt-15 is essential for these processes, the mechanism may involve membrane lipid desaturation, as both screens also identified FA desaturases and NHRs that promote desaturase expression. Additionally, yolk receptor endocytosis was impaired by loss of mdt-22 or mdt-30, although their modes of action are unclear (86).
Lipid molecules, and mdt-15-dependent production thereof, have been proposed to play a role in the regulation of longevity. Long-lived daf-2/insulin receptor mutants show altered expression of many genes due to the activation of downstream transcription factors such as DAF-16/FoxO (87,88). DAF-16 acts both cell autonomously and non-autonomously to regulate target gene expression (89). Interestingly, the cell non-autonomous upregulation of a DAF-16 target requires mdt-15 even though mdt-15 is not expressed in the tissue where this gene is induced; the authors thus speculated that mdt-15 may be required for the synthesis of a lipid molecule(s) that relays inductive signals to distant tissues (90). In line with its requirement for daf-16-dependent gene regulation, mdt-15 is also required for the long lifespan of daf-2 mutants (82,90); however, the specificity of this requirement is difficult to interpret, as mdt-15 is similarly essential for the normal lifespan of wild-type worms and several other long-lived mutants (82,90–92). Nevertheless, in the wild-type background, PUFA supplementation partially rescues the short lifespan of mdt-15-depleted worms (29), suggesting that mdt-15 indeed assures normal life span through its requirement for the synthesis of certain lipid molecules.
In addition to their role in FA desaturation, mdt-15 and nhr-49 are also required to express FA β-oxidation genes, particularly in response to fasting (29,93). However, not all mdt-15-dependent fasting-induced genes require nhr-49; instead, some nhr-49-independent β-oxidation genes may be activated by the MDT-15-interacting transcription factor SKN-1/Nrf2 (94), which is primarily known for its role in systemic detoxification ((95); see below). Additional MDT-15-binding transcription factors, especially NHRs (80), are likely involved in various aspects of mdt-15-dependent regulation of lipid metabolism, possibly performing redundant roles (96).
mdt-15 is required to express genes involved in lipid breakdown (e.g. β-oxidation genes) and in lipid synthesis (e.g. FA desaturases). What, then, is the overall effect of mdt-15 loss on fat storage? Worms subjected to mdt-15 depletion exhibit the Clear phenotype often associated with reduced fat storage (as seen e.g. in worms with reduced sbp-1/SREBP levels) (81). In agreement with this finding, mdt-15 depletion results in reduced staining with the lipid-labeling dye Oil Red O (80). In contrast, recent quantification of overall extractable fats in mdt-15-depleted worms and mdt-15 hypomorph mutants revealed that storage triglyceride abundance resembled that of wild-type worms (83). One way to reconcile these findings is that potentially the overall neutral lipid levels are similar in wild-type worms and in animals lacking mdt-15, yet the assembly of subcellular structures such as lipid droplets is impaired; this would explain the discrepancy between the morphological phenotypes and the extract-based lipid analysis.
How conserved is mdt-15-dependent regulation of lipid metabolism? In yeast, the MDT-15 ortholog Gal11 also regulates genes involved in β-oxidation (97). The interaction between MDT-15 and NHRs may also be conserved, as Gal11 interacts with Oaf1, a member of the Zinc-cluster type transcription factor family that is distantly related to NHRs (98,99). In mammals, the MDT-15 ortholog MED15 interacts physically and functionally with SREBP, the ortholog of SBP-1 (81), but mammalian MED15 is not known to interact with NHRs. Instead, many NHRs, including the lipid metabolism regulators HNF4α, PPARα and PPARγ, bind MED1 (20). Additionally, mammalian MED14 also binds SREBP and HNF4α (20). Thus, whereas C. elegans MDT-15 interacts physically and functionally with transcription factors that regulate lipid balance, mammalian MED15 may have evolved into a more specialized regulator of certain aspects of lipid metabolism, distributing its broad ancestral regulatory roles to other subunits. To investigate this possible functional divergence, it will be interesting to determine if C. elegans Mediator subunits other than mdt-15 are also required for metabolic homeostasis, and to comprehensively assess the function of mammalian MED15 in vivo.
Stress response regulation by C. elegans Mediator subunits
Besides its role in lipid metabolism, mdt-15 is also required for various stress responses. Gene expression studies revealed a large number of putative xenobiotic detoxification genes that depend on mdt-15 for expression, both in the absence of toxins and when worms are exposed to xenobiotic compounds (91). Accordingly, mdt-15 is required for resistance to the xenobiotics fluoranthene and RPW-24 and two compounds evoking oxidative stress (82,91,100). In contrast, mdt-15 is not required for thermotolerance or for resistance to the glycosylation inhibitor tunicamycin; similarly, the induction of detoxification genes in response to several pesticides is not blocked by loss of mdt-15 (83,91,101). This demonstrates that mdt-15 is required for specific adaptive responses rather than being a universal stress resistance factor, and argues against the possibility that knockdown of mdt-15 simply renders worms too sick to mount a response against all stresses. Instead, mdt-15's specificity may originate from the ability of the MDT-15 protein to selectively bind transcription factors that implement particular stress responses.
Available data suggest that the evolutionarily conserved detoxification regulator SKN-1/Nrf2 engages Mediator through MDT-15. Specifically, mdt-15 is required for detoxification gene induction and survival on the oxidative stressor arsenite (82), thus phenocopying loss of skn-1 (102). (As noted above, mdt-15 is also required to induced skn-1-dependent transcription of certain lipid metabolism genes (94).) Moreover, MDT-15 and SKN-1 physically interact in yeast two-hybrid assays via a central, structurally uncharacterized region in MDT-15; this region also contributes to NHR binding (82). In contrast, the N-terminal KIX-domain, which is required for NHR binding, is dispensable for SKN-1 interaction. Interestingly, the yeast MDT-15 homolog Gal11 binds the transcription factor Gcn4 via multiple hydrophobic bonds in a ‘fuzzy complex’, which may allow protein-protein interactions to be easily adapted and dissociated (103–105). Such a binding mode may also apply for MDT-15, perhaps especially for NHR binding, which apparently engages at least two surfaces. Further investigation of the interactions between MDT-15 and its partner transcription factors should clarify whether ‘fuzzy complex’ formation is evolutionarily conserved.
mdt-15 is also required for the oxidative stress response to tert-butyl hydroperoxide (t-BOOH), which differs from the arsenite response by engaging different target genes and by being largely skn-1-independent (82,102). Potentially, MDT-15-interacting NHRs are required for this transcriptional response, e.g. nhr-64, which is required for survival on t-BOOH (82), or nhr-8 and nhr-114, both of which engage in detoxification responses (106,107) and interact with MDT-15 in yeast-two hybrid assays (80,108). t-BOOH induced genes that strongly depend on mdt-15 but are refractory to loss of skn-1 will be useful tools to identify the factors that cooperate with mdt-15 in this regulatory context.
Mediator subunits other than mdt-15 are also required for stress responses. Two Mediator subunits were identified in a screen for cytoprotective genes conferring stress responses and/or longevity (109). mdt-26 is required to induce chaperones of the ER and mitochondrial UPR, as well as a detoxification gene responsive to sodium azide. mdt-26 knockdown causes decreased resistance to sodium azide, cadmium and paraquat (109). Additionally, mdt-26 is required for the normal lifespan of wildtype worms and for the longevity of daf-2 and eat-2 (a genetic mimic of dietary restriction) mutants (109,110). The same screen also identified putative roles for the kinase module subunit gene mdt-12, which is weakly required for detoxification gene induction, for resistance to paraquat and sodium azide, and for the extended lifespan of some mitochondrial mutants, but not daf-2 or eat-2 mutants (109). Another study showed that mdt-12 is essential to induce the oxidative stress response gene gcs-1 upon arsenite exposure, although it is not required for arsenite resistance per se (111). In sum, both mdt-26 and mdt-12 may have broad cytoprotective activities that deserve further attention, as relevant mechanisms remain undefined at this time. Moreover, these data support the notion that MDT-12 is a regulatory hub (see below) not just in developmental but also in physiological settings.
Mediator subunits participate in innate immune responses
Three tail module subunits of Mediator contribute to innate immune responses in C. elegans. mdt-15 is required for the innate immune response against the opportunistic bacterial pathogen Pseudomonas aeruginosa. Specifically, mdt-15 is essential to induce immune effectors downstream of the p38 MAPK PMK-1 upon P. aeruginosa infection and for pathogen resistance (100). Additionally, mdt-15, but not pmk-1, is essential for the resistance to P. aeruginosa-produced phenazine toxins, and mdt-15 is also required to induce PMK-1-independent immune effectors. Thus, mdt-15 apparently plays a dual role in the response to P. aeruginosa, mediating the response to bacterial infection by regulating PMK-1-dependent and -independent innate immunity genes, and mediating the response to bacterial toxins by regulating as yet undetermined target genes. In the future, it will be interesting to determine which MDT-15-interacting transcription factor(s) confers the PMK-1-dependent immune response. Together with the findings on MDT-15's role in the response to xenobiotic compounds, heavy metals and oxidative stress, these results suggest that MDT-15 is a key coregulator in the adaptation to environmental challenges, at least in C. elegans.
Two other Mediator tail module subunits, mdt-23/sur-2 and mdt-24/lin-25, are required for a distinct pathogen response, namely infection by Microbacterium nematophilum (112). In C. elegans, infection by these bacteria causes a swollen tail phenotype that reflects the normal immune response of the host (113). A screen for mutants that suppress infection-induced swelling revealed alleles of mdt-23 and mdt-24 (112). Interestingly, extracellular signal regulated kinase (ERK) MAPK pathway reduction-of-function mutants also suppress infection-induced swelling, thus phenocopying mdt-23 and mdt-24 mutants, and genetic epistasis experiments showed that mdt-23 and mdt-24 act downstream of MAPK in this immune response (112). As described above, mdt-23 and mdt-24 are also required downstream of EGFR-Ras-MAPK signaling in vulva development; thus, MAPK signaling pathways similarly engage mdt-23 and mdt-24 as downstream effectors in organogenesis and in an adult adaptive response. Additionally, the strong functional overlap between mdt-23 and mdt-24 suggests a shared underlying mechanism of action, perhaps involving physical interactions of these Mediator subunits with EGFR-regulated transcription factors such as LIN-1. By extension, given that mammalian MED23 is required downstream of MAPK signaling pathways (54), it would be worthwhile testing for similar roles for mammalian MED24.
REGULATORY HUB ACTIVITY OF mdt-12
Network theory can be used to analyze patterns of genetic interactions. Gene networks of various origins typically conform to a common format, wherein most genes interact with relatively few other genes, and a few genes form highly connected regulatory ‘hubs’ (114). A high-throughput screen for pairwise genetic interactions in C. elegans revealed that the Mediator subunit mdt-12 is such a regulatory hub (115). In this screen, ∼65 000 genetic interactions were directly tested by analyzing the phenotypes of 37 ‘query’ mutant strains (covering 31 genes) subjected to RNAi of 1750 ‘library’ genes (selected for their annotation as cell signaling-related genes). Strikingly, mdt-12 was one of six library genes that interacted genetically with multiple query genes, including genes involved in EGFR signaling, Notch signaling, Wnt signaling and cell death/migration (115). Thus, in this genetic network, mdt-12 is one of the six highly connected hubs that bridge multiple signaling pathways (Figure 2). Interestingly, mdt-12 and the other candidate hub genes enhanced the loss-of-function phenotype of genes involved in processes unrelated to cell signaling (115). This indicates that mdt-12 acts as a genetic buffer to moderate the effect of genetic perturbations in an organism. The results of this screen are supported by previous studies that implicate mdt-12 in multiple pathways, including EGFR signaling, Wnt signaling and cell cycle regulation (see above), as well as in overall animal fertility (50). Furthermore, mdt-12 may also be required for cytoprotective functions in adult animals (109,111), indicating that its role as a regulatory hub may also extend to control of adult physiology.
Figure 2.
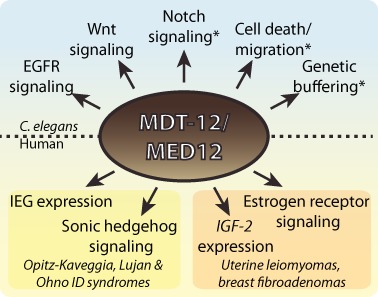
mdt-12 is a regulatory hub with links to human disease. C. elegans mdt-12 interacts genetically with multiple signaling pathways, indicating a role as a regulatory hub. In addition, mdt-12 interacts genetically with several additional cellular processes, implying a role as a general buffer against genetic perturbations. Similarly, human MED12 mutations are implicated in multiple developmental diseases or somatic tumors and are associated with perturbed cell signaling. Asterisks (*) indicate genetic interactions with pathways/processes that were identified in a genetic screen (115) but have not been further validated. IEG: immediate-early genes. ID: intellectual disability; IGF: insulin-like growth factor.
All hubs identified in this screen are components of transcriptional coregulator complexes or chromatin modifying complexes, suggesting that such proteins integrate inputs from multiple pathways to effect the correct cellular responses. However, mdt-12 was the only Mediator subunit identified as a hub, as mdt-23 and mdt-24, which were included as query genes, interacted with very few genes (115). This suggests that mdt-12 may occupy a unique role in Mediator as an integration site for various pathways. Systematic testing of pairwise genetic interactions between cell signaling pathway mutants and Mediator subunit mutants may be useful to determine whether additional Mediator subunits also serve as hubs in C. elegans.
The fact that MED12 is implicated in diverse human diseases may suggest that its role as a regulatory hub is evolutionarily conserved (Figure 2). Germline mutations in MED12 cause intellectual disability syndromes such as Opitz-Kaveggia syndrome, Lujan syndrome and Ohno syndrome (Maat–Kievit–Brunner type) (116), which are associated with disrupted immediate early gene expression (117) or sonic hedgehog signaling (118). Additionally, somatic MED12 mutations occur in over 50% of uterine leiomyomas and breast fibroadenomas (reviewed in (119)), and have also been found in some prostate cancer samples (120). Thus, by interacting with diverse signaling pathways, and hence conferring a risk for diverse pathologies, human MED12 may represent a regulatory hub as described for C. elegans mdt-12.
CONCLUSIONS
Mediator is an elaborate multiprotein complex that performs a broad variety of molecular functions and participates in a remarkable collection of cellular signaling pathways. Although much progress has been made in delineating its molecular functions, the biological, whole-organism activities of many individual Mediator subunits remain elusive. Studies on C. elegans Mediator have been extremely insightful in this regard, providing critical information on requirements for Mediator subunits in specific developmental and physiological pathways, many of which were later found to have parallels in mammalian and human biology. In the future, it will be important to comprehensively identify the specific activities of each individual Mediator subunit. With its powerful experimental toolbox, C. elegans should help reveal tissue-specific activities of Mediator subunits, a relevant line of investigation considering that several subunits (e.g. mdt-23 and mdt-24) act in both developmental and physiological regulatory contexts that engage distinct tissues. Moreover, chromatin immunoprecipitation and transcriptome studies should reveal genomic occupancy of and genes regulated by individual Mediator subunits in specific developmental and physiological contexts. Such studies are complicated by fact that whole animals are composed of many distinct tissues, but emerging techniques such as the purification of specific cell types from whole worms may provide cell populations of appropriate homogeneity (121). Lastly, the recent successful adaptation of the CRISPR/Cas9 genome editing technology in C. elegans (122) will enable the introduction of disease-associated point mutations in Mediator subunits in the native genomic context. Combined with existing knowledge on e.g. Mediator subunit action in developmental pathways that parallel signaling circuits deregulated in human cancers, such studies are well suited to assess whether and how disease-associated mutations in human Mediator subunits alter cognate pathway activity. Compared to similar methods in mammalian models, this represents a rapid and comparably inexpensive method to investigate pathological mutations. Thus, studies on C. elegans Mediator will continue to be a valuable tool in basic and translational biomedical research alike.
Acknowledgments
The authors thank Dr Donald G. Moerman for suggesting the idea of the article.
Footnotes
These authors contributed equally to the paper as first authors.
FUNDING
Canadian Institutes of Health Research [MOP-93713 to S.T.]; Natural Sciences and Engineering Research Council of Canada [RGPIN 386398-13 to S.T.]; Canada Foundation for Innovation (to S.T.); Vanier Canada Graduate Scholarship, a Natural Sciences and Engineering Research Council of Canada Canada Graduate Scholarship – Master's, and CFRI and UBC scholarships (to J.M.G.); CFRI and UBC scholarships (G.Y.S.G.); Canada Research Chair in Transcriptional Regulatory Networks. (to S.T.). Funding for open access charge: Operating Grant from the Natural Sciences and Engineering Research Council of Canada [RGPIN 386398-13 to S.T.]. The funders had no role in preparation of the manuscript or the decision to publish.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malovannaya A., Lanz R.B., Jung S.Y., Bulynko Y., Le N.T., Chan D.W., Ding C., Shi Y., Yucer N., Krenciute G., et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiegelman B.M., Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 4.O'Malley B.W., Qin J., Lanz R.B. Cracking the coregulator codes. Curr. Opini. Cell Biol. 2008;20:310–315. doi: 10.1016/j.ceb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik S., Roeder R.G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conaway R.C., Conaway J.W. Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poss Z.C., Ebmeier C.C., Taatjes D.J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koleske A.J., Young R.A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 9.Kim Y.J., Björklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 10.Kelleher R.J., Flanagan P.M., Kornberg R.D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan P.M., Kelleher R.J., Sayre M.H., Tschochner H., Kornberg R.D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 12.Carlsten J.O.P., Zhu X., Gustafsson C.M. The multitalented Mediator complex. Trends Biochem. Sci. 2013;38:531–537. doi: 10.1016/j.tibs.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Bourbon H.-M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai K.-L., Tomomori-Sato C., Sato S., Conaway R.C., Conaway J.W., Asturias F.J. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell. 2014;157:1430–1444. doi: 10.1016/j.cell.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guglielmi B., Van Berkum N.L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H.-M., Holstege F.C.P., Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esnault C., Ghavi-Helm Y., Brun S., Soutourina J., Van Berkum N., Boschiero C., Holstege F., Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 17.Soutourina J., Wydau S., Ambroise Y., Boschiero C., Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 18.Cooper K.F., Khakhina S., Kim S.K., Strich R. Stress-induced nuclear-to-cytoplasmic translocation of cyclin C promotes mitochondrial fission in yeast. Dev. Cell. 2014;28:161–173. doi: 10.1016/j.devcel.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S., Hölzel M., Knijnenburg T., Schlicker A., Roepman P., McDermott U., Garnett M., Grernrum W., Sun C., Prahallad A., et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borggrefe T., Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Spaeth J.M., Kim N.H., Boyer T.G. Mediator and human disease. Semin. Cell Dev. Biol. 2011;22:776–787. doi: 10.1016/j.semcdb.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napoli C., Sessa M., Infante T., Casamassimi A. Unraveling framework of the ancestral Mediator complex in human diseases. Biochimie. 2012;94:579–587. doi: 10.1016/j.biochi.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Markaki M., Tavernarakis N. Modeling human diseases in Caenorhabditis elegans. Biotechnol. J. 2010;5:1261–1276. doi: 10.1002/biot.201000183. [DOI] [PubMed] [Google Scholar]
- 24.Bourbon H.-M., Aguilera A., Ansari A.Z., Asturias F.J., Berk A.J., Björklund S., Blackwell T.K., Borggrefe T., Carey M., Carlson M., et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 25.McKay S.J., Johnsen R., Khattra J., Asano J., Baillie D.L., Chan S., Dube N., Fang L., Goszczynski B., Ha E., et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harbor Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- 26.Thompson O., Edgley M., Strasbourger P., Flibotte S., Ewing B., Adair R., Au V., Chaudhry I., Fernando L., Hutter H., et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 28.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 29.Taubert S., Van Gilst M.R., Hansen M., Yamamoto K.R. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steimel A., Suh J., Hussainkhel A., Deheshi S., Grants J.M., Zapf R., Moerman D.G., Taubert S., Hutter H. The C. elegans CDK8 Mediator module regulates axon guidance decisions in the ventral nerve cord and during dorsal axon navigation. Dev. Biol. 2013;377:385–398. doi: 10.1016/j.ydbio.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Yoda A., Kouike H., Okano H., Sawa H. Components of the transcriptional Mediator complex are required for asymmetric cell division in C. elegans. Development. 2005;132:1885–1893. doi: 10.1242/dev.01776. [DOI] [PubMed] [Google Scholar]
- 32.Wang J.-C., Walker A., Blackwell T.K., Yamamoto K.R. The Caenorhabditis elegans ortholog of TRAP240, CeTRAP240/let-19, selectively modulates gene expression and is essential for embryogenesis. J. Biol. Chem. 2004;279:29270–29277. doi: 10.1074/jbc.M401242200. [DOI] [PubMed] [Google Scholar]
- 33.Kwon J.Y., Kim-Ha J., Lee B.J., Lee J. The MED-7 transcriptional mediator encoded by let-49 is required for gonad and germ cell development in Caenorhabditis elegans. FEBS Lett. 2001;508:305–308. doi: 10.1016/s0014-5793(01)03072-1. [DOI] [PubMed] [Google Scholar]
- 34.Avraham R., Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell. Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 35.Yoon S., Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 36.Karnoub A.E., Weinberg R.A. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell. Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tidyman W.E., Rauen K.A. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternberg P.W. Vulval development. WormBook. 2005;2005:1–28. doi: 10.1895/wormbook.1.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Félix M.-A. Caenorhabditis elegans vulval cell fate patterning. Phys. Biol. 2012;9:045001. doi: 10.1088/1478-3975/9/4/045001. [DOI] [PubMed] [Google Scholar]
- 40.Félix M.-A., Barkoulas M. Robustness and flexibility in nematode vulva development. Trends Genet. 2012;28:185–195. doi: 10.1016/j.tig.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Singh N., Han M. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes Dev. 1995;9:2251–2265. doi: 10.1101/gad.9.18.2251. [DOI] [PubMed] [Google Scholar]
- 42.Tuck S., Greenwald I. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes Dev. 1995;9:341–357. doi: 10.1101/gad.9.3.341. [DOI] [PubMed] [Google Scholar]
- 43.Kwon J.Y., Lee J. Biological significance of a universally conserved transcription mediator in metazoan developmental signaling pathways. Development. 2001;128:3095–3104. doi: 10.1242/dev.128.16.3095. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X., Greenwald I. Spatial regulation of lag-2 transcription during vulval precursor cell fate patterning in Caenorhabditis elegans. Genetics. 2011;188:847–858. doi: 10.1534/genetics.111.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen N., Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell. 2004;6:183–192. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- 46.Stevens J.L., Cantin G.T., Wang G., Shevchenko A., Shevchenko A., Berk A.J. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 47.Nilsson L., Tiensuu T., Tuck S. Caenorhabditis elegans lin-25: a study of its role in multiple cell fate specification events involving Ras and the identification and characterization of evolutionarily conserved domains. Genetics. 2000;156:1083–1096. doi: 10.1093/genetics/156.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schutzman J.L., Borland C.Z., Newman J.C., Robinson M.K., Kokel M., Stern M.J. The Caenorhabditis elegans EGL-15 signaling pathway implicates a DOS-like multisubstrate adaptor protein in fibroblast growth factor signal transduction. Mol. Cell. Biol. 2001;21:8104–8116. doi: 10.1128/MCB.21.23.8104-8116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsubara Y., Kawasaki I., Urushiyama S., Yasuda T., Shirakata M., Iino Y., Shibuya H., Yamanashi Y. The adaptor-like protein ROG-1 is required for activation of the Ras-MAP kinase pathway and meiotic cell cycle progression in Caenorhabditis elegans. Genes Cells. 2007;12:407–420. doi: 10.1111/j.1365-2443.2007.01061.x. [DOI] [PubMed] [Google Scholar]
- 50.Moghal N., Sternberg P.W. A component of the transcriptional mediator complex inhibits RAS-dependent vulval fate specification in C. elegans. Development. 2003;130:57–69. doi: 10.1242/dev.00189. [DOI] [PubMed] [Google Scholar]
- 51.Fay D.S., Yochem J. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 2007;306:1–9. doi: 10.1016/j.ydbio.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T.T.J.P., van Leenen D., Holstege F.C.P. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez D., Hamidi N., Del Sol R., Benschop J.J., Nancy T., Li C., Francis L., Tzouros M., Krijgsveld J., Holstege F.C.P., et al. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2500–2505. doi: 10.1073/pnas.1307525111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X., Zhao M., Xia M., Liu Y., Yan J., Ji H., Wang G. Selective requirement for Mediator MED23 in Ras-active lung cancer. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2813–E2822. doi: 10.1073/pnas.1204311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whittaker S., Kirk R., Hayward R., Zambon A., Viros A., Cantarino N., Affolter A., Nourry A., Niculescu-Duvaz D., Springer C., et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci. Transl. Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 56.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schiano C., Casamassimi A., Rienzo M., de Nigris F., Sommese L., Napoli C. Involvement of Mediator complex in malignancy. Biochim. Biophys. Acta. 2014;1845:66–83. doi: 10.1016/j.bbcan.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Angers S., Moon R.T. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell. Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 59.Clevers H., Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Morris E.J., Ji J.-Y., Yang F., Di Stefano L., Herr A., Moon N.-S., Kwon E.-J., Haigis K.M., Näär A.M., Dyson N.J. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I., Freed E., Ligon A.H., Vena N., Ogino S., et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carrera I., Janody F., Leeds N., Duveau F., Treisman J.E. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korswagen H.C. Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: variations on a common signaling theme. Bioessays. 2002;24:801–810. doi: 10.1002/bies.10145. [DOI] [PubMed] [Google Scholar]
- 64.Clark S.G., Chisholm A.D., Horvitz H.R. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74:43–55. doi: 10.1016/0092-8674(93)90293-y. [DOI] [PubMed] [Google Scholar]
- 65.Ferreira H.B., Zhang Y., Zhao C., Emmons S.W. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev. Biol. 1999;207:215–228. doi: 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- 66.Hunter C.P., Harris J.M., Maloof J.N., Kenyon C. Hox gene expression in a single Caenorhabditis elegans cell is regulated by a caudal homolog and intercellular signals that inhibit wnt signaling. Development. 1999;126:805–814. doi: 10.1242/dev.126.4.805. [DOI] [PubMed] [Google Scholar]
- 67.Zhang H., Emmons S.W. A C. elegans mediator protein confers regulatory selectivity on lineage-specific expression of a transcription factor gene. Genes Dev. 2000;14:2161–2172. doi: 10.1101/gad.814700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H., Emmons S.W. The novel C. elegans gene sop-3 modulates Wnt signaling to regulate Hox gene expression. Development. 2001;128:767–777. doi: 10.1242/dev.128.5.767. [DOI] [PubMed] [Google Scholar]
- 69.Herman M. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development. 2001;128:581–590. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- 70.Zhao X., Yang Y., Fitch D.H.A., Herman M.A. TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development. 2002;129:1497–1508. doi: 10.1242/dev.129.6.1497. [DOI] [PubMed] [Google Scholar]
- 71.Euling S., Ambros V. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell. 1996;84:667–676. doi: 10.1016/s0092-8674(00)81045-4. [DOI] [PubMed] [Google Scholar]
- 72.Clayton J.E., van den Heuvel S.J.L., Saito R.M. Transcriptional control of cell-cycle quiescence during C. elegans development. Dev. Biol. 2008;313:603–613. doi: 10.1016/j.ydbio.2007.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Angus S.P., Nevins J.R. A role for Mediator complex subunit MED13L in Rb/E2F-induced growth arrest. Oncogene. 2012;31:4709–4717. doi: 10.1038/onc.2011.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Powell-Coffman J.A., Knight J., Wood W.B. Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev. Biol. 1996;178:472–483. doi: 10.1006/dbio.1996.0232. [DOI] [PubMed] [Google Scholar]
- 75.Kwon J.Y., Park J.M., Gim B.S., Han S.J., Lee J., Kim Y.J. Caenorhabditis elegans mediator complexes are required for developmental-specific transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14990–14995. doi: 10.1073/pnas.96.26.14990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shim E.Y., Walker A.K., Blackwell T.K. Broad requirement for the mediator subunit RGR-1 for transcription in the Caenorhabditis elegans embryo. J. Biol. Chem. 2002;277:30413–30416. doi: 10.1074/jbc.C200305200. [DOI] [PubMed] [Google Scholar]
- 77.Yin J.-W., Wang G. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 2014;141:977–987. doi: 10.1242/dev.098392. [DOI] [PubMed] [Google Scholar]
- 78.Meissner B., Warner A., Wong K., Dube N., Lorch A., McKay S.J., Khattra J., Rogalski T., Somasiri A., Chaudhry I., et al. An integrated strategy to study muscle development and myofilament structure in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000537. doi: 10.1371/journal.pgen.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmitz C., Kinge P., Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc. Natl. Acad. Sci. 2007;104:834–839. doi: 10.1073/pnas.0510527104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arda H.E., Taubert S., Macneil L.T., Conine C.C., Tsuda B., Gilst M., Sequerra R., Doucette-Stamm L., Yamamoto K.R., Walhout A.J.M. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol. Syst. Biol. 2010;6:367. doi: 10.1038/msb.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang F., Vought B.W., Satterlee J.S., Walker A.K., Jim Sun Z.-Y., Watts J.L., DeBeaumont R., Saito R.M., Hyberts S.G., Yang S., et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 82.Goh G.Y.S., Martelli K.L., Parhar K.S., Kwong A.W.L., Wong M.A., Mah A., Hou N.S., Taubert S. The conserved Mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans. Aging Cell. 2014;13:70–79. doi: 10.1111/acel.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hou N.S., Gutschmidt A., Choi D.Y., Pather K., Shi X., Watts J.L., Hoppe T., Taubert S. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2271–E2280. doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Svensk E., Ståhlman M., Andersson C.-H., Johansson M., Borén J., Pilon M. PAQR-2 regulates fatty acid desaturation during cold adaptation in C. elegans. PLoS Genet. 2013;9:e1003801. doi: 10.1371/journal.pgen.1003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winter J.F., Höpfner S., Korn K., Farnung B.O., Bradshaw C.R., Marsico G., Volkmer M., Habermann B., Zerial M. Caenorhabditis elegans screen reveals role of PAR-5 in RAB-11-recycling endosome positioning and apicobasal cell polarity. Nat. Cell Biol. 2012;14:666–676. doi: 10.1038/ncb2508. [DOI] [PubMed] [Google Scholar]
- 86.Balklava Z., Pant S., Fares H., Grant B.D. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 87.Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 88.Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 89.Libina N., Berman J.R., Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 90.Zhang P., Judy M., Lee S.-J., Kenyon C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 2013;17:85–100. doi: 10.1016/j.cmet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taubert S., Hansen M., Van Gilst M.R., Cooper S.B., Yamamoto K.R. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 2008;4:e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rogers A.N., Chen D., Mccoll G., Czerwieniec G., Felkey K., Gibson B.W., Hubbard A., Melov S., Lithgow G.J., Kapahi P. Life span extension via eIF4G inhibition Is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14:55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Van Gilst M.R., Hadjivassiliou H., Yamamoto K.R. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pang S., Lynn D.A., Lo J.Y., Paek J., Curran S.P. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 2014;5:5048. doi: 10.1038/ncomms6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taubert S., Ward J.D., Yamamoto K.R. Nuclear hormone receptors in nematodes: evolution and function. Mol. Cell. Endocrinol. 2011;334:49–55. doi: 10.1016/j.mce.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thakur J.K., Arthanari H., Yang F., Chau K.H., Wagner G., Näär A.M. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 2009;284:4422–4428. doi: 10.1074/jbc.M808263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Naar A.M., Thakur J.K. Nuclear receptor-like transcription factors in fungi. Genes Dev. 2009;23:419–432. doi: 10.1101/gad.1743009. [DOI] [PubMed] [Google Scholar]
- 99.Phelps C., Gburcik V., Suslova E., Dudek P., Forafonov F., Bot N., MacLean M., Fagan R.J., Picard D. Fungi and animals may share a common ancestor to nuclear receptors. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7077–7081. doi: 10.1073/pnas.0510080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pukkila-Worley R., Feinbaum R.L., McEwan D.L., Conery A.L., Ausubel F.M. The evolutionarily conserved Mediator subunit MDT-15/MED15 links protective innate immune responses and xenobiotic detoxification. PLoS Pathog. 2014;10:e1004143. doi: 10.1371/journal.ppat.1004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones L.M., Rayson S.J., Flemming A.J., Urwin P.E. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS ONE. 2013;8:e69956. doi: 10.1371/journal.pone.0069956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oliveira R.P., Abate J.P., Dilks K., Landis J., Ashraf J., Murphy C.T., Blackwell T.K. Condition-adapted stress and longevity gene regulation byCaenorhabditis elegansSKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Warfield L., Tuttle L.M., Pacheco D., Klevit R.E., Hahn S. A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3506–E3513. doi: 10.1073/pnas.1412088111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brzovic P.S., Heikaus C.C., Kisselev L., Vernon R., Herbig E., Pacheco D., Warfield L., Littlefield P., Baker D., Klevit R.E., et al. The acidic transcription activator Gcn4 binds the Mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell. 2011;44:942–953. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herbig E., Warfield L., Fish L., Fishburn J., Knutson B.A., Moorefield B., Pacheco D., Hahn S. Mechanism of mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol. Cell. Biol. 2010;30:2376–2390. doi: 10.1128/MCB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gracida X., Eckmann C.R. Fertility and germline stem cell maintenance under different diets requires nhr-114/HNF4 in C. elegans. Curr. Biol. 2013;23:607–613. doi: 10.1016/j.cub.2013.02.034. [DOI] [PubMed] [Google Scholar]
- 107.Lindblom T.H., Pierce G.J., Sluder A.E. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr. Biol. 2001;11:864–868. doi: 10.1016/s0960-9822(01)00236-6. [DOI] [PubMed] [Google Scholar]
- 108.Reece-Hoyes J.S., Pons C., Diallo A., Mori A., Shrestha S., Kadreppa S., Nelson J., DiPrima S., Dricot A., Lajoie B.R., et al. Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol. Cell. 2013;51:116–127. doi: 10.1016/j.molcel.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shore D.E., Carr C.E., Ruvkun G. Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. PLoS Genet. 2012;8:e1002792. doi: 10.1371/journal.pgen.1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Samuelson A.V., Carr C.E., Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Crook-McMahon H.M., Oláhová M., Button E.L., Winter J.J., Veal E.A. Genome-wide screening identifies new genes required for stress-induced phase 2 detoxification gene expression in animals. BMC Biol. 2014;12:64. doi: 10.1186/s12915-014-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nicholas H.R., Hodgkin J. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr. Biol. 2004;14:1256–1261. doi: 10.1016/j.cub.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 113.Hodgkin J., Kuwabara P.E., Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 2000;10:1615–1618. doi: 10.1016/s0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- 114.Barabási A.-L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 115.Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A.G. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 116.Graham J.M., Schwartz C.E. MED12 related disorders. Am. J. Med. Genet. A. 2013;161:2734–2740. doi: 10.1002/ajmg.a.36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hashimoto S., Boissel S., Zarhrate M., Rio M., Munnich A., Egly J.-M., Colleaux L. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science. 2011;333:1161–1163. doi: 10.1126/science.1206638. [DOI] [PubMed] [Google Scholar]
- 118.Zhou H., Spaeth J.M., Kim N.H., Xu X., Friez M.J., Schwartz C.E., Boyer T.G. MED12 mutations link intellectual disability syndromes with dysregulated GLI3-dependent Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19763–19768. doi: 10.1073/pnas.1121120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mehine M., Mäkinen N., Heinonen H.-R., Aaltonen L.A., Vahteristo P. Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil. Steril. 2014;102:621–629. doi: 10.1016/j.fertnstert.2014.06.050. [DOI] [PubMed] [Google Scholar]
- 120.Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.-P., White T.A., Stojanov P., Van Allen E., Stransky N., et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang S., Banerjee D., Kuhn J.R. Isolation and culture of larval cells from C. elegans. PLoS ONE. 2011;6:e19505. doi: 10.1371/journal.pone.0019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dickinson D.J., Ward J.D., Reiner D.J., Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]


