Abstract
Small RNA OxyS is induced during oxidative stress in Escherichia coli and it is an Hfq-dependent negative regulator of mRNA translation. OxyS represses the translation of fhlA and rpoS mRNA, which encode the transcriptional activator and σs subunit of RNA polymerase, respectively. However, little is known regarding how Hfq, an RNA chaperone, interacts with OxyS at the atomic level. Here, using fluorescence polarization and tryptophan fluorescence quenching assays, we verified that the A-rich linker region of OxyS sRNA binds Hfq at its distal side. We also report two crystal structures of Hfq in complex with A-rich RNA fragments from this linker region. Both of these RNA fragments bind to the distal side of Hfq and adopt a different conformation compared with those previously reported for the (A-R-N)n tripartite recognition motif. Furthermore, using fluorescence polarization, electrophoresis mobility shift assays and in vivo translation assays, we found that an Hfq mutant, N48A, increases the binding affinity of OxyS for Hfq in vitro but is defective in the negative regulation of fhlA translation in vivo, suggesting that the normal function of OxyS depends on the details of the interaction with Hfq that may be related to the rapid recycling of Hfq in the cell.
INTRODUCTION
Under variable stress responses, bacteria generate many sRNAs to adapt to changes in the environment (1,2). Upon oxidative stress, the bacterial sRNA OxyS is expressed and negatively regulates the translation of fhlA and rpoS mRNA (3–6). OxyS has been shown to repress the translation of fhlA through base-pairing interactions at two separate sites: (i) a seven-nucleotide sequence that overlaps with the Shine–Dalgarno sequences of fhlA and (ii) a nine-nucleotide sequence within the coding region (Cds) of fhlA. Kissing complexes have been proposed to form between OxyS and fhlA at these two sites, resulting in a stable antisense–target complex (3,7). The OxyS-fhlA base pairing at the coding sequence has been shown to facilitate the interaction at the ribosome binding site (RBS), thus repressing the fhlA translation (5). As for rpoS, evidence that OxyS and rpoS use complementary sequences for base pairing has not been reported, and the mechanism of OxyS repression is less well understood (8). The models based on sRNA competition for Hfq binding have been previously proposed to account for this process (6,8–10). Crucially, Hfq is required for OxyS sRNA repression of both fhlA and rpoS.
Hfq is a bacterial member of the Sm/LSm family. Escherichia coli Hfq (Ec Hfq) is a homohexameric protein with each subunit containing an N-terminal Sm domain (residues 1–65, Hfq65) and a flexible C-terminal extension (residues 66–102) (11–17). Structural studies have revealed that the Sm domain forms a ring-like hexameric structure that contains two distinct sides for RNA binding. The proximal side exhibits a preference for U-rich single-stranded RNA (ssRNA), whereas the distal side preferentially binds to A-rich ssRNA (18), and a single Hfq hexamer can simultaneously bind to A-rich and U-rich RNA substrates (18–20). Another group of positively charged residues on the lateral surface of Hfq constitutes the third, independent RNA-binding surface (20).
To date, several crystal structures of Hfq in complex with ssRNA have been determined. The structures of Staphylococcus aureus (Sa) Hfq in complex with AU5G (21), Salmonella typhimurium (St) Hfq in complex with U6 (22) and Ec Hfq in complex with AU6A (23) provide details of the interaction of U-rich ssRNA at the Hfq proximal side. In contrast, several structures of Hfq in complex with different poly(A) oligonucleotides reveal an (A-R-N)n tripartite or (A-L)n bipartite RNA-binding motif on the distal side of Hfq (19,24–26). sRNAs exhibit some variation in the Hfq-binding regions, which can be an AU-rich linker region or the 3′-poly(U) terminator (27–31). In addition, the Hfq-binding region in sRNAs often exhibits variations in its sequence, and both U-rich and A-rich sequences have been found to bind Hfq (8,28,32,33). Using minimal binding analysis, RNase footprinting and competition assays, an A-rich linker (nucleotides 65–74) between stem-loops b and c of OxyS was previously identified as the Hfq-binding region, and the Hfq–sRNA interaction likely results in the opening of stem-loop b of OxyS (8). However, due to the absence of atomic-level structural details, the questions of how the linker region of OxyS is recognized by Hfq and whether OxyS binds Hfq as poly(A) remain to be answered.
Here, using fluorescence polarization (FP) and tryptophan fluorescence quenching (TFQ) assays we verify that the A-rich linker region of OxyS sRNA binds Hfq at its distal side. Furthermore, we determined two crystal structures of Hfq in complex with two A-rich fragments from this linker region. In contrast to the previously reported (A-R-N)n or (A-L)n binding mode, in which the RNA fragment lies flat on top of the Hfq distal surface and follows a similar circular weaving path (19,24–26), our structures reveal that two RNA fragments of OxyS adopt a different conformation and only contact a small portion of the Hfq distal surface. Interestingly, using FP assays we found that the Hfq mutant N48A exhibits opposite effects on the binding of poly(A) A7 and ssRNAs from OxyS. The N48A mutant dramatically decreases the binding affinity of Hfq for A7 but unexpectedly enhances the interaction between Hfq and OxyS ssRNA. This enhancement is further verified by electrophoresis mobility shift assays (EMSAs) using full-length OxyS. However, in vivo translation assays demonstrated that, as observed for other mutants, N48A loses the ability to assist OxyS in the repression of fhlA mRNA translation.
MATERIALS AND METHODS
Hfq purification, crystallization and structure determination
Recombinant full-length Hfq and Hfq65 were overexpressed and purified from E. coli as previously described (19,23). The Hfq65 hexamer (0.2 mM) was mixed with 0.2–0.3-mM Aus (5′-AUAACUA-3′) or Ads (5′-AACUAAA-3′) ssRNA and then mixed with an equal volume of crystallization buffer (12% PEG4000 and 0.1-M citrate, pH 5.5, for Aus; 16% mPEG5000 and 0.1-M HEPES, pH 7.2, for Ads). The crystals were grown using hanging drop vapor diffusion. Crystals of both complexes belonged to the space group P212121 and diffracted to 2.00-Å (Hfq-Aus) and 1.97-Å (Hfq-Ads) resolution. X-ray diffraction data were collected at beamline BL17U at the Shanghai Synchrotron Radiation Facility (SSRF) and were merged and scaled using HKL2000/HKL3000 (Hfq-Aus) and MOSFLM (Hfq-Ads) and SCALA in the CCP4 suite (34–36). Data collection and refinement statistics are presented in Supplementary Table S1. Both Hfq-Aus and Hfq-Ads structures were determined by molecular replacement using Phaser (37) and the apo Ec Hfq structure (PDB ID: 1HK9) as the search model. The Rwork and Rfree of the Hfq-Aus structure were refined to 21.10 and 25.20%, respectively. The Rwork and Rfree of the Hfq-Ads structure were refined to 18.50 and 22.90%, respectively.
Coordinates
Coordinates and structure factors for the Hfq-Aus and Hfq-Ads complexes have been deposited in the Protein Data Bank under the accession codes 4QVC and 4QVD, respectively.
Nuclear magnetic resonance spectroscopy
Backbone resonances of Hfq65 were assigned as previously described (23). 1H-15N HSQC spectra were recorded on a Bruker DMX 600 spectrometer equipped with a cryoprobe to monitor the chemical shift perturbations. U-[15N]-labeled Hfq65 R16A/R17A protein was titrated with Aus and Ads ssRNA at 42°C in nuclear magnetic resonance (NMR) buffer (40-mM NaH2PO4, 40-mM NaCl and 1-mM ethylenediaminetetraacetic acid (EDTA) at pH 6.8) with 10% D2O as previously described (19,23). Experimental data were processed using NMRPipe (38) and Sparky. Full titration spectra are shown in Supplementary Figure S1.
Fluorescence polarization
Lyophilized 5′-FAM-labeled RNA oligomers were obtained from Takara Bio, Inc., and dissolved in diethylpyrocarbonate (DEPC)-treated water to a final concentration of 100 μM. Equilibrium dissociation constants (Kd) for different RNAs and different full-length Hfq constructs were determined by measuring the fluorescence polarization, as previously described (19,23).
Preparation of RNAs in vitro
Full-length OxyS and its variants used in this study all include the natural oligo-3′-poly(U)U8 tail, which has been shown to be important in Hfq binding by several recent studies (22,28,31). Full-length OxyS RNA containing 3′- poly(U)U8 tail (hereinafter referred to as OxySU8) and its variants were synthesized by in vitro transcription using T7 RNA polymerase and polymerase chain reaction-amplified DNA templates containing the T7 promoter and the transcribed sequences corresponding to OxySU8 and its variants OxySU8-A10dele (deletion of nucleotides 65–74) and OxySU8-A6U (A65U/A66U/A68U/A69U/A73U/A74U). Transcription products were purified by polyacrylamide gel electrophoresis and dialyzed into DEPC-treated water.
Fluorescence labeling of RNA
In vitro transcribed RNA was modified to introduce a thiol group at the 5′-end through cystamine modification of 5′-phosphate groups using the EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride; Thermo)/imidazole reaction (39). The −SH activated RNA was then labeled using DyLight 680 Maleimide (Thermo) as recommended by the manufacturer. Labeled RNA was isopropanol precipitated and further purified from polyacrylamide gels. The RNA concentration and labeling efficiency were determined by measuring the OD260 and OD680.
Electrophoresis mobility shift assay
EMSA assays were performed in a 10-μl reaction mixture containing 6.67-mM sodium phosphate, 50-mM NaCl, 0.33-mM EDTA and 2.5-mM MgCl2, pH 7.0. Prior to use, all RNAs were refolded by heating to 98°C for 30 s in reaction buffer without magnesium and then slowly cooled to room temperature. MgCl2 was added to a final concentration of 2.5 mM prior to the experiment. The final concentration of OxySU8 was 5 nM, and the final concentrations of the full-length Hfq hexamer are as indicated (Figure 6E and F). Reactions were incubated at room temperature for 40 min and resolved on 0.5 x TB 6% native 19:1 polyacrylamide gels. Gels were scanned in an Odyssey Infrared Imaging System using the 700-nm channel for detection. Each experiment was performed on the same gel and was repeated at least three times. Fluorescence intensities were integrated to calculate the bound fraction of labeled RNA and these intensities were fit to a 1:1 binding model as previously reported (Supplementary Figure S2) (23).
Figure 6.
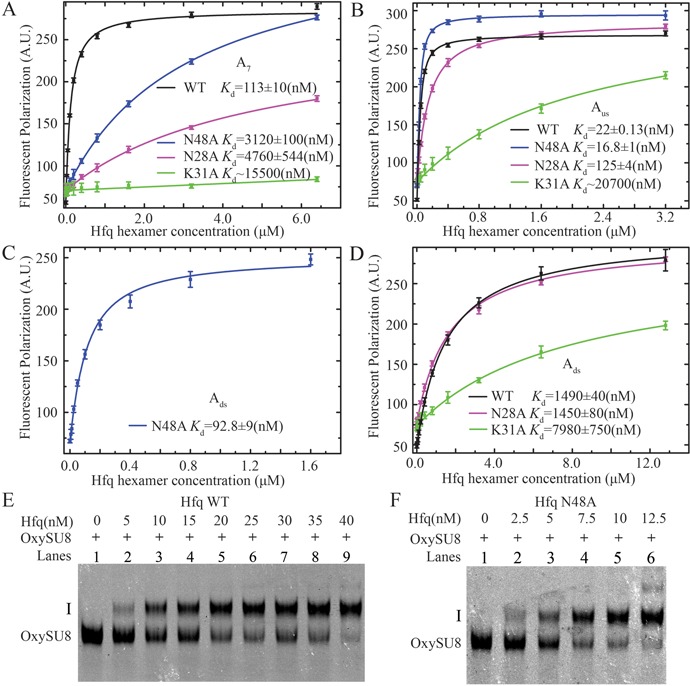
The effects of Hfq mutations (N28A, K31A and N48A) on RNA binding affinities. (A) N28A (magenta), N48A (blue) and K31A (green) mutations dramatically decrease the binding affinity for A7 to Hfq (compared with a Kd of ∼113 nM for wild-type Hfq; (19)). (B) The binding affinity of Aus RNA for N28A and K31A is both lower compared with that for wild type. In contrary, the N48A mutant exhibits higher affinity for Aus. (C) The N48A mutation dramatically increases the affinity for Ads (∼16-fold) compared with wild type. (D) The binding affinity Ads for the K31A mutant is significantly lower. The N28A mutation does not affect the binding of Ads. In EMSA assays, compared to wild-type Hfq (E), the N48A mutant caused a prominent mobility shift of full-length OxySU8 at lower concentrations (F).
Tryptophan fluorescence quenching
Tryptophan fluorescence quenching measurements were performed using an RF-5301PC SHIMADZU spectrofluorophotometer (40). The emission spectrum with excitation at 293 nm was scanned from 320 to 400 nm. One micromolar of each Hfq hexamer Trp mutant protein at 20-μl system in TFQ buffer (20-mM HEPES, pH 8.0, 200-mM NaCl, 0.5-mM EDTA) was scanned. RNAs were dissolved into TFQ buffer to 80-μM concentration and quenching studies were performed using final 1- and 4-μM RNA. Peak height was read from spectra for each Hfq Trp mutant. Quenching percentage was calculated as recently reported (40,41). Each titration was done at least three times.
Western blotting
Overnight cultures of bacteria harboring the GFPuv reporter and full-length Hfq plasmids were diluted 100-fold in Luria Broth (LB) media treated with 50-μM H2O2 and grown at 37°C with appropriate antibiotics in the presence of L-arabinose (0.0225%) and Isopropyl β-D-1-thiogalactopyranoside (IPTG) (50 μM) for an additional 8 h with agitation. Western blots were processed as previously described (19).
RESULTS
The OxyS linker binds to the distal side of Hfq
To better understand the binding properties between OxyS and Hfq, we selected two A-rich fragments, Aus and Ads, which are named to represent the upstream (nucleotides 66–72, 5′-AUAACUA-3′) and downstream (nucleotides 68–74, 5′-AACUAAA-3′) sequences of the OxyS A-rich linker region (Figure 1A), and performed FP assays to investigate their interactions with Hfq. Intriguingly, although the RNA sequence of Aus largely overlaps with that of Ads and both fragments are 7-nt long, the binding of Aus to wild-type full-length Hfq (Hfq WT) (Kd = 22 nM) is much stronger than that of Ads (Kd = 1490 nM) by ∼70-fold (Figure 1B and C). To determine whether this substantial difference in binding affinity results from the binding of the two RNA fragments to different sides of Hfq, two Hfq mutants (F42S on the proximal side and Y25A on the distal side) were prepared for FP assays. Our results indicated that the binding of Aus and Ads to Hfq was significantly decreased by the Y25A mutation, whereas F42S did not exhibit clear effect on RNA binding (Figure 1B and C), indicating that both Aus and Ads bind to the distal side of Hfq.
Figure 1.

The A-rich fragments from the linker region of OxyS bind to the distal side of Hfq. (A) A schematic diagram showing the secondary structure of OxySU8. This structure is based on the experimentally verified secondary structure of OxyS (4), which is in close agreement with the predicted secondary structure of OxySU8 RNA using Mfold (43). The two segments selected for cocrystallization with Hfq, namely Aus and Ads, represent nucleotides 66–72 and nucleotides 68–74 of OxyS, respectively. OxySU8-A10dele (deletion of nucleotides 65–74) and OxySU8-A6U (A65U/A66U/A68U/A69U/A73U/A74U) mutants as indicated. Based on the Mfold prediction (43), the secondary structures of both mutants are similar to that of wild-type OxySU8. (B, C) Fluorescence polarization assay to determine the binding affinities of Aus and Ads for wild-type Hfq and mutants. A mutation on the distal side, Y25A, dramatically decreased the binding affinity of both Aus and Ads, whereas the proximal side mutation, F42S, did not exhibit a prominent effect. (D, E, F) TFQ experiments for Hfq Trp mutants by OxySU8, OxySU8-A10dele and OxySU8-A6U. F42W represents the proximal RNA binding site, and Y25W and K31W represent the distal RNA binding sites. The black bar represents the percent quenching by 1-μM RNA, whereas the gray bar above the black bar represents the quenching by 4-μM RNA.
To determine whether these interactions reflect the binding of full-length OxyS, we further utilized TFQ assays to investigate the binding of full-length OxyS to Hfq (40,42). Two OxySU8 variants, OxySU8-A10dele and OxySU8-A6U, were transcribed in vitro. The secondary structures of both variants are highly similar to that of the wild-type OxySU8 based on the Mfold prediction (43). Three Trp mutants (Y25W, K31W and F42W) were also generated to evaluate the binding effects of OxySU8 on the distal and proximal sides of Hfq. As shown in Figure 1D, E and F, wild-type OxySU8 significantly quenches the fluorescence of Hfq mutants F42W, Y25W and K31W, indicating that full-length OxyS RNA can interact with both proximal and distal sides of Hfq, a finding that is consistent with previous studies (44). In contrast, fluorescence quenching of Hfq is largely weakened for Y25W by ∼1.6-fold and for K31W by ∼1.7-fold (Figure 1E and F) when the A-rich linker of OxyS is either deleted or mutated, whereas a clear reduction in quenching was not observed for the F42W mutant, suggesting that the A-rich region of full-length OxyS binds Hfq at the distal side. Collectively, our FP and TFQ results confirmed that the Hfq binding behaviors of A-rich sequences are similar between short RNA fragments and full-length OxyS.
Crystal structures of Hfq65 in complex with Aus and Ads ssRNA
A previous study showed that the binding affinity of poly(A) A7 to Hfq with a Kd of ∼113 nM (19) was between those of Aus and Ads. Considering that the A-rich sequences from the OxyS linker region do not follow a strict (A-R-N)n binding motif, it is unclear whether the binding of these sequences to Hfq would adopt a similar interaction mode. Attempts were made to crystallize the Sm domain of Ec Hfq (Hfq65) in complex with linker fragments of OxyS. Interestingly, although attempts to crystallize Hfq65 in complex with a 10-nt OxyS fragment (Kd = 13 nM, nucleotides 65–74, 5′-AAUAACUAAA-3′) (Supplementary Figure S3) were unsuccessful, two crystal structures of Hfq in complex with the 7-nt fragments Aus and Ads were successfully determined.
Both Hfq65-Aus and Hfq65-Ads structures were determined by molecular replacement using the apo Ec Hfq structure (PDB ID: 1HK9) as the search model. The Hfq65-Aus structure was refined to Rwork and Rfree values of 21.10 and 25.20%, respectively, at 2.00-Å resolution. In the Hfq65-Aus crystal structure, the asymmetric unit contains six Hfq65 subunits and one RNA fragment, which exhibits continuous electron density for only three of seven ribonucleotides (A68, A69 and C70) (Figure 2A and B). The riboses of A68 and A69 adopt a C2′-endo sugar pucker, whereas the sugar pucker of C70 is C3′-endo, and each base adopts an anti-conformation (Figure 2B). The structure of the Hfq65-Ads complex was refined to 1.97-Å resolution with Rwork and Rfree values of 18.50 and 22.90%, respectively. The asymmetric unit contains an Hfq65 hexamer and exhibits electron density for only four consecutive 3′-nucleotides of Ads (U71, A72, A73 and A74) (Figure 2C and D). The riboses of U71, A72 and A73 all adopt a C2′-endo pucker, whereas the 3′-end A74 adopts a C3′-endo pucker, with all four bases adopting an anti-conformation. For both crystal structures, the reason that full oligonucleotides could not be modeled remains unclear, but most likely is due to the structure disorder. Superposition of Aus- and Ads-bound Ec Hfq65 hexamers with apo Ec Hfq (PDB ID: 1HK9) reveals no significant conformational changes, with RMSD (root mean square deviation) values of 0.46 and 0.48 Å (Supplementary Figure S4A and B), respectively. Data collection and refinement statistics are summarized in Supplementary Table S1.
Figure 2.
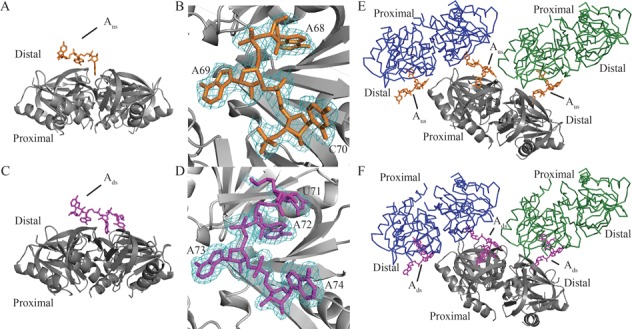
Global structures of the Hfq-Aus and Hfq-Ads complexes. (A) Each Hfq65 hexamer (gray) binds to one Aus (orange) RNA fragment at the distal side in the structure of the Hfq65-Aus complex. (B) Clear electron density (difference maps 2F0−Fc densities are shown as a cyan mesh contoured at 1.0σ) is observed for three of seven nucleotides (A68, A69 and C70; orange) in Aus. (C) Each Hfq hexamer binds to one Ads (magenta) RNA fragment in the structure of the Hfq65-Ads complex. (D) Electron density (difference maps 2F0−Fc densities are shown as a cyan mesh contoured at 1.0σ) is observed for four of seven nucleotides of Ads (U71, A72, A73 and A74; magenta). Three closely packed asymmetric units of Hfq65-Aus (E) and Hfq65-Ads crystals (F) are shown. One hexamer is presented as gray carbon and the remaining two hexamers as blue or green Cα traces. Aus (orange) and Ads (magenta) are represented as sticks. In the complex structures, C70 from Aus and A74 from Ads both point outward from one Hfq distal side and bind at the ‘R-site’ on the distal side of a neighboring Hfq65 hexamer. Data collection and refinement statistics are summarized in Supplementary Table S1.
Recognition of Aus and Ads ssRNA by the distal side of Hfq65
Poly(A) (A7 and A15) has been previously demonstrated to bind to the distal side of Ec Hfq at a similar (A-R-N)n repeat motif (19,24). However, in our Hfq-Aus and Hfq-Ads complex structures, both ssRNA fragments adopt a very similar conformation (Figure 3A), which differs from that of A7 in the Hfq-A7 complex (Figure 2E and F and Figure 3B and C). Our crystal structures reveal that both Aus and Ads interact with the distal side of two different Hfq hexamers, and in both structures, each Hfq hexamer interacts with two ssRNA fragments (Figure 2E and F).
Figure 3.
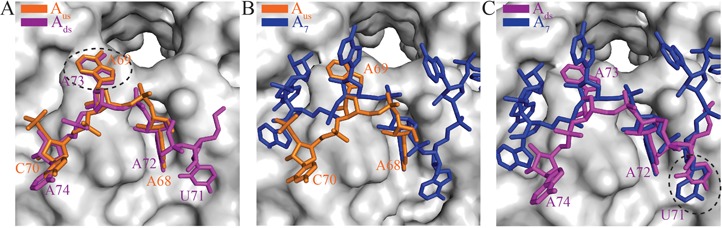
A distinct RNA recognition mode in the Hfq-Aus and Hfq-Ads complex structures compared with the Hfq-A7 structure. Hfq is shown as a gray surface. (A) Aus and Ads bind to Hfq in a similar conformation. Aus and Ads are shown as orange and magenta sticks, respectively. A new nucleotide binding pocket close to the central pore of Hfq (formed near the interface of two adjacent Hfq subunits) is circled with a dotted line. (B) Aus and (C) Ads binding at the distal side of Hfq differs from that of A7 (blue sticks). Nucleotide U71 of Ads binds to the adenosine selective site (‘A-site’) in the ‘A-R-N’ motif and is highlighted with a dotted circle in panel (C).
Specifically, C70 of Aus and A74 of Ads insert into a distal R-site located on a neighboring Hfq hexamer in our crystal structure. C70 of Aus binds to a classic R-site in a manner that is similar to that of the adenosine of poly(A) on the distal side of an adjacent Hfq65 hexamer (Figure 4A and Supplementary Figure S5A), despite previous modeling studies that suggested that the R-site cannot accommodate a pyrimidine (24). The C70 base stacks against Y25(2) and participates in van der Waals interactions with L32′(2) and L26′(2), and its N4 atom participates in polar interaction with the side chain of T61(2) (where ′ denotes the residues from an adjacent Hfq65 subunit and (2) denotes residues from an adjacent Hfq65 hexamer). The O2 atom of C70 engages in a hydrogen bond with the Nδ atom of residue N28′(2). Moreover, a water molecule is observed that bridges the hydrogen bonds between the N3 and O2 atoms of C70 and Oγ of S60(2), and an additional hydrogen bond is formed between the ribosyl 2′-hydroxyl group of C70 and the carbonyl oxygen of G29(2). Recently, it was reported that this R-site could also accommodate Cytidine-5'-triphosphate (CTP) (45). This phenomenon is also observed for A74 of Ads, the base of A74 stacks against Y25(2), participates in van der Waals interactions with L32′(2) and L26′(2) and forms hydrogen bonds between its aromatic nitrogen atoms (N1 and N6) and the Oγ atoms of S60(2) and T61(2) (Figure 4B).
Figure 4.
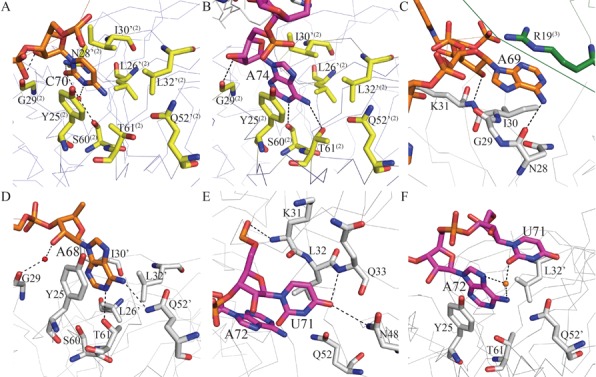
Interactions with RNA on the distal face of Hfq. The carbon atoms of Aus (5′-AUAACUA-3′, nucleotides 66–72) and Ads (5′-AACUAAA-3′ nucleotides 68–74) are colored orange and magenta, respectively. Hfq is shown as gray Cα traces except for the residues involved in RNA binding, which are shown as sticks. The carbon atoms of these residues are colored gray in one Hfq and yellow or green in two adjacent Hfq molecules. The prime sign ‘denotes residues from an adjacent Hfq65 subunit in the same hexamer. The superscripts (2) and (3) denote residues from the neighboring Hfq65 hexamers. (A, B) C70 of Aus and A74 of Ads bind to the R-site of an adjacent Hfq hexamer. (C) A69 of Aus binds to a pocket on the distal side of Hfq close to the central pore. The adenosine ribosyl 2′-hydroxyl group of A69 interacts with the backbone amide of K31. The exocyclic N6 atom forms a hydrogen bond with the carbonyl oxygen groups of N28. (D) A68 of Aus binds to the R-site of Hfq. (E) U71 of Ads binds to the A-site of Hfq via hydrogen bonds to Nδ of N48 and the backbone atom of Q33. (F) U71 is coordinated with A72 by two non-discriminating water-mediated hydrogen bonds.
In contrast to the corresponding adenine in A7, which does not exhibit interactions with Hfq and represents a typical N-site nucleotide in the Hfq-A7 structure, both A69 of Aus and A73 of Ads bind Hfq in close proximity to the central pore. In both cases, the adenine base participates in van der Waals interactions with R19 from a nearby Hfq hexamer. The adenosine ribosyl 2′-hydroxyl group interacts with the backbone amide of K31, and the exocyclic N6 atom of the base forms a hydrogen bond with the carbonyl oxygen groups of N28. These adenine bases also participate in van der Waals interactions with residue I30 (Figure 4C and Supplementary Figure S5B). To further determine whether the interactions between ssRNA and R19 from a different Hfq hexamer exist in solution or are simply a result of crystal packing, we generated an R19W mutant and analyzed this mutant using the TFQ assay. Compared to A7, Aus and Ads were quenched much more significantly by ∼2.4-fold and ∼2.8-fold, respectively (Supplementary Figure S6), indicating that R19 contributes to the interaction between ssRNA and Hfq. Thus, although we cannot exclude the possibility that this binding is a consequence of crystal packing, our results suggest that R19 may play an interesting role in mediating the ssRNA–Hfq interaction.
In the two crystal structures, A68 of Aus and A72 of Ads bind to the canonical R-site at the distal side of Hfq as observed for their counterpart(s) in poly(A) (Figure 4D and Supplementary Figure S5C): the adenine rings insert into the crevice between neighboring Hfq65 subunits and stack against the side chain of Y25 and participate in van der Waals interactions with residues L26′, I30′ and L32′; the adenine exocyclic atoms N6 and N1 form hydrogen bonds with Nϵ of Q52′ and Oγ of T61, respectively; and the adenine ribosyl 2′-hydroxyl forms a water-mediated hydrogen bond with the carbonyl oxygen of G29, which is slightly different from the direct hydrogen bond observed in the structure of the Hfq65 poly(A) complex (24).
Unexpectedly, U71 of Ads binds to a former A-site in the Hfq65-A7 complex (Figure 4E), which is composed of residues from β-strands 2 and 4 and was previously named for its binding specificity for adenosine nucleotides (24). The O4 atom of the U71 base forms hydrogen bonds with the backbone amide of Q33 and the Nδ atom of N48. In addition, as observed for the adenosine nucleotide, the U71 base stacks against the side chain of L32, and the 5′- phosphate group of U71 interacts with the backbone amide of K31. Moreover, U71 is coordinated with A72 by two non-discriminating water-mediated hydrogen bonds (O2 of the uracil base to N7 and N6 of A72, respectively) (Figure 4F). We speculate that the interactions between U71 and A72 may be important to position the uridine in this A-site with the energetically favorable anti-conformation.
Aus and Ads primarily perturb the distal side of Hfq in solution NMR
To confirm the interaction details between Hfq and OxyS fragments, solution NMR was used to investigate the binding of Hfq65 to Aus and Ads (Figure 5 and Supplementary Figure S1). The mutant Hfq65 R16A/R17A was used in the NMR titration to avoid protein aggregation upon RNA binding (19,23). Our chemical shift perturbation experiments indicate that Hfq residues are affected by Aus and Ads binding in a similar fashion (Figure 5). As expected, around 17 residues, such as Y25, N28, I30, K31, Q33, S60 and T61, which are involved in A-rich RNA binding on the distal side, experienced large chemical shift changes (Figure 5A). Interestingly, several residues on the proximal side including Q8, Q41, F42, V43 and H57, which have been reported to form polar interactions with uracils (19,23), also demonstrate clear chemical shift changes or disappear upon RNA titration. However, in terms of the number of perturbed residues and the magnitude of the perturbation, RNA binding affects the Hfq distal surface more prominently than the proximal side, which is consistent with our FP assay, which was performed in solution. Considering that the proximal side residues are not directly involved in the binding of Aus and Ads in our crystal structures, the related minor perturbations and the slight decrease in the binding affinity of ssRNAs for Hfq with an F42S mutation on the proximal face, as demonstrated by the FP assay (Figure 1B and C), are perhaps caused by the subtle structural changes induced by a cross-talk between the distal and proximal binding sites which may be important for Hfq regulation (23). These influences on the Hfq proximal side may also result from weak binding of excessive ssRNA in the experimental systems.
Figure 5.
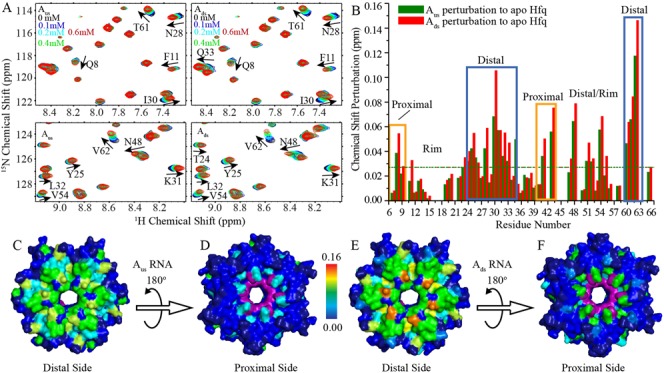
NMR chemical shift perturbations of Hfq by Aus and Ads. Hfq65 R16A/R17A protein (0.1 mM) were titrated with Aus and Ads, respectively. (A) Selected regions of 1H-15N HSQC spectra of Hfq upon Aus (left) and Ads (right) titration. (B) Chemical shift differences between the first and last titration points for Aus and Ads are presented as green and red column bars, respectively. (C, E) Both Aus and Ads binding to Hfq result in prominent chemical shift perturbations on the distal side of Hfq. (D, F) Residues Q8, Q41 and V43 on the proximal side of Hfq are also perturbed by Aus and Ads titration. Hfq is colored according to chemical shift changes using a blue to red gradient. The resonances of F42 and H57, which disappeared upon Aus and Ads titration, are colored purple. Unassigned residues are colored dark blue.
Key residues involved in Hfq binding of Aus and Ads
As previously indicated, careful structural comparison of the observed nucleotides in Hfq-Aus, Hfq-Ads and Hfq-A7 reveals that the side chain of N28 is only involved in binding to Aus and A7 but not Ads. In addition, N48 from the Hfq A-site interacts with U71 via its side chain in Hfq-Ads but not with the observed nucleotides in Hfq-Aus and Hfq-A7. To determine whether these interactions are crucial for the recognition of ssRNA by Hfq, two mutations (N28A and N48A) were generated for FP investigation (Figure 6). K31A, which causes a 100-fold loss in the binding affinity of Hfq for poly(A) (18,19,24,46), was also analyzed in our FP assay, although a direct interaction with ssRNA was not observed for this residue in our crystal structures. As previously reported, the K31A mutation disrupts the binding of all three ssRNAs (Figure 6A, B and D). N28A decreases the binding of A7 and Aus by ∼40 and ∼6-fold compared with Hfq wild type (WT) but does not affect the binding of Ads, which is consistent with our crystal structures (Figure 6A, B and D). Unexpectedly, the results of the FP assay indicated that N48A causes two distinct Hfq-binding effects on OxyS ssRNAs and A7. This mutant decreases the binding affinity of Hfq for A7 by ∼27-fold (Figure 6A), whereas it increases the binding affinities for Ads and Aus by ∼16-fold (Kd = 1480 to 92.8 nM) and ∼1.3-fold (Kd = 22 to 16.8 nM) (Figure 6B and C). To confirm this result, we further performed EMSA assays to investigate the interaction between Hfq and full-length OxyS (Figure 6E and F). OxyS has been previously shown to form more than one discrete complex (i.e. I and II) with Hfq and the complex I was suggested to consist of a 1:1 sRNA:Hfq hexamer stoichiometric ratio (8,44,47). The EMSA assay was therefore performed at a concentration range of 0–40 nM for wild-type Hfq and 0–12.5 nM for the N48A mutant such that only complex I would be predominantly formed. Fluorescence intensities were integrated to calculate the bound fraction of the labeled RNA, and these intensities were fit assuming a 1:1 binding stoichiometry between one Hfq hexamer and one molecule of RNA (Supplementary Figure S2). Our results indicated that N48A (Kd = ∼4.6 ± 0.7 nM) exhibits higher RNA binding affinity than wild-type Hfq (Kd = ∼17 ± 1.0 nM). In addition, we generated an N48W mutant and performed the TFQ assay to demonstrate that the full-length OxySU8 RNA interacts with N48 of Hfq in solution (Supplementary Figure S7). Overall, we report for the first time that the Hfq N48A mutant exhibits distinctly opposite effects on the binding of poly(A) and A-rich RNA sequences.
An appropriate binding affinity is important for Hfq to regulate OxyS function in vivo
Our in vitro results indicated that several Hfq mutants including N28A, K31A and N48A demonstrated distinct effects on Hfq binding of OxyS fragments. To evaluate whether these mutants could influence the repression of fhlA mRNA translation by OxyS sRNA in vivo, a reporter system consisting of the fhlA 5′- end fragment from −514 upstream of the AUG initiation codon to +60 that contained the Cds site (i.e. the fhlA coding sequence from +34 to +42, which is involved in a direct OxyS–fhlA interaction) fused to the coding sequence of GFPuv (3,7) (Figure 7A) was generated with a GSSG spacer. We examined the role of OxyS on this fhlA-GFPuv fusion in the hfq− strain or using wild-type Hfq or the N28A, K31A, N48A or Y25A mutants. All growth media were treated with 50-μM H2O2 to ensure that the bacteria were cultured under oxidative stress. Our results (Figure 7B) demonstrate that wild-type Hfq (lane 2) can effectively facilitate OxyS-repression of fhlA-GFPuv translation, whereas in the hfq− strain (lane 1), OxyS cannot efficiently repress fhlA-GFPuv translation. For the Y25A mutant (lane 3), the translation level of fhlA-GFPuv is very similar to that in the hfq− strain. In addition, neither the N28A (lane 4) nor the K31A (lane 5) mutant could repress GFPuv expression as wild-type Hfq, which is primarily due to the disruption of key hydrogen bond between Hfq and OxyS. However, the results of the in vivo translation assay are not entirely consistent with the in vitro FP results, in which the K31A mutant exhibited dramatically decreased binding affinity compared with wild-type Hfq, whereas its in vivo repression of fhlA-GFPuv translation is not as drastic as that observed for Y25A. This discrepancy has been observed in previous studies (19). The discrepancy between the in vivo repression level and the in vitro binding assay is possibly due to the changes in the RNA concentrations. In vivo, the system is much more complicated than in vitro, and the expression of mutant Hfq may affect sRNA accumulation, which would in turn influence the mRNA translation efficiency (48). Intriguingly, the N48A mutant, which binds OxyS fragments more tightly than wild-type Hfq, also exhibits a significant decrease in the expression level of GFPuv (lane 6). In previous studies, the observation of a defective sRNA–mRNA regulation is often related to the disruption rather than the enhancement of the key interactions between Hfq and sRNA (19,48,49). The interesting behavior of the N48A mutant raises the possibility that N48 may play a special role in regulating the interaction between Hfq and OxyS such that limited levels of Hfq in the cell can be efficiently recycled to mediate the regulation of mRNA translation by sRNAs.
Figure 7.
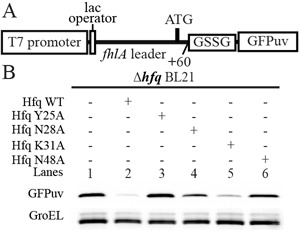
An appropriate RNA binding affinity is important for Hfq regulation in vivo. (A) A schematic diagram of the reporter system used for in vivo translation assays. The DNA sequence encoding the fluorescent protein GFPuv is fused to the leader sequence of fhlA and is thus under the regulation of OxyS and Hfq. The level of GFPuv expression in this system will consequently reflect the translation level of the mRNA with this fhlA leader. (B) Deletion of hfq increases the GFPuv expression level (lane 1) compared with wild-type Hfq (lane 2). GFPuv expression in the presence of the Y25A mutant (lane 3) is similar to that in the hfq− strain. Similarly, neither the N28A (lane 4) nor the K31A mutant (lane 5) was capable of suppressing GFPuv expression as wild-type Hfq. This finding is presumably due to the deficiency of these mutants in binding OxyS. The N48A mutant, which binds to OxyS more tightly than wild-type, also does not suppress the expression level of GFPuv (lane 6). GFPuv was stained with an anti-GFP antibody and GroEL was used as a loading control.
DISCUSSION
OxyS A-rich sequences adopt a binding mode that differs from the (A-R-N)n mode at the Hfq distal side
It is well known that poly(A) and many mRNAs bind to the distal side of Hfq and adopt an (A-R-N)n binding motif (24,50). Previous studies have shown that the linker region of OxyS RNA is crucial for Hfq binding (8). Recent studies showed that mutations of proximal side residues disrupted Hfq–OxyS interactions, whereas mutations of distal side residues exhibited a largely reduced effect (10). A more recent study using SAXS and SANS presented a model of the Hfq–OxyS complex, which reveals that OxyS predominantly binds Hfq at the proximal side and partially interacts with the distal side (44). In this study, using FP and TFQ assays, and the determined crystal structures, we identified that the A-rich sequence of OxyS binds Hfq at its distal side. Our atomic-level structures are consistent with the previously reported structural model in which OxyS can partially interact with the Hfq distal surface (44).
In the present crystal structures, Aus and Ads adopt a very similar conformation, with the exception that two RNA fragments do not overlap in the observed nucleotides (A68, A69 and C70 for Aus and U71, A72, A73 and A74 for Ads). Structural comparison reveals that the binding locations of the observed nucleotides on the Hfq distal surface are essentially identical in the two crystal structures, raising a concern whether both structures or only one structure is biologically correct. We propose two possibilities to account for the determined crystal structures: (i) all of the aforementioned nucleotides may simultaneously bind to two Hfq hexamers: nucleotides A68, A69 and C70 bind the first Hfq hexamer with the base of C70 inserting into the R-site of a second Hfq hexamer and nucleotides U71, A72, A73 and A74, in turn, interact with the second hexamer. (ii) These nucleotides belong to two consecutive binding motifs that may not bind Hfq simultaneously but function together to increase the chance of mutual recognition between Hfq and OxyS. However, due to the lack of a structure of Hfq in complex with a greater number of observed nucleotides, more systematic investigations are definitely needed to determine which model is biological relevant.
The sRNA conformation in the two structures differs from the previously reported A-R-N or A-L binding mode, in which the RNA fragment lies flat on top of the Hfq distal surface and follows a similar circular weaving path (19,24–26). In our structures, both A69 of Aus and A73 of Ads bind Hfq in close proximity to the central pore, which differs from the N-site in the crystal structures of Hfq-A7 and Hfq-A15. More interestingly, the rearrangement of this nucleotide binding site leads to a structural change of the downstream nucleotides (C70 of Aus and A74 of Ads), allowing the bases of C70 and A74 to stretch away from the Hfq hexamer and insert into an R-site on the distal side of another adjacent Hfq hexamer. In contrast, A69 of Aus and A73 of Ads bind to the Hfq distal side close to the central pore and participate in van der Waals interactions with a third hexamer. Although we cannot exclude the possibility that these inter-hexamer interactions are caused by crystal packing, our FP and TFQ assays indicated that, compared to A7, Aus and Ads exhibited different binding behaviors toward Hfq in solution (Figure 6 and Supplementary Figure S6), supporting the notion that the RNA binding mode that we observed in the crystal structures of Hfq-Aus and Hfq-Ads is indeed different from the previously known (A-R-N)n or (A-L)n mode.
N48 may play an important role in balancing the OxyS–Hfq interaction
To evaluate the crucial protein–nucleic acid interactions observed in our crystal structures, we generated several mutants at the Hfq-RNA binding interface and analyzed them using the FP assay. Unexpectedly, among these mutants, N48A does not weaken the Hfq–ssRNA interaction; rather, it dramatically increases the binding affinity of Hfq for Ads by ∼17-fold. This result is unexpected because a hydrogen bond is observed between the N48 side chain and the U71 base in the Hfq-Ads crystal structure. The enhancement of this interaction is further confirmed in the EMSA assay of full-length OxyS and the Hfq N48A mutant, suggesting that the Hfq–OxyS interaction may also be strengthened by this mutation in the cell. One hypothesis to rationalize this apparent discrepancy is that the original A-site may not represent an optimal binding site for U71, although hydrogen bonds are observed between the U71 base and Hfq Q33 and N48, and alanine substitution of N48 remodels this site such that it can better accommodate the U71 base. In addition, the electron density for U71 is not as well defined as that for the other three adenines. Therefore, it is also possible that the conformation of this uracil is flexible, and the N48A mutation may stabilize the conformation of this uracil by generating new interactions.
Although showing an opposite effect on RNA binding compared with the other mutants, N48A exhibits similar defective regulation of mRNA translation in vivo as the other mutants, raising the question of whether an appropriately moderate sRNA binding capacity is important for the regulatory role of Hfq in mRNA translation, whereas either reduced or enhanced sRNA binding to Hfq is primarily related to defective sRNA–mRNA regulation. We speculate that the biological significance of this ‘correct’ RNA binding at the Hfq distal side may be related to the rapid recycling of Hfq. A strong Hfq binding with sRNAs in vivo will decrease the chance of Hfq for recycling so that repress Hfq function as an mRNA-translation regulator (51,52). Alternatively, considering that N48A dramatically disrupts the interaction between Hfq and poly(A)A7 (Figure 6A), which represents the Hfq binding motif of mRNA, it is also possible that the low in vivo expression level is caused by the loss of direct fhlA mRNA binding.
OxyS uses two different mechanisms to regulate the translation of fhlA and rpoS
The small RNA OxyS is induced by E. coli in response to oxidative stress. OxyS represses the translation of fhlA and rpoS mRNA, which encode the formate hydrogen lyase system activator and the σs subunit of RNA polymerase, respectively (3–5). The RNA chaperone protein Hfq binds to OxyS to facilitate sRNA repression of mRNA translation (8). The regulation mechanism of fhlA mRNA by OxyS in the presence of Hfq has been better characterized. Two complementary regions between OxyS and fhlA exist, and kissing complexes form at these two base-pairing sites. The interaction generated within the 5′- leader region of fhlA sequesters the RBS preventing translation (7). In general, it was assumed that the simultaneous binding of sRNA and mRNA to Hfq can increase the local concentration of RNAs and their base-pairing opportunity. Our crystal structures provide the basis for recognition of OxyS A-rich sequence by Hfq. Additional in vivo translation assays indicated that mutants harboring mutations at the RNA-binding interface of Hfq are defective in the repression of fhlA translation. In contrast to the crystal structure of Hfq-A7, in which two A7 molecules symmetrically bind on top of the distal side of Hfq, our structures indicated that OxyS fragments only contact a small portion of the Hfq distal face. We propose that the lower occupancy of OxyS on the distal side of Hfq exposes a sufficient area for fhlA binding to allow two RNAs to bind and form a functional entity through base pairing.
OxyS has been reported to negatively regulate rpoS gene expression without apparent base pairing and this regulation depends on Hfq (8). The mechanism of OxyS regulation of the translation of rpoS mRNA remains less well understood. Recently, in addition to regulation by base pairing, another regulatory mechanism by Hfq competition was proposed and supported by in vitro and in vivo studies for Hfq-dependent sRNAs (9,51–54). In the cell, due to the limited availability of Hfq, sRNAs must compete with each other to fulfill their roles in the regulation of mRNA translation (9,10,51,52). It has been proposed that this competition may occur not only between sRNAs but also between sRNA and mRNA (9). For rpoS, the (A-A-N)4 motif in the 5′-untranslated region (UTR) has been demonstrated to bind Hfq and plays a critical role in sRNA regulation of rpoS translation (50). Very recently, Peng et al. exhibits a structural model in which rpoS wraps around the Hfq protein and can bind to its distal face and lateral rim (55). In this study, our structures reveal that sRNA fragments of OxyS bind to the distal surface of Hfq and likely share a similar binding interface on Hfq with the (A-A-N)4 motif of rpoS, suggesting that competition for binding on the distal face of Hfq may occur between OxyS and rpoS. Considering that a base-pairing interaction has not been detected between OxyS and rpoS (4,8), this competition may be the major issue occurring on the Hfq distal side. Furthermore, using EMSA, we demonstrated that full-length OxyS can successfully compete with an rpoS fragment of the same length (rpoScp108: nucleotides 331–438, containing an (A-A-N)4 and an A6 element) for Hfq binding in vitro (Supplementary Figure S8). Collectively, our structural and biochemical results support the notion that competitive displacement can occur, not only between OxyS and other sRNAs but also between OxyS and rpoS, and, in turn, can affect the translation of rpoS mRNA. In summary, the linker region of OxyS that binds to Hfq may utilize two different mechanisms to regulate the translation of fhlA and rpoS.
ACCESSION NUMBERS
PDB IDs: 4QVC, 4QVD.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Prof. Zhiyong Zhang, Prof. Ke Ruan, Prof. Jianye Zang, Bin Wen, Na Wang, Yonghui Zhang, Hongyu Bao, Pengzhi Wu, Lingna Yang, Ling Xu and Jia Gao for helpful discussions; Dr Debiao Zhao, Yiyang Jiang, Yang Zou, Zhonghua Liu and Chongyuan Wang for assistance in reflection data collection; Dr Yun-Xing Wang for kindly providing the plasmid of T7 RNA polymerase; F. Delaglio and A. Bax for providing the software NMRPipe; T.D. Goddard and D. Kneller for Sparky; A.T. Brünger for CNS; and W.L. DeLano for PyMol. We thank the staff at beam lines 17U of Shanghai Synchrotron Radiation Facilities (SSRF) for assistance with X-ray data collection.
FUNDING
The National Basic Research Program of China (973 Program) [2011CB911104, 2011CB966302]; Chinese National Natural Science Foundation [31270782, 31330018]; the Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010101]; Research Program of the Chinese Academy of Sciences [KJZD-EW-L05]; The Fundamental Research Funds for the Central Universities [WK2070000020]. Funding for open access charge: The National Basic Research Program of China (973 Program) [2011CB911104, 2011CB966302]; Chinese National Natural Science Foundation [31270782, 31330018]; the Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010101]; Research Program of the Chinese Academy of Sciences [KJZD-EW-L05]; The Fundamental Research Funds for the Central Universities [WK2070000020].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gottesman S., McCullen C.A., Guillier M., Vanderpool C.K., Majdalani N., Benhammou J., Thompson K.M., FitzGerald P.C., Sowa N.A., FitzGerald D.J. Small RNA regulators and the bacterial response to stress. Cold Spring Harb. Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Lay N., Schu D.J., Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altuvia S., Zhang A., Argaman L., Tiwari A., Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang A., Altuvia S., Tiwari A., Argaman L., Hengge-Aronis R., Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altuvia S., Weinstein-Fischer D., Zhang A., Postow L., Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 6.Battesti A., Majdalani N., Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argaman L., Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 8.Zhang A., Wassarman K.M., Ortega J., Steven A.C., Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 9.Moon K., Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Updegrove T.B., Wartell R.M. The influence of Escherichia coli Hfq mutations on RNA binding and sRNA*mRNA duplex formation in rpoS riboregulation. Biochim. Biophys. Acta. 2011;1809:532–540. doi: 10.1016/j.bbagrm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Sauter C., Basquin J., Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arluison V., Folichon M., Marco S., Derreumaux P., Pellegrini O., Seguin J., Hajnsdorf E., Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 13.Vecerek B., Rajkowitsch L., Sonnleitner E., Schroeder R., Blasi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer E. Special focus hfq. RNA Biol. 2013;10:590–591. doi: 10.4161/rna.24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weichenrieder O. RNA binding by Hfq and ring-forming (L)Sm proteins: a trade-off between optimal sequence readout and RNA backbone conformation. RNA Biol. 2014;11:537–549. doi: 10.4161/rna.29144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan R.G., Link T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013;10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikulecky P.J., Kaw M.K., Brescia C.C., Takach J.C., Sledjeski D.D., Feig A.L. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Wang L., Wu J., Gong Q., Shi Y. Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA. Nucleic Acids Res. 2013;41:5938–5948. doi: 10.1093/nar/gkt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer E., Schmidt S., Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher M.A., Pearson R.F., Moller T., Valentin-Hansen P., Brennan R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauer E., Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W., Wang L., Zou Y., Zhang J., Gong Q., Wu J., Shi Y. Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 2011;25:2106–2117. doi: 10.1101/gad.16746011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link T.M., Valentin-Hansen P., Brennan R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Someya T., Baba S., Fujimoto M., Kawai G., Kumasaka T., Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012;40:1856–1867. doi: 10.1093/nar/gkr892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horstmann N., Orans J., Valentin-Hansen P., Shelburne S.A., III, Brennan R.G. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 2012;40:11023–11035. doi: 10.1093/nar/gks809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel J., Luisi B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otaka H., Ishikawa H., Morita T., Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller T., Franch T., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 30.Beisel C.L., Updegrove T.B., Janson B.J., Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa H., Otaka H., Maki K., Morita T., Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lease R.A., Woodson S.A. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Geissmann T.A., Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leslie A.G.W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsl. Protein Crystallogr. 1992;26 [Google Scholar]
- 35.Bailey S. The Ccp4 suit-programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 37.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 39.T H.G. Bioconjug. Tech. (2nd edn) 2008;27:1012–1013. [Google Scholar]
- 40.Robinson K.E., Orans J., Kovach A.R., Link T.M., Brennan R.G. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014;42:2736–2749. doi: 10.1093/nar/gkt1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovach A.R., Hoff K.E., Canty J.T., Orans J., Brennan R.G. Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes. RNA. 2014;20:1548–1559. doi: 10.1261/rna.044032.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panja S., Schu D.J., Woodson S.A. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41:7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henderson C.A., Vincent H.A., Casamento A., Stone C.M., Phillips J.O., Cary P.D., Sobott F., Gowers D.M., Taylor J.E., Callaghan A.J. Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA. 2013;19:1089–1104. doi: 10.1261/rna.034595.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murina V., Lekontseva N., Nikulin A. Hfq binds ribonucleotides in three different RNA-binding sites. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1504–1513. doi: 10.1107/S090744491301010X. [DOI] [PubMed] [Google Scholar]
- 46.Sun X., Wartell R.M. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 47.Updegrove T.B., Correia J.J., Chen Y., Terry C., Wartell R.M. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA. 2011;17:489–500. doi: 10.1261/rna.2452111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang A.X., Schu D.J., Tjaden B.C., Storz G., Gottesman S. Mutations in Interaction Surfaces Differentially Impact E. coli Hfq Association with Small RNAs and Their mRNA Targets. J. Mol. Biol. 2013;425:3678–3697. doi: 10.1016/j.jmb.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salim N.N., Faner M.A., Philip J.A., Feig A.L. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 2012;40:8021–8032. doi: 10.1093/nar/gks392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soper T., Mandin P., Majdalani N., Gottesman S., Woodson S.A. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hussein R., Lim H.N. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fender A., Elf J., Hampel K., Zimmermann B., Wagner E.G. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 54.Wagner E.G. Cycling of RNAs on Hfq. RNA Biol. 2013;10:619–626. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng Y., Curtis J.E., Fang X., Woodson S.A. Structural model of an mRNA in complex with the bacterial chaperone Hfq. Proc. Natl. Acad. Sci. U.S.A. 2014;111:17134–17139. doi: 10.1073/pnas.1410114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


