Abstract
Methylation of ribose sugars at the 2′-OH group is one of the major chemical modifications in rRNA, and is catalyzed by snoRNA directed C/D box snoRNPs. Previous biochemical and computational analyses of the C/D box snoRNAs have identified and mapped a large number of 2′-OH ribose methylations in rRNAs. In the present study, we systematically analyzed ribose methylations of 18S rRNA in Saccharomyces cerevisiae, using mung bean nuclease protection assay and RP-HPLC. Unexpectedly, we identified a hitherto unknown ribose methylation at position G562 in the helix 18 of 5′ central domain of yeast 18S rRNA. Furthermore, we identified snR40 as being responsible to guide snoRNP complex to catalyze G562 ribose methylation, which makes it only second snoRNA known so far to target three ribose methylation sites: Gm562, Gm1271 in 18S rRNA, and Um898 in 25S rRNA. Our sequence and mutational analysis of snR40 revealed that snR40 uses the same D′ box and methylation guide sequence for both Gm562 and Gm1271 methylation. With the identification of Gm562 and its corresponding snoRNA, complete set of ribose methylations of 18S rRNA and their corresponding snoRNAs have finally been established opening great prospects to understand the physiological function of these modifications.
INTRODUCTION
RNA modifications are present in all three kingdoms of life and detected in all classes of cellular RNAs. RNA modifications are diverse, with >100 types of chemical modifications identified to date (1). These chemical modifications provide RNA higher complexity and expand its topological potential, which otherwise is limited by four bases (2).
Ribosomal RNA (rRNA) contains two types of covalent modifications, either methylation on the sugar (Nm) or bases (mN), or base isomerization (conversion of uridine into pseudouridines (Ψ)). Pseudouridylations and ribose methylations are catalyzed by site-specific H/ACA and C/D box snoRNPs, respectively (3,4). The RNA component (snoRNA) of both types of snoRNPs is responsible for the site selection by base pairing with the rRNA substrate, whereas the protein component catalyzes the modification reaction: Nop1 in C/D box and Cbf5 in H/ACA box snoRNPs (5–7). Contrastingly, base methylations are performed by snoRNA independent, ‘protein-only’, methyltransferases (MTases) (8).
In Saccharomyces cerevisiae, 18S rRNA of the small subunit contains 14 Ψs, 17 Nms and 4 mNs, whereas 25S rRNA of the large subunit contains 30 Ψs, 37 Nms and 6 base methylations (9). Homo sapiens contains ∼91 Ψs, 105 Nms and 10 mNs (9). Mapping of these modifications has revealed that these modifications cluster in the functionally conserved regions of the ribosomes like intersubunit and peptidyl transferase center (10). Interestingly, in vitro reconstitution of bacterial ribosomes has highlighted the importance of these chemical modifications in the functioning and synthesis of ribosomes. 30S subunits assembled with in vitro transcribed 16S rRNA, lacking chemical modifications showed only 50% tRNA-binding capacity, whereas chemical modifications of the rRNA of large ribosomal subunit, 23S is necessary for the assembly of the 50S (11,12). Seven modifications around PTC in domain V of the 23S rRNA (2445–2523) turned out to be indispensable for in vitro reconstitution of functional particles (12).
Since in vitro reconstitution of eukaryotic ribosomes is so far not feasible, to further analyse and explore the function of these modifications in ribosome biogenesis and functioning, it is quite important to identify the machinery responsible for these chemical modifications; snoRNA in case of snoRNPs and base methyltransferases for snoRNA independent modifications.
Methylation of 2′-OH of ribose sugar to a 2′-O-methyl is a characteristic modification in both mRNAs and non-coding RNAs (ncRNA) including tRNAs, rRNAs and siRNAs (13). Interestingly, ribose methylation favors a 3′-endo conformation of the ribose and since 3′-endo conformations are known to stabilize A-form helices, methylation of ribose increases the rigidity of the RNA by promoting base stacking (14,15). Furthermore, ribose methylation provides RNA stability against hydrolysis by bases and nucleases. Intriguingly, the analysis of the chemical composition of RNA from thermophiles has revealed that these organisms contain a higher amount of ribose methylation, supporting their role in stabilization of RNA (16,17).
C/D box snoRNA protein (snoRNPs) complexes catalyse ribose methylation in the rRNA of eukaryotes including yeast S. cerevisiae, and comprise of four common core proteins: Fibrillarin/Nop1, Nop58, Nop56 and Snu13 along with a site-specific C/D box snoRNA (6). C/D box snoRNAs are characterized by conserved and distinguishing sequence elements called boxes C/C′ (5′-RUGAUGA-3′), D/D′ (5′-CUGA-3′), and guide sequences that base pair to the RNA target (15). The methylation guide sequences are positioned upstream of the box D/D′ element and consists of 10–21 nucleotides (3). The guide sequences direct ribose methylation to the nucleotide base paired to the fifth nucleotide up-stream of the box D or D′ sequence (box D+5 rule) (3). Nop1 is a S-adenosyl methionine (SAM) dependent methyltransferase and catalyzes the 2′-O-methylation reaction. Snu13 binds to the kink-turn (loop–stem structure that includes the canonical C/D elements in the loop portion) in the C/D box snoRNA. Nop56 and Nop58 are characterized by extensive coiled-coil domains, likely responsible for hetero-dimerization and providing stability to the snoRNA (6).
Although the precise role of ribose methylations in rRNA remained a mystery in single cell organisms like yeast, multicellular organisms display specific phenotypes upon loss of specific or functionally related ribose methylations (18). Recent studies have also emphasized their role in murine and human cancers development, especially in breast and prostate cancer. Both Nop1 and snoRNAs are overexpressed in cancer tissues and this overexpression has been shown to be crucial for tumorigenicity and suppression of the elevated snoRNA pathway considerably concedes tumorigenicity, via activation of p53 (19).
In the present study, we analyzed ribose methylations of the yeast 18S rRNA by RP-HPLC in combination with mung bean nuclease assay and identified a new ribose methylation at position 562 in the helix 18 of 5′ central domain of the yeast 18S rRNA. Furthermore, we also identified snR40 as the corresponding snoRNA to guide the respective snoRNP to the newly identified G562 methylation site.
MATERIALS AND METHODS
Growth conditions and yeast media
Yeast strains were grown at 30°C either in YPD media (1% yeast extract, 2% peptone, 2% glucose) or in SCD-Ura media. For antibiotic analysis, 5 μl of a paromomycin solution with different concentrations (50–400 mg/ml) were spotted on filter discs, which were then placed on YPD plates containing the strains to be tested.
Plasmids and strains
All strains used in the present study are listed in Supplementary Table S1. Plasmid pJN22 was constructed by ‘gap repairing’ using the primers snr40FP and snr40RP listed in Supplementary Table S2. For introducing specific point mutations, PCR site-directed mutagenesis using Single-Primer Reactions IN Parallel (SPRINP) using high-fidelity Pfu-DNA polymerase (Promega) was used (20). Primers used for constructing pJN22a and pJN22b are listed in Supplementary Table S2.
Preparation of ribosomal RNA
18S rRNA for the RP-HPLC analysis was prepared using sucrose gradient analysis. Ribosomal subunits were separated on a 20–50% sucrose density gradient and the 18S rRNA was then extracted from the fractions collected from 40S subunit. Separation of the subunits were performed by sucrose gradient ultracentrifugation in a SW28 rotor (Beckman Coulter, Inc.) for 20 h at 21 000 rpm and 4°C as explained previously.
40S subunits were collected with the Density Gradient Fractionation System (Teledyne Isco) and precipitated with 2.5 vol. of 100% ethanol at −20°C for 16 h. Precipitated 40S subunits were dissolved in water and 18S rRNA was purified using the RNeasy Kit (QIAGEN) following the protocol for RNA cleanup. RNA was eluted in two steps with 65 μl water each. The quality of 18S rRNA was then analyzed on 1% agarose gel.
Mung bean nuclease protection assay
Mung bean nuclease protection assay was performed exactly as described previously (21). Complementary synthetic deoxyoligonucleotides were used for hybridization and protection of specific sequence of 18S rRNA. Two thousand picomoles of the synthetic deoxyoligonucleotides complementary to yeast 18S rRNA were incubated with 100 pmol of total rRNA and were digested by mung bean nuclease (MBN Kit: M0250S, NEB) and 0.05 mg/ml RNase A (Sigma–Aldrich). Protected fragments were purified by denaturing 8 M urea-PAGE (13%) and were eluted passively using with 0.3 M NaAc on a shaker, overnight at 4°C. Eluted fragments were then precipitated using 100% EtOH.
High performance liquid chromatography
For HPLC analysis, 100 pmol 18S rRNA was digested with nuclease P1 and bacterial alkaline phosphatase (Sigma–Aldrich) according to the method of Gehrke and Kuo (22). Nucleosides were analyzed by RP-HPLC on a Supelcosil LC-18-S HPLC column (25 cm x 4.6 mm, 5 μm) equipped with a precolumn (4.6 × 20 mm) at 30°C on an Agilent 1200 HPLC system. A gradient elution using buffer A (10 mM NH4H2PO4, 2.5% methanol at pH 5.3) and buffer B (10 mM NH4H2PO4, 20% methanol at pH 5.1) were used as described previously (23). In our RP-HPLC set up, Gm residues have a retention time of ∼30 min. Amount of Gm was calculated using their UV extinction coefficient and normalized to guanosine residues as explained previously (24).
RNA isolation and northern Blotting
Total RNA was isolated using phenol/chloroform extraction as previously described (25). Ten micrograms of total RNA was separated on 1.5% agarose for rRNA processing analysis or on 6 M urea-PAGE (10%) gel for snoRNA analysis, and transferred to a positively charged nylon membrane (Hybond N+, GE Healthcare) using vacuum blotting. Fifty picomoles of the corresponding oligonucleotide was radioactively labeled at the 5′ end using 6 μl γ-[32P] ATP (∼3.3 pmol/μl, Hartmann-Analytics) and 1 μl T4 polynucleotide kinase (Roche) in the supplied buffer for 1 h at 37°C and purified with G-25 column. Hybridization was performed in 20 ml hybridization buffer (GE Healthcare) overnight at 42°C and signal were visualized by phosphoimaging using a Typhoon 9400 (GE Healthcare).
Polysome profiling
Polysome profiles were performed exactly as described previously (26). Yeast strains were grown in YPD media (100 ml) at 30°C. Cycloheximide (100 μg/ml) was added before cell disruption. Yeast cells (2 × 109) were harvested by centrifugation at 4°C. Cells were then washed twice with 10 ml of polysome buffer A (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA and 1 mM Dithiothreitol (DTT)), resuspended in 0.5 ml of buffer A and disrupted by vortexing with an equal volume of glass beads. Equivalent amounts of absorbing material were layered on a 10–50% (w/v) sucrose gradient in buffer A. A gradient was made using Gradient Master 107 (Biocomp). Samples were then centrifuged at 19 000 rpm for 17 h at 4°C in a SW40 rotor using Beckman ultracentrifuge (L-70: Beckman). Gradients were fractionated in an ISCO density gradient fractionar and the absorbance profile at 254 nm was analyzed in ISCO UA-5 absorbance monitor.
RESULTS
Systematic mung bean nuclease assay of 18S rRNA reveals hitherto unidentified ribose methylation
Biochemical and computational analyses of ribose methylations in 18S rRNA of S. cerevisiae have identified and mapped most of the residues, along with their corresponding snoRNPs (3,27). Nevertheless, Rudi Planta′s lab in early 1970s provided evidence for a fifth Gm residue in 18S rRNA, which if correct remained to be mapped (28).
To clarify if there are still ribose methylations that have not been annotated until now, we systematically analyzed the entire 18S rRNA with mung bean nuclease assays and RP-HPLC. As illustrated in Figure 1A, using mung bean nuclease assay, we first isolated 45 distinct, overlapping fragments of 50 nucleotides in size, spanning whole 18S rRNA. All these fragments were digested to nucleosides, and the composition of each fragment was next analyzed separately by RP-HPLC.
Figure 1.
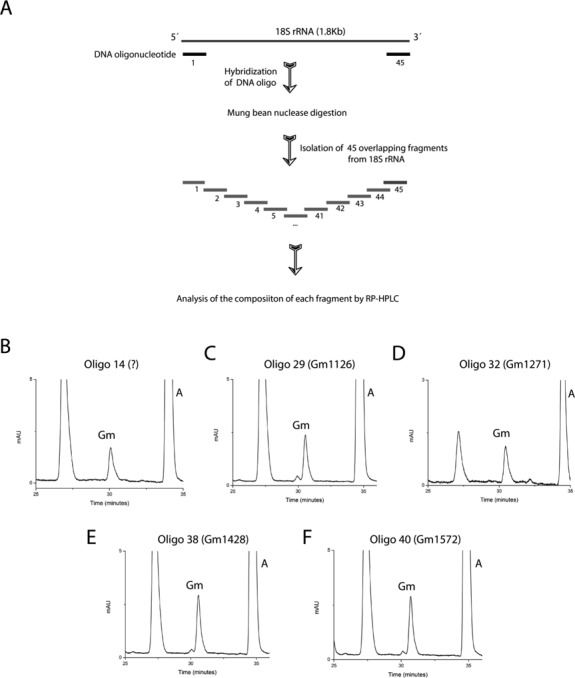
Systematic mung bean nuclease assay of ribose methylations of the 18S rRNA. (A) Graphic illustration of mung bean nuclease protection assay used in the present study for the analysis of ribose methylations of 18S rRNA in S. cerevisiae. 45 distinct overlapping fragments were isolated using mung bean nuclease assay from the 18S rRNA and were systematically analysed by RP-HPLC for the presence of 2′-O-ribose methylated guanosine (Gm). RP-HPLC chromatogram of five distinct fragments isolated using oligo 14 (B), 29 (C), 32 (D), 38 (E) and 40 (F) exhibited the presence of five Gm residues. Four out of five of these Gm residues corresponding to Gm1126 (oligo 29), Gm1271 (oligo 32), Gm1428 (oligo 38) and Gm1572 (oligo 40) were also previously mapped. Existence of Gm in the fragments isolated using oligo 14 (B) highlighted the presence of a new Gm residue in the fragment covering nucleotides from 527 to 576 of the 18S rRNA.
As evident in Figure 1, using this approach we could not only identify Gm residue in the fragments 29, 32, 38 and 40, corresponding to position 1126, 1271, 1428 and 1572, respectively, but could also identify a new Gm residue in the fragment 14 encompassing nucleotides from 527 to 576 of the 18S rRNA. Apart from Gm residues we could also map other 2′-O-ribose methylated nucleotides (data not shown).
Mapping of precise location of new Gm residue in the 18S rRNA
To further locate the precise G within the fragment 14 that contains 2′-O-ribose methylation, four overlapping fragments corresponding to the regions displayed in Figure 2A were isolated. Mung bean nuclease assay using oligo-558, oligo-561, oligo-562 and oligo-564, each 50 nucleotides in size were performed to isolate these fragments. The compositions of these fragments were next analyzed using RP-HPLC and their respective chromatograms suggested G562 to contain 2′-O-ribose methylation (Figure 2B, C, D and E). A very slight amount of Gm residue was also observed in 18S rRNA fragment isolated using oligo 561(Figure 2C), which most likely is derived from adjacent Gm562. Interestingly, previous 2D electrophoresis analysis of T1 nuclease digests in combination with dinucleotides analyses of alkali digests from Rudi Planta′s lab of the yeast 18S rRNA had predicted the methylation site to be GmU, which is in complete agreement with our mapping results: Gm562 is followed by U at 563 position (28).
Figure 2.
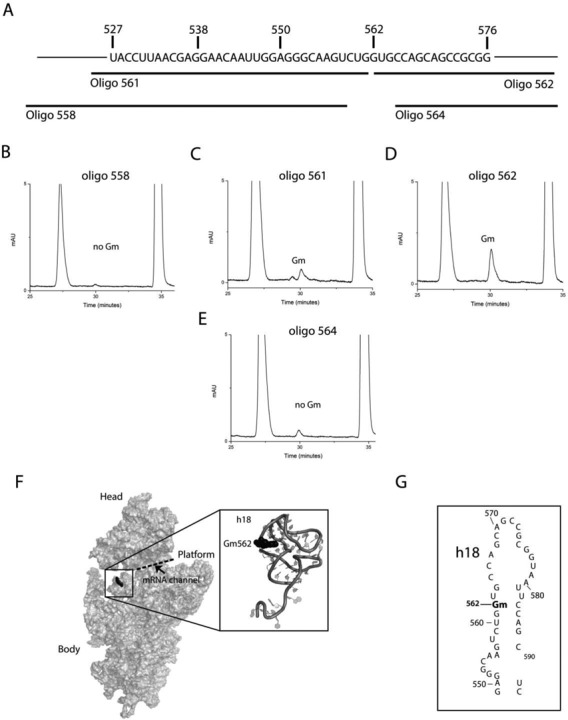
Mapping of precise location of new Gm residue in the 18S rRNA. To identify precise location of the Gm residue within fragment covering nucleotides from 527 to 576 of 18S rRNA, shown in panel (A), four overlapping fragments using oligo-558, oligo-561, oligo-562 and oligo-564, each 50 nucleotides in size were isolated using mung bean nuclease assay. Nucleosides composition of these fragments was next analyzed using RP-HPLC. RP-HPLC chromatogram of oligo-558 (B), oligo-561(C), oligo-562 (D) and oligo-564 (E). The compositions of these fragments suggested G562 to contain 2′-O-ribose methylation in the 18S rRNA. Insignificant amount of Gm residue was also observed in 18S rRNA fragment isolated using oligo-561, which most likely is derived from neighboring Gm562. (F) 3D cartoon of the yeast 18S rRNA highlighting the location of Gm562 (black spheres) with a zoomed in view of helix 18. PDB file 3U5B was used for the representation of 18S ribosomal RNA. The cartoon was made by PyMol software (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.). (G) 2D sequence map of the helix 18 of the yeast 18S rRNA.
Sequence analysis of the methylation guide sequence of C/D box snoRNAs suggested involvement of snR40 in Gm562 methylation
Most of the ribose methylations in yeast are mediated by C/D box snoRNA-directed complexes (3,8,27). C/D box snoRNA provide substrate specificity, whereas Nop1 catalyzes the methylation reaction. These snoRNAs contain a short segment (10–20 nucleotides) complementary to the target RNA and as mentioned above the methylation site depends on its relative distance to conserved box D (or box D′) sequence. This distance is precisely conserved to five nucleotides and the ribose sugars of the fifth nucleotide upstream to the D or D′ box are methylated, irrespective of the nature of the base (A, G, C or U).
To identify the snoRNA involved in the methylation at Gm562, we started with reanalyzing the snoRNA sequences, especially the 10–20 nucleotides involved in assigning substrate specificity of all already known C/D box snoRNAs, shown previously to be involved in ribose methylation of G (3). We hypothesized that since the relative distance to the D or D′ box is always fixed to five nucleotides, only a snoRNA involved in Gm is more likely to be involved in methylation at Gm562, unless we have encountered an exception to the rule. Sequence analyses of these 10–20 nucleotides among snoRNA involved in Gm methylation of both 18S and 25S rRNA suggested the possibility of snR40 to be the most likely candidate for the Gm562 (Figure 3A).
Figure 3.
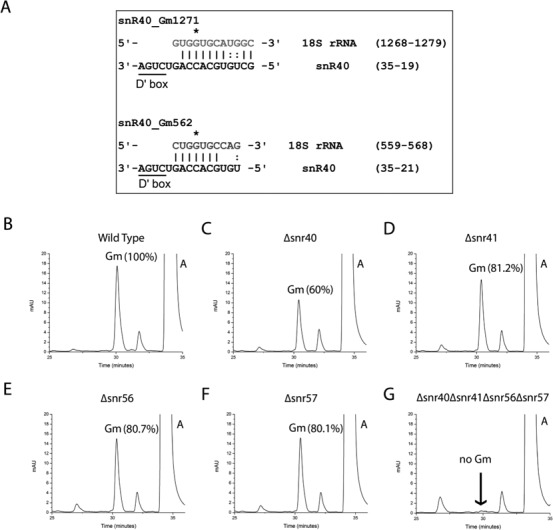
Sequence and RP-HPLC analysis of the C/D box snoRNAs for the identification of snoRNA involved in Gm562. To identify snoRNA involved in the methylation of Gm562, we reanalyzed methylation guide sequences involved in assigning substrate specificity to the C/D box snoRNPs of all already known C/D box snoRNAs. (A) Our sequence analysis revealed the likelihood of snR40 to be the snoRNA involved in guiding methylation of Gm562 residue. Sequence analysis of the methylation guide sequence of snR40 shows significant complementarity to the region surrounding Gm562 in the 18S rRNA. Ribose methylated G is marked with star and is always five nucleotides upstream to the D/D′ box. We next checked the involvement of snR40 in the methylation of Gm562. Nucleoside composition of 18S rRNA isolated from snR40 deletion mutant was analyzed, and compared with the single deletion mutants of three other snoRNAs; snr41, snr56 and snr57. RP-HPLC chromatogram of WT (B), Δsnr40 (C), Δsnr41 (D), Δsnr56 (E) and Δsnr57 (F) demonstrated that where single deletions of snr41, snr56 and snr57 led only to 20% decrease in Gm amount compared to wild type 18S rRNA, deletion of snr40 led to 40% decrease in the Gm amount. This clearly indicated that loss of snr40 influences methylation of more than one Gm residues and supported involvement of snR40 in Gm562 methylation. (G) RP-HPLC chromatogram of the quadruple mutant (Δsnr40Δsnr41Δsnr56Δsnr57). Deletion of all four snoRNAs (snR40, snR41, snR56 and snR57) leads to complete loss of Gm residues from the 18S rRNA. Together with composition analysis of 18S rRNA from single deletion mutant, analysis with quadruple mutant further supported the involvement of snR40 in methylation of Gm562.
To check the involvement of snR40 in the methylation of Gm562, we next analyzed composition of 18S rRNA from snR40 deletion mutant, and compared it with the single deletion mutants of three other snoRNAs; snR41, snR56 and snR57, which have been previously shown to be involved in methylation of rest of the previously mapped Gm residue at position 1126, 1428 and 1572 of the 18S rRNA, respectively (3,27). Interestingly, as seen in Figure 3, single deletions of snR41, snR56 and snR57 led only to 20% decrease in Gm amount, whereas deletion of snR40 led to 40% decrease in the Gm amount. This clearly indicated that loss of snR40 influences methylation of more than one Gm residues.
To further validate participation of snR40 in methylation of Gm562, we constructed a quadruple mutant, where we deleted all four snoRNAs involved in Gm methylation of 18S rRNA: snR40, snR41, snR56 and snR57, and analyzed the 18S rRNA from this quadruple mutant. As seen in Figure 3G deletion of all four snoRNA led to complete loss of Gm residues from the 18S rRNA, which further augments the importance of snR40 in Gm562 methylation.
Deletion of snR40 leads to complete loss of methylation at Gm562
To substantiate the specific involvement of snR40 in the methylation of Gm562, we isolated the fragment 14 (527–576) corresponding to Gm562 from all four single deletion mutants of snoRNAs (Δsnr40, Δsnr41, Δsnr56 and Δsnr57), using mung bean nuclease digestion assay. As evident in Figure 4, RP-HPLC analysis of the fragment 14 from respective deletion mutant explicitly demonstrated that only deletion of snR40 leads to complete loss of Gm at position 562, whereas loss of other snoRNAs do not influence the methylation of this residue. This clearly showed that snR40 is involved in methylation of Gm at potion 562 in the helix 18 of 5′ central domain of 18S rRNA.
Figure 4.

Deletion of snr40 leads to complete loss of methylation at Gm562. To demonstrate the specific involvement of snR40 in the methylation of Gm562, 18S rRNA fragment corresponding to Gm562 was isolated from all four single deletion mutants of snoRNAs (Δsnr40, Δsnr41, Δsnr56 and Δsnr57), using oligo 14 by mung bean nuclease digestion assay. Fragment from all deletion mutants was subsequently analyzed using RP-HPLC. RP-HPLC chromatograms of fragment corresponding to Gm562 isolated from WT (A), Δsnr40 (B), Δsnr41 (C), Δsnr56 (D) and Δsnr57 (E) explicitly demonstrated that only deletion of snr40 leads to complete loss of Gm562. Deletion of all rest of three snoRNAs do not affect the methylation of Gm562.
Episomal expression of snR40 restores the methylation at Gm562 in Δsnr40 deletion mutant
To further corroborate the role of snR40 in mediating Gm562 methylation and to rule out any secondary mutations as the cause of loss of ribose methylation at Gm562 in Δsnr40 deletion mutant, we performed a complementation study, where we episomally expressed snR40 using a plasmid pJN22 in Δsnr40 deletion mutant and checked if this restores methylation at Gm562. As observed in Figure 5A and B, RP-HPLC analysis of the 18S rRNA fragment corresponding to Gm562 from Δsnr40 deletion strain carrying plasmid pRS426 (empty plasmid) and pJN22 (snR40) exhibited that episomal expression of SNR40 indeed restored the methylation at Gm562.
Figure 5.
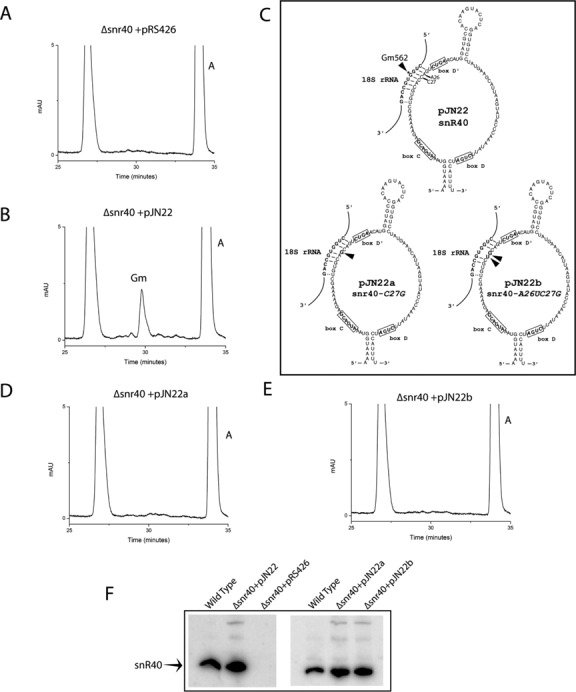
Episomal expression of snR40 in Δsnr40 deletion strain restores the Gm562 methylation. To substantiate the role of snR40 in mediating Gm562 methylation and to exclude any secondary mutations as the cause of loss of ribose methylation at Gm562 in Δsnr40 deletion mutant, we episomally expressed snR40 using a plasmid pJN22 in Δsnr40 deletion mutant and tested if this restores methylation at Gm562. RP-HPLC chromatogram of the fragment corresponding to Gm562 in Δsnr40 deletion strain carrying plasmid pRS426 (empty plasmid) (A) and snr40 deletion strain carrying plasmid pJN22 (snR40) evidently demonstrated specific involvement of snR40 in Gm562 methylation. Additionally to demonstrate direct involvement of snR40, especially methylation guide sequence of snR40, we point mutated C27G of snR40 alone and in combination A26U C27G. (B) Graphical representation of 2D structure of wild type snR40 expressed from pJN22 and mutant snR40, snr40-C27G and snr40-A26UC27G expressed from pJN22a and pJN22b respectively. Next, we analyzed the status of Gm562 in Δsnr40 deletion strain expressing pJN22a and pJN22b by mung bean nuclease assay and RP-HPLC. RP-HPLC chromatogram of fragments corresponding to Gm562 in Δsnr40+pJN22a (D) and Δsnr40+pJN22b (E) revealed that obstructing base paring, especially between C27 and G562 alone and in combination with A26 U563 within the guide sequence leads to complete loss of Gm562 methylation (C). To validate expression of both wild type snR40 from plasmid pJN22 and mutant snr40 from plasmid pJN22a and pJN22b, we performed northern blotting using 32P-labeled oligo specific to snR40. (F) Northern blot analysis confirmed the expression of both wild type and mutant snR40 from plasmid pJN22, pJN22a and pJN22b, respectively.
Furthermore, to demonstrate direct involvement of the methylation guide sequence of snR40, especially the fifth nucleotide upstream of D′ box of snR40 (C27) that base pairs with the G562 of 18S rRNA in guiding Gm562 methylation, we point mutated C27G of snR40 (pJN22a) alone and in combination A26U C27G (pJN22b) (Figure 5C). Both point-mutated snR40 were expressed from plasmids, pJN22a and pJN22b, respectively. As observed in Figure 5D and E, exchange of C27G and A26U C27G completely abolished Gm562 methylation. Northern blot analysis confirmed the expression of both point-mutated snr40 from the plasmid pJN22a and pJN22b (Figure 5F). Since snR40 uses same guide sequence for methylation at Gm562 and Gm1271, we also tested if the methylation at Gm1271 is affected in both point mutated snR40 snoRNAs. As shown in Supplementary Figure S1A and B, exchange of both C27U and A26U C27G also led to complete loss of ribose methylation at Gm1271. Taken together our result clearly underlined the unambiguous involvement of snR40 in the methylation of Gm562 and indispensability of C27 residue of snR40 for the methylation reaction both at Gm562 and Gm1271.
Loss of Gm562 is dispensable for growth, rRNA processing and translation
snR40 was previously shown to be involved in ribose methylation of Gm1271 of 18S rRNA of the small subunit and Um898 of 25S rRNA of the large subunit of ribosome (27). With the identification of its new target in small subunit at G562, we next investigated if the loss of this snoRNA involved in ribose methylations at three positions leads to any growth, rRNA processing and translation defects. Surprisingly, deletion of snR40 did not exhibit any growth phenotype at 19, 30 and 37°C (Figure 6A). Similarly, as shown in Figure 6B, C and E, polysomes profile and northern blotting revealed that loss of snR40 also did not effect the processing of rRNA and steady state levels of 40S, 60S and polysomes. Antibiotic sensitivity analyses are crucial tool for analyzing any change in conformation of rRNA upon the loss of modification (2,29,30). Therefore, we also checked if loss of Gm methylations mediated by snR40 lead to any such antibiotic hypersensitivity. Since both Gm methylations mediated by snR40 are located in the decoding center we first analyzed if deletion of snr40 influence paromomycin sensitivity. As seen in Figure 6F, Δsnr40 did not show any paromomycin hypersensitivity compared with wild type. Similarly, we also did not observe any hypersensitivity to anisomycin and cycloheximide (data not shown).
Figure 6.
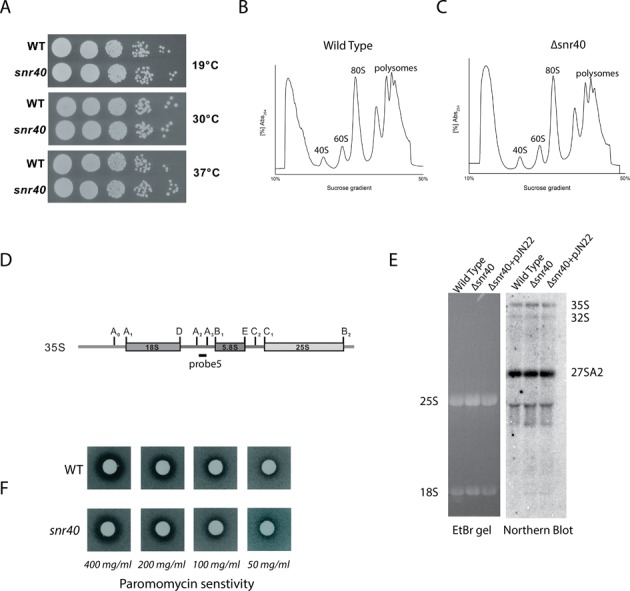
Growth, polysome and rRNA processing analysis for Δsnr40. (A) Ten-fold serial dilutions of the strains were spotted onto solid YPD plates and were incubated at different temperatures. (B) Polysome profile of isogenic wild type (B) and Δsnr40 (C). (D) Illustration for the 35S primary transcript. 35S rRNA contains 18S, 5.8S and 25S rRNA sequences separated by ITS1 and ITS2. Processing of 35S rRNA to mature rRNA involves endonucleolytic and exonucleolytic steps at specific sites. (E) Ethidium bromide (EtBr) stained gel-carrying mature 25S and 18S rRNA from wild type, Δsnr40 and Δsnr40 + pJN22 strains separated on 1% agarose gel along with Northern blot analysis of the rRNA processing in WT, Δsnr40 and Δsnr40+pJN22. The membrane was hybridized with radioactive (32P) labeled probe 5 for ITS1. (F) Paromomycin sensitivity test was performed by spotting 5 μl paromomycin solution on filter discs, which were then applied on YPD plates containing Δsnr40 strain.
DISCUSSION
Rudi Planta's lab demonstrated for the first time in 1973 that 18S rRNA of yeast contains five 2′-O-ribose methylated guanosine (Gm) (28). Surprisingly, subsequent biochemical and computational analyses of 18S rRNA led to the identification of only four Gm residues at position 1126, 1271, 1428 and 1572 (3,27). Thus, the presence of any other Gm residue in the 18S rRNA remained dubious. In the present study, using mung bean nuclease assay in combination with RP-HPLC, we not only validated presence of fifth Gm residue in the 18S rRNA but also mapped its precise location to position 562 in the 18S rRNA, in the decoding center. Our present analysis of ribose methylations also validated locations of all other previously identified ribose methylations in the 18S rRNA.
Additionally, using in silico sequence analysis of C/D box snoRNAs together with biochemical analysis we also successfully identified the snoRNA involved in methylation of Gm562. We explicitly demonstrated that 2′-O-ribose methylation of G562 is mediated by snR40. snR40 was previously shown to be involved in ribose methylation of both 18S rRNA of small subunit at Gm1271 and 25S rRNA of large subunit at Um898 (27). With the identification of Gm562 as another target of snR40, it is just second snoRNA known so far to mediate three ribose methylations; Gm562, Gm1271 and Um898. Initially, it appears surprising, since C/D box snoRNAs cannot afford to have more than two targets due to presence of only maximum of two site-specific sequences upstream to D and D' box (3,31). Intriguingly, it is the sequence similarity and flexibility of methylation guide sequence that facilitates snR40 to mediate modification at third site. Using specific point mutations, C27U and A26U C27G in the guide sequence upstream to D′ box, we clearly demonstrated that snR40 uses same D' box to target methylation at both G562 and G1271. Interestingly Um898 remained unaffected in both snR40 point mutants demonstrating that these two point mutants specifically effect the Gm methylations at 562 and 1271 of the 18S rRNA (data not shown). Therefore, although snR40 targets three residues, the D+5 rule proposed for site directed methylations of C/D box snoRNA still holds true (3). Targeting three residues by a single snoRNA could also provide a mean of regulation of the functioning of both subunits of ribosomes, but so far we could not see any effect of deletion of snR40 on growth, rRNA processing and translation. Future studies using ribosome profiling and structural probing should be employed to reveal any subtle changes in the translation and local structure of ribosome upon loss of these modifications mediated by snR40.
Sequence alignment of helices 17 and 18 of 18S rRNA (proximal to Gm562 of yeast) among various eukaryotes showed that residue G562 is highly conserved in both lower and higher eukaryotes including humans (Supplementary Figure S2A). Furthermore, sequence analysis of humans U32A, which is a human homolog of yeast snR40, revealed that like snR40, U32A can also base pair with the region around G611 (G562 in yeast) in humans, making it very likely the case that humans also contain 2′-O-ribose methylation at G611 (Supplementary Figure S2B). Future experiment should be directed to unravel the presence of any Gm methylation at G611 in humans and involvement of U32A in directing this methylation.
Recently, we identified a partial ribose methylation at position A100 in the 18S rRNA underlining a new source of ribosome heterogeneity (24). In the present study, we also analyzed Gm methylations of the 18S rRNA for any quantitative differences leading to ribose heterogeneity. As shown in Figure 3, single deletions of snr41, snr56 and snr57 led only to 20% decrease in the amount of Gm derived from 18S rRNA, whereas deletion of snr40 led to 40% decrease. Since there are five Gm residues in the 18S rRNA, 20% reduction of the amount of total Gm derived from 18S rRNA in single deletion mutant of snr41, snr56 and snr57 and 40% in snr40 deletion clearly demonstrated that Gm methylations in the 18S rRNA do not show any partial methylation leading to ribosome heterogeneity as observed for Am100 (24). This was further confirmed by RP-HPLC quantification of single Gm residues isolated using mung bean nuclease assay (data not shown).
Lately using RP-HPLC, we established the complete set of rRNA base methylation of the 25S rRNA along with their corresponding base methyltransferase in yeast (21,23,26,30). Here, again utilizing RP-HPLC in combination with mung bean nuclease digestion assay for the analysis of ribose methylations, we exhibited the potential and advantage of RP-HPLC in precision and sensitivity for the analyses of variety of modifications in RNA over other biochemical methods. As this manuscript was under peer-review, a new sequencing-based method (RiboMeth-seq) based on the resistance of methylated 2′-OH group to alkaline cleavage has been established for the transcriptome-wide analysis of ribose methylations. This study also confirmed the presence of 2′-O-ribose methylation at G562, identified in the present study (32).
In summary, we identified and mapped a new Gm residue to position 562 in 18S rRNA, previously predicted by Rudi Planta′s lab. Our analysis also validated the positions of other previously identified ribose methylations in the 18S rRNA. Furthermore, we explicitly demonstrated the involvement of snR40 in mediating methylation of Gm562. Our systematic biochemical analysis together with the identification of Gm562 and its corresponding snoRNA, has finally established the complete set of ribose methylations of the 18S rRNA and their corresponding snoRNAs, which will allow future analyses of the precise role of these chemical modifications in ribosome structure and hence function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
J.Y. would like to thank China Scholarship Council for her doctoral scholarship. S.S. would like to acknowledge EMBO for Long-term fellowship (ALTF 644-2014) and European Commission (EMBOCOFUND2012, GA-2012-600394) support from Marie Curie Action. Authors would like to thank Peter Watzinger for his excellent technical support for mung bean nuclease assay and all other lab members for their fruitful discussions.
Footnotes
These authors contributed equally to the paper as first authors.
Present address: Sunny Sharma, RNA Molecular Biology, Universite Libre de Brussels, Rue Profs Jeener & Brachet, 12B-6041 Charleroi – Gosselies, Belgium.
FUNDING
DFG: Deutsche Forschungsgemeinschaft [En134-9]. Funding for open access charge: DFG: Deutsche Forschungsgemeinschaft [En134-9].
Conflict of interest statement. None declared.
REFERENCES
- 1.Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. MODOMICS: a database of RNA modification pathways—2012 update. Nucleic Acids . 2012 doi: 10.1093/nar/gks1007. doi:10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter-Roshek J.L., Petrov A.N., Dinman J.D. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiss-László Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., KISS T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 4.Ganot P., Bortolin M.L., KISS T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 5.Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 6.Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013 doi: 10.1038/nature12581. doi:10.1038/nature12581. [DOI] [PubMed] [Google Scholar]
- 7.Lafontaine D.L., Bousquet-Antonelli C., Henry Y., Caizergues-Ferrer M., Tollervey D. The box H+ ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decatur W.A., Fournier M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 9.Piekna-Przybylska D., Decatur W.A., Fournier M.J. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decatur W.A. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2002;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- 11.Krzyzosiak W., Denman R., Nurse K., Hellmann W., Boublik M., GEHRKE C.W., Agris P.F., Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987;26:2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- 12.Green R., Noller H.F. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 13.Limbach P.A., Crain P.F., McCloskey J.A. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi C., Pan T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011;44:1380–1388. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motorin Y., Helm M. RNA nucleotide methylation. WIREs RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 16.Noon K.R., Guymon R., Crain P.F., McCloskey J.A., Thomm M., Lim J., Cavicchioli R. Influence of temperature on tRNA modification in archaea: methanococcoides burtonii (optimum growth temperature [Topt], 23°C) and Stetteria hydrogenophila (Topt, 95°C) J. Bacteriol. 2003;185:5483–5490. doi: 10.1128/JB.185.18.5483-5490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCloskey J.A., Graham D.E., Zhou S., Crain P.F., Ibba M., Konisky J., Söll D., Olsen G.J. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higa-Nakamine S., Suzuki T., Uechi T., Chakraborty A., Nakajima Y., Nakamura M., Hirano N., Suzuki T., Kenmochi N. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Research. 2011;40:391–398. doi: 10.1093/nar/gkr700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su H., Xu T., Ganapathy S., Shadfan M., Long M., Huang T.H.-M., Thompson I., Yuan Z.-M. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2013;33:1348–1358. doi: 10.1038/onc.2013.89. [DOI] [PubMed] [Google Scholar]
- 20.Edelheit O., Hanukoglu A., Hanukoglu I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009;9:61. doi: 10.1186/1472-6750-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S., Yang J., Watzinger P., Kötter P., Entian K.-D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–9076. doi: 10.1093/nar/gkt679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehrke C.W., Kuo K.C. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 1989;471:3–36. doi: 10.1016/s0021-9673(00)94152-9. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S., Watzinger P., Kötter P., Entian K.-D. Identification of a novel methyltransferase, Bmt2, responsible for the N-1-methyl-adenosine base modification of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2013;41:5428–5443. doi: 10.1093/nar/gkt195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchhaupt M., Sharma S., Kellner S., Oswald S., Paetzold M., Peifer C., Watzinger P., Schrader J., Helm M., Entian K.-D. Partial methylation at Am100 in 18S rRNA of Baker's yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS ONE. 2014;9:e89640. doi: 10.1371/journal.pone.0089640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEntee C.M., Hudson A.P. Preparation of RNA from unspheroplasted yeast cells (Saccharomyces cerevisiae) Anal. Biochem. 1989;176:303–306. doi: 10.1016/0003-2697(89)90313-8. [DOI] [PubMed] [Google Scholar]
- 26.Sharma S., Yang J., Duttmann S., Watzinger P., Kotter P., Entian K.D. Identification of novel methyltransferases, Bmt5 and Bmt6, responsible for the m3U methylations of 25S rRNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:3246–3260. doi: 10.1093/nar/gkt1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe T.M., Eddy S.R. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 28.Klootwijk J., PLANTA R.J. Analysis of the methylation sites in yeast ribosomal RNA. Eur. J. Biochem. 1973;39:325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- 29.Liang X.H., Liu Q., Fournier M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peifer C., Sharma S., Watzinger P., Lamberth S., Kötter P., Entian K.-D. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 2013;41:1151–1163. doi: 10.1093/nar/gks1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watkins N.J., Bohnsack M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. WIREs RNA. 2011;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 32.Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. 2014 doi: 10.1002/anie.201408362. doi:10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


