Abstract
The Ku heterodimer serves in the initial step in repairing DNA double-strand breaks by the non-homologous end-joining pathway. Besides this key function, Ku also plays a role in other cellular processes including telomere maintenance. Inactivation of Ku can lead to DNA repair defects and telomere aberrations. In model organisms where Ku has been studied, inactivation can lead to DNA repair defects and telomere aberrations. In general Ku deficient mutants are viable, but a notable exception to this is human where Ku has been found to be essential. Here we report that similar to the situation in human Ku is required for cell proliferation in the fungus Ustilago maydis. Using conditional strains for Ku expression, we found that cells arrest permanently in G2 phase when Ku expression is turned off. Arrest results from cell cycle checkpoint activation due to persistent signaling via the DNA damage response (DDR). Our results point to the telomeres as the most likely source of the DNA damage signal. Inactivation of the DDR makes the Ku complex dispensable for proliferation in this organism. Our findings suggest that in U. maydis, unprotected telomeres arising from Ku depletion are the source of the signal that activates the DDR leading to cell cycle arrest.
INTRODUCTION
Repair of DNA double strand breaks (DSBs) is critical for maintenance of genome integrity. Failure to repair DSBs properly can result in chromosomal abnormalities, sensitivity to genotoxins, and cell death. Cells rely on either homologous recombination (HR) or non-homologous end-joining (NHEJ) to repair such lesions (1). While HR ensures accurate repair, NHEJ is often error prone. In a number of organisms, including mammalian cells, NHEJ is the preferred mechanism of DSB repair (2,3). Crucial for NHEJ is the Ku70/Ku80 heterodimer or Ku complex, which binds and stabilizes the ends of broken DNA molecules. Ku70/Ku80 also facilitates the recruitment of downstream factors that mediate NHEJ (4).
The Ku complex also has key roles in a number of other fundamental cellular processes such as telomere maintenance, transcription and apoptosis (5). However, while the function of the Ku complex in NHEJ is well conserved among eukaryotes, the other roles are more species-specific. A paradigmatic case is the role of Ku in telomere maintenance (6). The Ku heterodimer is required for proper telomere function in multiple species, and deletion of either Ku subunit results in telomere defects (7). However, the precise defects engendered by Ku deletion seem to depend upon the particular telomere biology of the species in question. For instance, in budding yeast, loss of Ku leads to modest telomere shortening and elevated levels of telomeric recombination (8–10), while in Arabidopsis thaliana loss of Ku results in massive telomeric expansion and a rampant increase in telomeric recombination (11,12). In metazoans, the consequence is even more unpredictable. In chicken cells, loss of Ku has no effect on telomeres (13). But in mice there are conflicting reports of either slight telomere expansion or significant telomere shortening for similar strains (14,15). In any event, the most striking effect of Ku deficiency arises in human cells, where the Ku complex is essential for viability and is required to prevent unrestrained telomere loss (16,17). Why Ku is essential in cells from one organism and dispensable in others is unknown. Understanding these differences will advance our general knowledge of how telomeres differ among species.
Ustilago maydis, a basidiomycete fungus developed as a model organism to study DNA repair (18), has certain features of its recombinational repair system more in common with metazoans than the standard yeast systems and also, unlike standard yeasts, has telomeres with the same 6-bp repeat as in metazoans. Recently we showed that Brh2, a key component of the HR pathway and the U. maydis ortholog of the breast cancer susceptibility protein BRCA2, promotes the maintenance of normal telomere lengths in telomerase positive cells (19), just like in mammals (20). As a follow-up from that study we were interested in examining how Ku deficiency might influence telomere lengths. However, we encountered an unanticipated difficulty in not being able to generate mutants deleted of the Ku structural genes. In this investigation, we explored the reason for the failure to obtain such mutants and found that Ku is essential for cell viability in U. maydis. We explored the molecular mechanism for this essential function and came to the conclusion that it results from the ability of the Ku complex to suppress unscheduled activation of the DNA damage response (DDR) at telomeres. This unscheduled activation causes a stringent cell cycle arrest and thus a loss of the cells’ ability to proliferate. These findings suggest that telomere regulation in U. maydis shares features with the human system and could serve as a paradigm to gain insights on human telomere biology.
MATERIALS AND METHODS
Strains and growth conditions
Ustilago maydis strains are derived from FB1 genetic background (21) and are listed in Supplementary Table S1. Cells were grown in rich medium (YPD) or minimal medium (MMD) (22). Controlled expression of genes under the nar1 promoter, and FACS analysis were performed as described previously (23,24).
Strains and plasmid constructions
Plasmid pGEM-T easy (Promega) was used for cloning, subcloning and sequencing of genomic fragments and fragments generated by polymerase chain reaction (PCR). Oligonucleotides used in this study are described in Supplementary Table S2. To construct the different strains, transformation of U. maydis protoplasts with the indicated constructions was performed by standard procedures (25). Integration of the disruption cassette into the corresponding loci was verified in each case by diagnostic PCR and subsequent Southern blot analysis.
To produce the uku80nar1 allele, fragments from the promoter (5′ fragment) and the Open Reading Frame (ORF) region (3′ fragment) were ligated to pRU2 (26) digested with NdeI and EcoRI. The 5′ fragment (flanked by EcoRI and PacI) was produced by PCR using the primers Ku80-2 and Ku80-3. The 3′ fragment (flanked by NdeI and PacI) was obtained by PCR amplification with primers Ku80-4 and Ku80-5. The resulting plasmid pUKU80nar1 was integrated, after digestion with PacI, by homologous recombination into the uku80 locus.
To produce the uku70nar1 allele, a pair of fragments (one from the promoter region and the other encoding the Uku70 N-terminal region) were amplified using the primer pairs Ku70-2/Ku70-3 and Ku70-4/Ku70-5, respectively. These fragments were ligated to pRU2 (26) that had been digested with NdeI and EcoRI. The resulting plasmid pUKU70nar1 was integrated, after digestion with PacI, by homologous recombination into the uku70 locus.
Deletion of mre11, rec1, exo1 and rad51 genes was done by gene replacement following published protocols (27). Briefly, a pair of DNA fragments flanking the corresponding ORF were amplified and ligated to antibiotic resistance cassettes via SfiI sites. The 5′ and 3′ fragments were amplified using the appropriate oligonucleotide pairs. Each fragment was about 1 kbp in length.
For C-terminal fusion of proteins to fluorescent markers, the adaptation of the SfiI-dependent gene replacement strategy for C-terminal tag was used (28). To produce Pot1-cherry and Mre11-3GFP, the 5′ and 3′ fragments of Pot1 and Mre11 were generated by PCR, digested with SfiI and ligated to a cassette carrying a cherry-encoding gene and a triple GFP-encoding gene, respectively. Rad51-GFP, Cut11-RFP and Chk1-GFP fusions were already described (29).
To construct the mre11H228N mutant allele, we used a two-step mutagenesis protocol involving overlapping PCR. The His228 residue was replaced with Arg. This change destroyed a DraIII restriction site that was used to track the mutation by PCR amplification of genomic DNA.
Genetic screen for suppressors
Around 109 cells of a strain carrying the uku70nar1 allele grown in MMD were irradiated with UV (600 J/m2) until only 10% of the cells survived. These cells were plated in YPD plates and proliferating colonies were re-isolated. The lack of expression of the uku70nar1 allele in these clones in restrictive conditions was confirmed by RT-PCR as described in Supplementary Figure S3. Twenty independent clones that showed proliferation in YPD and no expression of uku70nar1 allele were selected for further analysis.
Protein analysis
Protein extraction and western blotting were performed using standard procedures (30). To detect the phosphorylated and non-phosphorylated forms of Cdk1, commercial antibodies were used as described (31). To detect the phosphorylated forms of Chk1, immunoprecipitates from cell extracts from strains carrying a myc-tagged form of Chk1 were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) in 8% acrylamide/0.1% bisacrylamide, pH 9.2, gels (32). Blots were incubated with anti-myc-horseradish peroxidase and visualized using a chemiluminescent substrate.
Telomere analysis
Standard telomere southern analysis was performed using established protocols with the modifications described previously (19). Two-dimensional gels were performed as described previously (33), with some modifications during DNA extraction. Briefly, 200 ml of strain cultures grown in the appropriate medium until OD600 = 1.0 were collected and treated with Novozyme to produce protoplasts as described (25). Protoplasts were resuspended in NIB buffer (17% glycerol, 50 mM MOPS buffer, pH 7.5, 150 mM K acetate, 2 mM MgCl2, 500 μM spermidine, 150 μM spermine) to break the cells and isolate the nuclei. After this, the genomic DNA was extracted with the Qiagen Genomic Extraction kit, precipitated with isopropanol and washed with 70% ethanol. DNA was finally resuspended in TE (10:1) and digested.
Microscopy
Images were obtained using a Nikon Eclipse 90i fluorescence microscope with a Hamamatsu Orca-ER camera driven by Metamorph (Universal Imaging, Downingtown, PA, USA). Images were further processed with Adobe Photoshop CS software.
RESULTS
Downregulation of the uku70 and uku80 genes in U. maydis results in G2 phase cell cycle arrest
Attempts to delete the genes encoding Ku70 or Ku80 in U. maydis (uku70 and uku80, respectively; Supplementary Figure S1) by standard disruption procedures were unsuccessful, raising the notion that Ku70/80 complex is essential for viability. This was unanticipated since Ku structural genes in a number of fungal species, including ascomycete yeasts as well as basidomycetes such as Cryptococcus neoformans and Coprinopsis cinerea (34,35) have been deleted with no major effects on proliferation. In fact, the only organism yet reported where an essential role of the Ku proteins has been described is human (16,17). To address this possibility, we inactivated one uku70 or uku80 allele in a diploid strain, replacing it with a hygromycin-resistance cassette to generate the corresponding null allele. When the meiotic progeny of these strains were analyzed after sporulation, no hygromycin-resistant cells were found, indicating that both uku70 and uku80 are essential genes in U. maydis (Supplementary Figure S2).
To characterize the Ku-encoding genes in U. maydis in more detail we constructed conditional alleles of both the ku70 and ku80 genes by exchanging their native promoters with the regulable nar1 promoter (Supplementary Figure S3A). This promoter is active when cells are grown in nitrate as the nitrogen source (MMD, minimal medium), but is repressed when cells are grown in ammonium or amino acids as nitrogen source (YPD, rich medium). Repressing uku70 or uku80 expression (Supplementary Figure S3B) resulted in a drastic growth impairment of the strains on solid medium (Figure 1A) as well as in liquid medium (Supplementary Figure S4A). The decrease in growth is more clearly observed in the uku70nar1 strain, most likely because of some leakiness in the uku80nar1 allele. We wondered whether this growth defect correlated with some defect in cell cycle. In U. maydis, the cell cycle stage can be easily monitored by analysis of the cell morphology (36). Microscopic examination of the conditional strains growing in YPD (restrictive condition) revealed the accumulation of cells with large buds. These cells carried a single nucleus, and an intact nuclear envelope (Figure 1B, inset) as determined by staining with the DNA dye DAPI and nuclear envelope marker Cut11-RFP. Since bud formation occurs in U. maydis during the G2 phase and entry into mitosis is accompanied by nuclear membrane extrusion (36), these microscopic observation strongly suggest that these cells were arrested at the G2 phase. FACS analysis (Figure 1C) indicates that cells of the conditional strains grown in restrictive conditions have 2C DNA content, corroborating the assumption that Ku-conditional cells were arrested at the G2 phase in restrictive conditions.
Figure 1.

Ku complex is required for proliferation in U. maydis. (A) Growth of conditional strains in solid medium. Serial 10-fold dilutions of FB1 (WT), UCS33 (uku70nar1) and UCS30 (uku80nar1) cultures were spotted on solid rich medium (YPD) and minimal medium with nitrate (MMD). YPD plates were incubated for 2 days and the nitrate plates for 4 days at 28°C. (B) Morphology of wild-type (UMP132), uku70nar1 (UCS34) and uku80nar1 (UCS37) cells incubated for 8 h in permissive (MMD) and restrictive (YPD) conditions. All cells were shown at the same magnification, Bar: 15 μm. Note that the uku70nar1 and uku80nar1 mutants in YPD showed buds substantially more enlarged and elongated, which are characteristic of G2 arrested cells (white stars). Inset showed the nucleus of a representative cell. Nuclear membrane (red) was visualized using Cut11-RFP and DNA (blue) was visualized upon staining with DAPI. (C) FACS analysis of FB1 (WT), UCS33 (uku70nar1) and UCS30 (uku80nar1) cell DNA content after 8 h of incubation in permissive (minimal medium with nitrate, MMD) or restrictive conditions (rich medium, YPD). 1C and 2C indicate haploid and diploid DNA content. The shift to a DNA content higher than 2C observed in conditional cells incubated in YPD was due to mitochondrial DNA staining (31).
In summary, these results indicate that down-regulating Ku expression blocks cell proliferation and causes cells to arrest at the G2/M transition.
Disabling the DNA damage response suppresses the essentiality of Ku
As it is known that Ku plays an important role in the NHEJ pathway of DNA repair (37), it seemed possible that the essentiality of Ku proteins in U. maydis might be related to a DNA repair defect. However, we considered this unlikely because the HR pathway appears to be the predominant mode of repair of DSBs in U. maydis and because cells deleted for dnl4, the gene encoding DNA ligase IV that is required for NHEJ (37), were not affected in their ability to proliferate (Supplementary Figure S5).
In Saccharomyces cerevisiae yku70 mutants are unable to proliferate at high temperature as a consequence of a G2/M cell cycle arrest (38). However, deletion of the gene encoding the 5′-3′ exonuclease Exo1 strongly suppresses this growth defect (38). We deleted the gene encoding Exo1 in U. maydis (39), but observed no suppression or alleviation of the growth inhibition upon uku70 down regulation (Supplementary Figure S6).
To understand why Ku proteins are essential in U. maydis, we performed a genetic screen to isolate extragenic suppressors of the uku70nar1 conditional allele. Following mutagenesis we isolated 20 independent mutants that were able to grow in conditions of uku70 repression. The recovered mutants exhibited variable sensitivity to either ultraviolet irradiation or the DNA replication stressor hydroxyurea (HU) (Supplementary Figure S7), a phenotype we had observed before with the chk1 and atr1 mutants encoding components of the DDR pathway (29,32,40). Indeed, we found that some of the mutant isolates were rescued (in terms of DNA damage sensitivity) with a plasmid carrying chk1 or a plasmid carrying atr1 (Supplementary Figure S7). These observations suggest a connection between loss of Ku and the DDR pathway.
To confirm that disabling the DDR pathway in U. maydis suppresses the essentiality of the Ku proteins for proliferation, we deleted either the chk1 or the atr1 gene in the Ku conditional strains. Although the deletion of chk1 or atr1 has some effect on the ability of control cells to grow (more pronounced in the case of atr1 mutants, see Supplementary Figure S4B and C), the absence of either Chk1 or Atr1 in cells depleted of Ku proteins enabled these cells to proliferate (Figure 2A, Supplementary Figure S4B and C). Moreover, in these double mutant strains the G2 cell cycle arrest was suppressed as evident from the analysis of the morphology of the cells (a mixture of budded and non-budded cells) as well as from FACS analysis (Figure 2B and C).
Figure 2.

Disabling the DNA damage response suppressed the requirement of Ku for proliferation of U. maydis cells. (A) Serial 10-fold dilutions of FB1 (WT), UMP122 (chk1Δ), UCS1 (atr1Δ), UCS33 (uku70nar1), UCS35 (uku70nar1 chk1Δ), UCS40 (uku70nar1 atr1Δ), UCS30 (uku80nar1), UCS39 (uku80nar1 chk1Δ) and UCS44 (uku80nar1 atr1Δ) cultures were spotted on solid rich medium (YPD) and minimal medium with nitrate (MMD). YPD plates were incubated for 2 days and the nitrate plates for 4 days at 28°C. (B) Morphology of uku70nar1 (UCS33), uku80nar1 (UCS30), uku70nar1 chk1Δ (UCS35), uku70nar1 atr1Δ (UCS40), uku80nar1 chk1Δ (UCS39) and uku80nar1 atr1Δ (UCS44) cells incubated for 8 h in restrictive conditions (YPD). All cells were shown at the same magnification, Bar: 15 μm. (C) FACS analysis of FB1 (WT), UCS33 (uku70nar1), UCS35 (uku70nar1 chk1Δ), UCS40 (uku70nar1 atr1Δ), UCS30 (uku80nar1), UCS39 (uku80nar1 chk1Δ) and UCS44 (uku80nar1 atr1Δ) after 8 h of incubation in permissive (minimal medium with nitrate, MMD) or restrictive conditions (rich medium, YPD). 1C and 2C indicate haploid and diploid DNA content.
Absence of Ku blocks cell proliferation by triggering the DNA damage checkpoint
In U. maydis activation of the DDR pathway results in a G2 cell cycle arrest (32). The results here showing the same arrest upon Ku depletion and suppression of Ku essentiality by loss of Chk1 or Atr1 imply that the absence of Ku activates the DNA damage checkpoint. Activation of the DDR is marked by phosphorylation of Chk1 and by its relocalization into the nucleus. To test whether the DDR is activated by Ku depletion we used fluorescence microscopy to track accumulation of GFP-tagged Chk1 in the nucleus of live cells, and we monitored increased phosphorylation of Chk1 by looking for a mobility shift of myc-tagged Chk1 following SDS-gel electrophoresis and western blot analysis (32). Upon depletion of the Ku proteins, we observed that Chk1-GFP accumulated in the nucleus of almost all the cells (Figure 3A and B) and that myc-tagged Chk1 exhibited reduced electrophoretic mobility indicative of hyperphosphorylation (Figure 3C). These results support the notion that Ku depletion activates the DDR.
Figure 3.
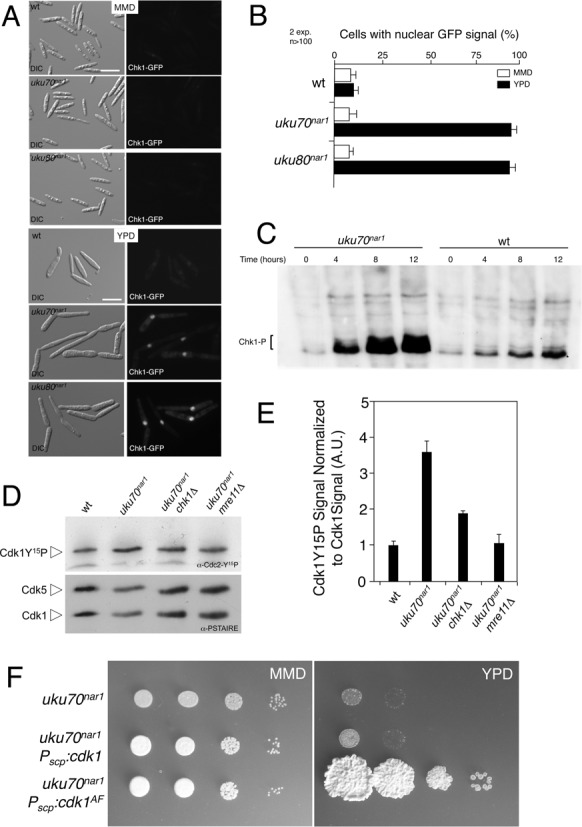
Down-regulation of Ku genes triggers the DNA damage response. (A) Cell images of UMP111 (wt), UCS43 (uku70nar1) and UCS42 (uku80nar1) strains carrying a Chk1-3GFP fusion grown for 8 h in permissive (MMD) and restrictive (YPD) conditions. All cells were shown at the same magnification (Bar: 15 μm). (B) Quantification of the cellular response to DNA damage as the percentage of cells carrying a clear nuclear GFP fluorescence signal. (C) In vivo phosphorylation of Chk1 in response to Ku70 protein depletion. UMP124 (wt) and UMP231 (uku70nar1) cells carrying an endogenous Chk1-3MYC allele were incubated in YPD (restrictive conditions) for the indicated time. Protein extracts were immunoprecipitated with a commercial anti-MYC antibody and the immunoprecipitates were subjected to SDS-PAGE and immunoblotting using anti-MYC antibody. Phosphorylated Chk1 migrated as a smear (brackets) most likely because the presence of multiple species of phosphorylated protein. (D) Levels of Cdk1 inhibitory phosphorylation upon Ku70 depletion. Protein extracts from the indicated strains (FB1, wt; UCS33, uku70nar1; UCS35, uku70nar1 chk1Δ; UMP218, uku70nar1 mre11Δ) that were grown in restrictive conditions (YPD) for 8 h were separated by SDS-PAGE. Immunoblots were probed successively with an antibody that recognizes phosphorylated Cdk1 (α-Cdc2-Y15P) and anti-PSTAIRE. (E) Levels of Cdk1 phosphorylation were determined by quantifying the level of antibody signal using a ChemiDoc unit (Bio-Rad). Signal from the phosphopeptide-specific antibodies was normalized to the amount of phosphorylation in the control strain (FB1). Differences in loading of samples were corrected by dividing each phosphopeptide-specific antibody signal by the Cdk1 (α-PSTAIRE) antibody signal. Two independent experiments were used to calculate the mean and s.d. (F) Growth of conditional strains in solid medium. Serial 10-fold dilutions of UCS33 (uku70nar1), UMP221 (uku70nar1 Pscp:cdk1), UMP222 (uku70nar1 Pscp:cdk1AF) cultures were applied to solid rich medium (YPD) and minimal medium with nitrate (MMD). YPD plates were incubated for 2 days at 28°C.
It was shown previously that activation of Chk1 in U. maydis results in the inhibition of Cdc25, the protein phosphatase that removes the Wee1-mediated inhibitory phosphorylation of Cdk1 (31,41). As a result of this inhibition, Tyr15-phosphorylated Cdk1 accumulates and the cells are unable to enter mitosis (32,42). We analyzed the level of inhibitory phosphorylation in cells depleted of Ku70, and as expected, observed an increase in the Tyr15-phosphorylation of Cdk1 and a reduction in Tyr15-phosphorylation when chk1 was removed (Figure 3D and E).
All together these results suggest that the likely reason for the essentiality of Ku proteins in U. maydis is the activation of the DDR pathway, which results in a permanent G2 cell cycle arrest. If so, then bypassing the cell cycle arrest should render the Ku proteins dispensable for proliferation. One way to bypass arrest is to substitute a Cdk1 variant that is refractory to inhibitory Tyr15-phosphorylation, such as encoded by the cdk1AF allele (31). Because high level expression of cdk1AF is lethal in U. maydis (31), we introduced the uku70nar1 conditional allele into a strain carrying an ectopic copy of the cdk1AF allele under the control of the weak constitutive scp promoter (43), which produces low levels of Cdk1AF, but sufficient to bypass the Wee1-mediated inhibitory phosphorylation of Cdk1 (31). We also introduced the uku70nar1 conditional allele into a control strain carrying an ectopic copy of a wild-type cdk1 allele under the control of the same promoter. It was evident that the down-regulation of uku70 expression in the strain carrying the cdk1AF allele (Supplementary Figure S8A) did not result in the loss of cell proliferation (Figure 3F). Microscopic examination of this strain grown in restrictive conditions showed no accumulation of cells with long buds, again consistent with bypass of the G2 arrest. Although some cells showed defects in cell separation (Supplementary Figure S8B), this is a previously reported consequence of cdk1AF expression, which is unrelated to uku70 repression (31).
The essential function of Ku depends on the MRN complex but not on the 9–1–1 complex
The activation of DDR upon Ku depletion suggests that some type of DNA damage is produced in the cell when Ku is absent. To clarify the nature of this damage, we disabled two different complexes devoted to signaling different forms of DNA damage—Mre11-Rad51-Nbs (MRN) complex, which senses DSBs, and 9–1–1 complex, which senses single stranded DNA that is produced as a consequence of DNA replication stress or resection (1). To disable the 9–1–1 complex we deleted the rec1 gene, encoding one of the three components of 9–1–1 (44). Mutants in rec1 were described before and showed no defects in proliferation (45). To disable the MRN complex we disrupted the mre11 gene encoding one of the MRN components (Supplementary Figure S9) (46). Like the rec1 mutant, the mre11 mutant is viable. When mutant combinations with the uku70nar1 allele were generated we found that loss of Mre11 but not Rec1 substantially suppressed the inviability of uku70nar1 cells (Figure 4A). Consistent with the growth phenotypes, accumulation of GFP-Chk1 in the nucleus in the uku70nar1 mutant was not dependent on Rec1 but completely dependent on Mre11 (Figure 4B and C). In addition, Chk1 phosphorylation was also abrogated upon Ku depletion in the absence of Mre11 (Figure 4D) and levels of inhibitory phosphorylation in Cdk1 were similar to those found in control cells (Figure 3D and E). Supporting these findings, we identified mre11 as one of the genes inactivated in the collection of mutants able to suppress the lethality of Ku70 depleted cells by showing that a plasmid carrying a wild-type mre11 allele could complement the UV and HU phenotype (clone 20 in Supplementary Figure S7). Our results contrast with those obtained in S. cerevisiae yku70 mutant cells, where the observed thermosensitive growth defect was suppressed by deletion of exo1, but was exacerbated by deletion of mre11 (38).
Figure 4.
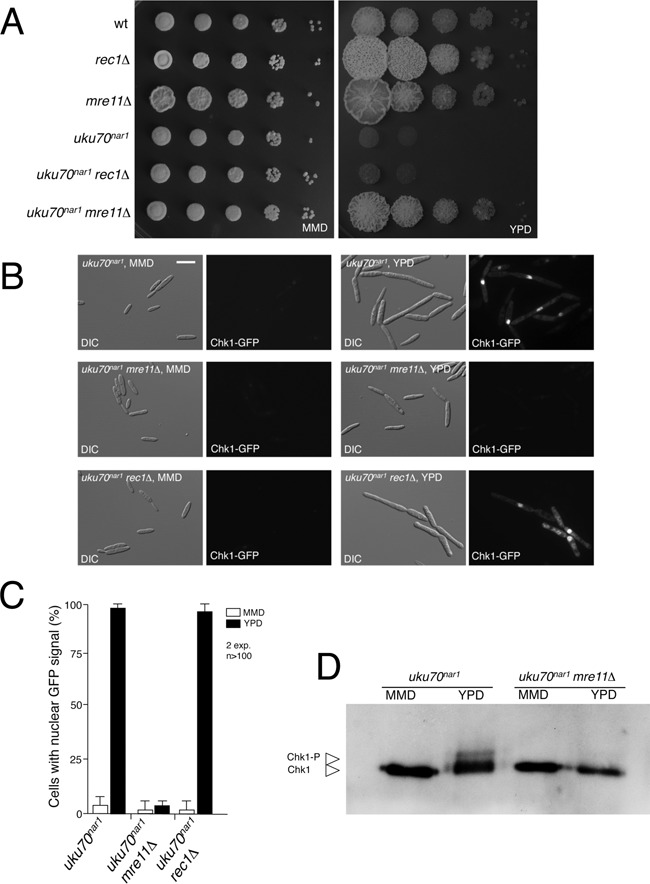
The MRN complex is required for the essential role of Ku proteins. (A) Serial 10-fold dilutions of FB1 (WT), UM210 (rec1Δ), UMP219 (mre11Δ), UCS33 (uku70nar1), UMP220 (uku70nar1 rec1Δ), UMP218 (uku70nar1 mre11Δ), cultures were applied to solid rich medium (YPD) and minimal medium with nitrate (MMD). YPD plates were incubated for 2 days and the nitrate plates for 4 days at 28°C. (B) Cell images of UMP111 (wt), UCS43 (uku70nar1), UMP223 (uku70nar1 rec1Δ) and UMP224 (uku70nar1 mre11Δ) carrying a Chk1-GFP fusion grown for 8 h in permissive (MMD) and restrictive (YPD) media. All cells were shown at the same magnification (Bar: 15 μm). (C) The percentages of cells carrying a clear nuclear GFP fluorescence signal in the indicated strains were quantified. (D) In vivo phosphorylation of Chk1. UMP231 (uku70nar1) and UMP228 (uku70nar1 mre11Δ) cells carrying an endogenous Chk1-3MYC allele were incubated in YPD (restrictive conditions) or MMD (permissive conditions) for 8 h. Protein extracts were immunoprecipitated with a commercial anti-MYC antibody and the immunoprecipitates were subjected to SDS-PAGE and immunoblotting using the anti-MYC antibody.
In summary, since the MRN complex, which detects DSBs, is disabled by deletion of mre11, we conclude that the essential role of Ku is tied to protecting DNA ends.
Loss of Ku causes telomere aberrations
Given the strong indication that Ku depletion results in a form of DNA damage recognized by the MRN complex, we monitored formation of Rad51 foci to assess the nature of the damage since Rad51 forms discrete nuclear foci in response to DSBs. We expressed a Rad51-GFP fusion and used live-cell imaging to monitor focus formation in Ku depleted cells. Focus formation was evident in the majority of cells in the population and these foci were located at the periphery of nucleus (Supplementary Figure S10). In light of the observations (i) that telomeres in fungi tend to concentrate in a single nuclear domain around the nuclear periphery (47,48), (ii) that MRN is known to play a conserved function in telomere regulation (49–51) and (iii) that the Ku complex is also associated with telomeres and is implicated in telomere regulation (6), we considered the possibility that telomeres might be the sites of origin of the DDR activation.
Therefore, we examined the co-localization of telomeres and Rad51 foci. As a marker for telomeres we expressed the telomere binding protein Pot1 (19) tagged with cherry fluorescent protein (Pot1-cherry) and found that it formed 1–2 foci per cell, usually located at the periphery of nucleus (Supplementary Figure S11). We introduced the Pot1-cherry fusion into cells carrying the Rad51-GFP fusion as well as the uku70nar1 and uku80nar1 alleles. In control cells, Rad51-GFP foci were rarely detected, and these foci rarely co-localized with the Pot1-cherry foci. In contrast, a high percentage of Ku depleted cells showed Rad51-GFP foci, and these foci co-localized at high frequency with Pot1-cherry foci (Figure 5A and B). These results implicate telomeres as the source of the DDR signal in Ku depleted cells.
Figure 5.
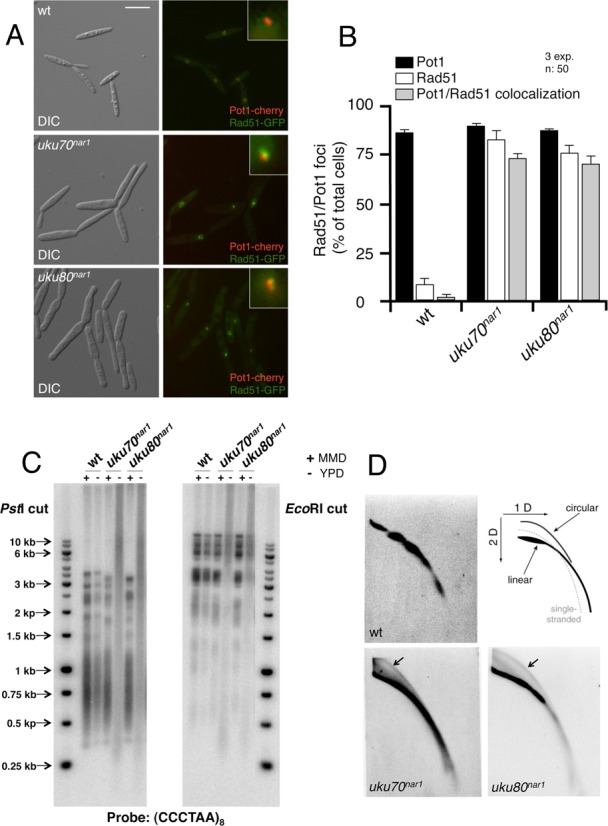
Depletion of the Ku proteins produced cells with altered telomeres. (A) Live cell analysis of Rad51-GFP and its co-localization with Pot1-cherry, a telomere marker. Control (wt, UCS57) and Ku conditional (uku70nar1, UCS45; uku80nar1, UCS46) cells were incubated for 8 h in restrictive conditions (YPD), and images were taken with the corresponding set of filters. Pictures showed DIC and merged image from GFP and cherry fluorescence of representative samples. Each inset shows a magnified image of a nucleus. Bar: 15 μm. (B) Quantification of the percentages of cells showing Rad51 and Pot1 foci as well as those with co-localization of Rad51 and Pot1. (C) Southern analysis of telomeric restriction fragments. DNAs from the indicated strains (FB1, wt; UCS33, uku70nar1; UCS30, uku80nar1) grown in restrictive (−,YPD) or permissive (+, MMD) conditions for 18 h was isolated, digested with either PstI or EcoRI and hybridized with a radioactively labeled telomere-specific probe. (D) Ku conditional cells contain elevated levels of extrachromosomal t-circles. Two-dimensional neutral/neutral gel electrophoresis analyses of restriction enzyme-digested genomic DNA from the indicated strains (FB1, wt; UCS33, uku70nar1; UCS30, uku80nar1) grown in restrictive conditions (YPD) for 8 h. After electrophoresis, the DNAs in each gel were transferred to a nitrocellulose memrbane and hybridized with a radioactively labeled telomere-specific probe. A diagram illustrating the expected arcs for linear genomic DNA, open-circular DNA (‘t-circle’) and for single-stranded DNA is shown in the upper right corner. Extrachromosomal t-circles in the uku70nar1 and uku80nar1 blots are marked by arrows.
We then directly investigated telomere aberrations by telomere restriction fragment (TRF) analysis. In Ku depleted cells, we observed a loss of normal TRF distribution pattern, accompanied by a substantial increase in the heterogeneity of TRF lengths, which ranged from <0.5 Kb to >15 Kb (Figure 5C). The heterogeneous telomere lengths observed upon Ku depletion as well as the accumulation of Rad51 at telomeres suggest that depleting Ku in U. maydis leads to aberrant structures at telomeres. Supporting this view, we observed in 2D gels abnormally high levels of t-circles in Ku depleted cells (Figure 5D). These structures have been proposed to result from the resolution of telomere-loop junctions (t-loop junctions) by recombination proteins (52).
Disabling the DNA damage response does not suppress the formation of altered telomeres
An increase in aberrant telomere structures by recombination could be the cause or the consequence of the observed DDR activation upon Ku depletion in U. maydis. To distinguish between these possibilities we analyzed the telomere lengths in strains with the uku70nar1 conditional allele as well as with deletions of chk1 or atr1. Even though the absence of Chk1 or Atr1 suppressed the requirement of Ku proteins for proliferation, it did not abolish the heterogeneity in telomere lengths (Figure 6A). Hence, an intact DDR pathway is not required for elevated telomere recombination in cells depleted of Ku.
Figure 6.
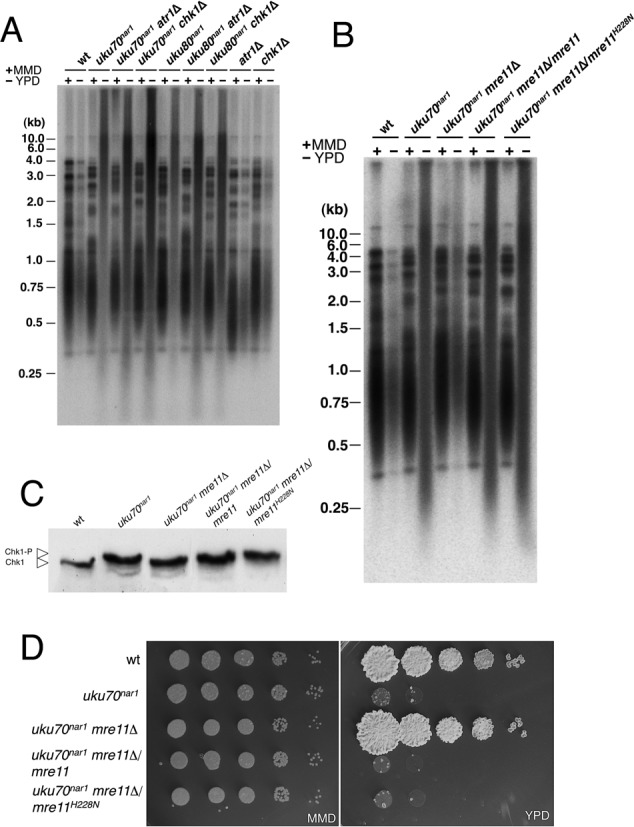
Disabling the DNA damage response does not suppress the formation of altered telomeres. (A) DNAs from the indicated strains (FB1 (WT), UCS33 (uku70nar1), UCS35 (uku70nar1 chk1Δ), UCS40 (uku70nar1 atr1Δ), UCS30 (uku80nar1), UCS39 (uku80nar1 chk1Δ), UCS44 (uku80nar1 atr1Δ), UMP122 (chk1Δ) and UCS1 (atr1Δ)) grown in restrictive (−, YPD) or permissive (+, MMD) conditions for 18 h were isolated, digested with PstI and hybridized with a radioactively labeled telomere-specific probe. (B) DNAs from the indicated strains (FB1 (WT), UCS33 (uku70nar1), UMP218 (uku70nar1 mre11Δ), UMP235 (uku70nar1 mre11Δ/mre11) and UMP236 (uku70nar1 mre11Δ/mre11H228N) grown in restrictive (−, YPD) or permissive (+, MMD) conditions for 18 h were isolated, digested with PstI and hybridized with a radioactively labeled telomere-specific probe. (C) In vivo phosphorylation of Chk1. UMP124 (wt), UMP231 (uku70nar1), UMP228 (uku70nar1 mre11Δ), UMP237 (uku70nar1 mre11Δ/mre11) and UMP238 cells carrying an endogenous Chk1-3MYC allele were incubated in YPD (restrictive conditions) for 8 h. Protein extracts were immunoprecipitated with a commercial anti-MYC antibody and the immunoprecipitates were subjected to SDS-PAGE and immunoblotted with the anti-MYC antibody. (D) Serial 10-fold dilutions of FB1 (WT), UCS33 (uku70nar1), UMP218 (uku70nar1 mre11Δ), UMP235 (uku70nar1 mre11Δ/mre11) and UMP236 (uku70nar1 mre11Δ/mre11H228N) cultures were applied to solid rich medium (YPD) and minimal medium with nitrate (MMD). YPD plates were incubated for 2 days and the nitrate plates for 3 days at 28°C.
Telomeres were also examined in strains deleted of rad51, which encodes a key component in the HR pathway. However, the lack of Rad51 did not restore normal telomere lengths (Supplementary Figure S12A) or suppress the requirement of Ku70 protein for proliferation (Supplementary Figure S12B).
The MRN complex is necessary for recombination-mediated telomere maintenance in mammals and yeasts (49–51). Since mre11 deletion suppressed the requirement of Ku for viability, we also analyzed telomere length in the double mre11Δ uku70nar1 mutant. The absence of Mre11 not only suppressed the proliferation defects but also the altered telomere lengths in the uku70 mutant (Figure 6B). This result agrees with studies in mammalian cells, where it has been proposed that Mre11 has a double role in the response to dysfunctional telomeres, participating in DDR pathway activation as well as in telomere 3′ end processing that triggers recombination processes (53).
In mammalian cells the nuclease activity of Mre11 is required for telomere 3′ end processing but is dispensable for checkpoint signaling (53). Since we cannot distinguish whether increased recombination at telomeres is the cause of DDR activation upon Ku depletion, or whether increased recombination at telomeres and DDR activation are independent processes, both triggered by Ku depletion, we thought that inactivating the Mre11 nuclease activity might help distinguish between these possibilities. We therefore constructed a mutant allele (mre11H228N) with a single amino acid change in nuclease motif III (Supplementary Figure S13) analogous to the S. cerevisiae Mre11H125N mutation, which is known to inactivate the nuclease activity (46,54,55). Either a wild-type copy of mre11 or the nuclease deficient mre11H228N allele was introduced into an uku70nar1 mre11Δ strain. In both cases cell viability was lost upon Ku depletion (Figure 6D) and activation of Chk1 was restored (Figure 6C). These results show that Mre11 nuclease activity is not required for the DDR. However, we also observed that Mre11 nuclease is not necessary for the generation of altered telomere lengths following Ku depletion (Figure 6B). In the earlier mammalian study, the Mre11 nuclease activity was implicated in the removal of 3′ overhangs at uncapped telomeres (Deng et al., 2009). Hence, our results suggest that the telomere defects in the U. maydis uku70 mutant do not involve this reaction.
Mre11 forms foci at telomeres upon Ku depletion
Regardless whether increased recombination at telomeres and DDR activation are linked or independent processes, it is clear that Mre11 plays a prominent role in response to Ku depletion. This in turn suggests that Ku might normally inhibit the activity of the MRN complex at telomeres. One potential mechanism is that Ku inhibits the ability of MRN complex to interact with telomeres. We examined the association of Mre11 at telomeres by live-cell imaging using GFP-tagged protein and Pot1-cherry as a marker for telomeres. Upon Ku depletion, the majority of cells showed colocalization of Mre11-GFP and Pot1-cherry (Figure 7). To determine whether colocalization was dependent on a functional DDR pathway, we analyzed foci in cells deleted of chk1. In this case, upon Ku depletion the Mre11-GFP protein still colocalized with Pot1-cherry. Furthermore, there was no decrease in the number of telomere-associated Mre11 foci. These results suggest that the Ku complex inhibits either recruitment or activation of the MRN complex at telomeres in U. maydis.
Figure 7.

Mre11 forms foci at telomeres upon Ku depletion. (A) Live cell analysis of Mre11-GFP and its co-localization with Pot1-cherry. UMP230 (wt), UMP231 (uku70nar1) and UMP234 (uku70nar1 chk1Δ) cells were incubated for 8 h in restrictive conditions (YPD) and images were taken with the corresponding set of filters. Pictures showed DIC and merged images from GFP and cherry fluorescence of representative samples. Each inset shows a magnified image of a nucleus. Bar: 15 μm. (B) Quantification of the percentages of cells displaying co-localized Mre11 and Pot1 foci.
DISCUSSION
In this work we report the surprising observation that the Ku complex is required for cell viability in U. maydis. Our studies suggest that the apparent essentiality of the Ku complex is due to its necessity in preventing a chronic activation of the DDR pathway, which results in permanent G2 cell cycle arrest. Disabling the DDR pathway or circumventing the DDR-responsive cell cycle arrest circuitry by using specific cell cycle mutants renders the Ku complex dispensable for proliferation. Rad51 was observed to form foci at telomeres upon Ku depletion. In addition, there was a substantial increase in the formation of aberrant telomeres and t-circles upon Ku depletion, suggesting elevated recombination at telomeres. However, we observed that in the absence of an active DDR pathway (i.e. deleting chk1 or atr1), U. maydis cells tolerate the altered telomeres observed upon Ku depletion, indicating that the proliferation defects in Ku mutants cannot be explained by the telomere aberrations alone. In conclusion, our results point to unprotected telomeres as the most likely source of the signal that activates the DDR pathway in the absence of Ku, explaining the essential role of the Ku complex in U. maydis.
The Ku complex has been proposed to protect telomeres from inappropriate degradation and fusion in a number of organisms, from yeast to human (5–7). Indeed, inactivation of Ku resulted in altered telomere length in most of the experimental systems analyzed. Despite dramatic alterations in the length of telomeres in numerous species lacking Ku, these cells are able to proliferate.
In budding yeast, yku70 mutants showed proliferative defects associated with a G2/M cell cycle arrest when grown at high temperatures, which can be alleviated with additional mutations in genes involved in DDR such as CHK1 and MEC1, suggesting that, as we have described here, the loss of Ku proteins in budding yeast also triggers the DDR (38). Also, similar to what we have described here, in spite of the ability to suppress the growth defect, deletion of the CHK1 gene does not abrogate the telomere defects associated with the lack of yKu70. However, in striking contrast to our results, in S. cerevisiae, deletion of EXO1 but not MRE11 suppressed the growth defects of yku70. Thus, there are clear similarities as well as obvious differences between the roles of the Ku complex in the standard budding yeast and their roles in U. maydis.
In human cells, conditional depletion of Ku86 resulted in lack of proliferation accompanied by high levels of t-circles and a precipitous loss of telomere repeats from chromosome ends (17). Human Ku86 depletion resulted in p53 activation, so most likely, the depletion also activates the DDR (16). In light of our observations, it will be interesting to determine if the loss of viability of the human Ku86 mutant also involves checkpoint-mediated cell cycle arrest.
Depleting Ku in U. maydis triggers two processes—cell cycle arrest as a consequence of the DDR and unscheduled recombination at telomeres. We found that Mre11, a component of the MRN complex, was required for both processes, which is in agreement with a previous report in mammalian cells (53). In addition we observed the formation of Mre11 foci at telomeres upon down-regulation of Ku expression. These foci are not a consequence of DDR activation since they are also produced in the absence of Chk1. On the basis of these observations, we propose that in U. maydis, Ku prevents excessive recruitment or activation of the MRN complex at telomeres. We imagine that in the absence of Ku an abnormally high level of MRN is recruited to telomeres, thereby signaling checkpoint activation and triggering unscheduled recombination. In mammalian cells, the nuclease activity of Mre11 is dispensable for DDR activation, but required for a telomere 3′ overhang processing reaction (53). Our analysis in U. maydis with the equivalent nuclease-deficient Mre11 variant suggests that this activity is not only dispensable for DDR, but also not required for the aberrant telomeric recombination. Hence, removal of the 3′ telomere overhang may not be involved in generation of telomere defects in Ku depleted cells.
How can the Ku complex inhibit access of MRN to telomeres in U. maydis? One possibility is that Ku plays an independent and direct role, protecting telomeres from unscheduled recombination caused by uncharacterized nucleases as was described in the case of Exo1 in S. cerevisiae (56). A second possibility is that the Ku complex in U. maydis collaborates with the shelterin complex to protect the telomere. In mammalian cells it has been reported that Ku70 is able to interact with TRF2 (57), and it has been suggested that the effects of Ku at human telomeres could be mediated through its interaction with shelterin (17). Shelterin components are evident in the U. maydis genome (19,58), but so far no characterization of their roles has been undertaken. Future research will be needed to address these possibilities.
Assuming that the signal that activates DDR in U. maydis upon Ku complex depletion emanates from unprotected telomeres, it is unclear why in U. maydis there would be no failsafe mechanism in place to prevent or attenuate DDR activation. No previous study from any other system has reported that the absence of Ku triggers irreversible DDR in spite of dramatic effects on telomere homeostasis. Recently it has been proposed that in human cells, deprotected telomeres are able to adopt an intermediate-state that induces a differential DDR, which does not lead to an immediate cell cycle arrest unless the problem is finally not resolved (59). It is conceivable that a mechanism for this flexibility in the response is not present in U. maydis, and therefore even a moderate telomere deprotection could induce a full-blown activation of DDR resulting in the cell cycle arrest. In other words, U. maydis could be hypersensitive to telomere defects. It is worth noting that in contrast to other well-known systems, U. maydis lacks a Chk2/Rad53 ortholog and therefore relies only on Chk1 as a downstream transducer of the Ataxia Telangiectasia Mutated (ATM) and Ataxia Telangiectasia and Rad9 related (ATR) checkpoint kinases (32,40). It is possible that the postulated inflexibility in the DDR activation from telomere defects is the consequence of a DDR pathway less subject to fine-tuning.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Prof. Pedro San-Segundo (IBFG, Salamanca) for stimulating discussions.
Footnotes
These authors contributed equally to this work.
FUNDING
Ministerio de Economia y Competitividad (MICINN); Gobierno de España [BIO2011-27773, BIO2014-55398-R]; Weill Cornell Cancer Center (to N.F.L.); Alice Bohmfalk Charitable Trust (to N.F.L.); National Institutes of Health [GM042482, GM079859 to W.K.H.]. Funding for open access charge: MICINN; Gobierno de España [BIO2011-27773].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deriano L., Roth D.B. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 3.Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 4.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downs J.A., Jackson S.P. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 6.Fisher T.S., Zakian V.A. Ku: a multifunctional protein involved in telomere maintenance. DNA Rep. 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira M.G., Miller K.M., Cooper J.P. Indecent exposure: when telomeres become uncapped. Mol. Cell. 2004;13:7–18. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- 8.Baumann P., Cech T.R. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell. 2000;11:3265–3275. doi: 10.1091/mbc.11.10.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulton S.J., Jackson S.P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gravel S., Wellinger R.J. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 2002;22:2182–2193. doi: 10.1128/MCB.22.7.2182-2193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riha K., Shippen D.E. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:611–615. doi: 10.1073/pnas.0236128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zellinger B., Akimcheva S., Puizina J., Schirato M., Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 13.Wei C., Skopp R., Takata M., Takeda S., Price C.M. Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 2002;30:2862–2870. doi: 10.1093/nar/gkf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d'Adda di Fagagna F., Hande M.P., Tong W.M., Roth D., Lansdorp P.M., Wang Z.Q., Jackson S.P. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 15.Espejel S., Franco S., Rodriguez-Perales S., Bouffler S.D., Cigudosa J.C., Blasco M.A. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 2002;21:2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G., Nelsen C., Hendrickson E.A. Ku86 is essential in human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Ghosh G., Hendrickson E.A. Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holloman W.K., Schirawski J., Holliday R. The homologous recombination system of Ustilago maydis. Fungal Genet. Biol. 2008;45(Suppl. 1):S31–S39. doi: 10.1016/j.fgb.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu E.Y., Kojic M., Holloman W.K., Lue N.F. Brh2 and Rad51 promote telomere maintenance in Ustilago maydis, a new model system of DNA repair proteins at telomeres. DNA Rep. 2013;12:472–479. doi: 10.1016/j.dnarep.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badie S., Escandell J.M., Bouwman P., Carlos A.R., Thanasoula M., Gallardo M.M., Suram A., Jaco I., Benitez J., Herbig U., et al. BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat. Struct. Mol. Biol. 2010;17:1461–1469. doi: 10.1038/nsmb.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banuett F., Herskowitz I. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5878–5882. doi: 10.1073/pnas.86.15.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holliday R. Ustilago maydis. In: King RC, editor. Handbook of Genetics. NY: Plenum Press; 1974. pp. 575–595. [Google Scholar]
- 23.Brachmann A., Weinzierl G., Kamper J., Kahmann R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 2001;42:1047–1063. doi: 10.1046/j.1365-2958.2001.02699.x. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Muse T., Steinberg G., Perez-Martin J. Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot. Cell. 2003;2:494–500. doi: 10.1128/EC.2.3.494-500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukuda T., Carleton S., Fotheringham S., Holloman W.K. Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 1988;8:3703–3709. doi: 10.1128/mcb.8.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmann H.A., Kahmann R., Bolker M. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 1996;15:1632–1641. [PMC free article] [PubMed] [Google Scholar]
- 27.Brachmann A., Konig J., Julius C., Feldbrugge M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics. 2004;272:216–226. doi: 10.1007/s00438-004-1047-z. [DOI] [PubMed] [Google Scholar]
- 28.Becht P., Konig J., Feldbrugge M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 2006;119:4964–4973. doi: 10.1242/jcs.03287. [DOI] [PubMed] [Google Scholar]
- 29.Mielnichuk N., Sgarlata C., Perez-Martin J. A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J. Cell Sci. 2009;122:4130–4140. doi: 10.1242/jcs.052233. [DOI] [PubMed] [Google Scholar]
- 30.Garrido E., Voss U., Muller P., Castillo-Lluva S., Kahmann R., Perez-Martin J. The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAPK protein. Genes Dev. 2004;18:3117–3130. doi: 10.1101/gad.314904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sgarlata C., Perez-Martin J. Inhibitory phosphorylation of a mitotic cyclin-dependent kinase regulates the morphogenesis, cell size and virulence of the smut fungus Ustilago maydis. J. Cell Sci. 2005;118:3607–3622. doi: 10.1242/jcs.02499. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Martin J. DNA-damage response in the basidiomycete fungus Ustilago maydis relies in a sole Chk1-like kinase. DNA Rep. 2009;8:720–731. doi: 10.1016/j.dnarep.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Ayuda-Duran P., Devesa F., Gomes F., Sequeira-Mendes J., Avila-Zarza C., Gomez M., Calzada A. The CDK regulators Cdh1 and Sic1 promote efficient usage of DNA replication origins to prevent chromosomal instability at a chromosome arm. Nucleic Acids Res. 2014;42:7057–7068. doi: 10.1093/nar/gku313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goins C.L., Gerik K.J., Lodge J.K. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet. Biol. 2006;43:531–544. doi: 10.1016/j.fgb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Nakazawa T., Ando Y., Kitaaki K., Nakahori K., Kamada T. Efficient gene targeting in DeltaCc.ku70 or DeltaCc.lig4 mutants of the agaricomycete Coprinopsis cinerea. Fungal Genet. Biol. 2011;48:939–946. doi: 10.1016/j.fgb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Steinberg G., Perez-Martin J. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 2008;18:61–67. doi: 10.1016/j.tcb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Weterings E., Chen D.J. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 38.Maringele L., Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao N., Kojic M., Holloman W.K. Role of Blm and collaborating factors in recombination and survival following replication stress in Ustilago maydis. DNA Rep. 2009;8:752–759. doi: 10.1016/j.dnarep.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Sena-Tomas C., Fernandez-Alvarez A., Holloman W.K., Perez-Martin J. The DNA damage response signaling cascade regulates proliferation of the phytopathogenic fungus Ustilago maydis in planta. Plant Cell. 2011;23:1654–1665. doi: 10.1105/tpc.110.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sgarlata C., Perez-Martin J. The cdc25 phosphatase is essential for the G2/M phase transition in the basidiomycete yeast Ustilago maydis. Mol. Microbiol. 2005;58:1482–1496. doi: 10.1111/j.1365-2958.2005.04925.x. [DOI] [PubMed] [Google Scholar]
- 42.Mielnichuk N., Perez-Martin J. 14-3-3 regulates the G2/M transition in the basidiomycete Ustilago maydis. Fungal Genet. Biol. 2008;45:1206–1215. doi: 10.1016/j.fgb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 43.Bolker M., Bohnert H.U., Braun K.H., Gorl J., Kahmann R. Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI) Mol. Gen. Genet. 1995;248:547–552. doi: 10.1007/BF02423450. [DOI] [PubMed] [Google Scholar]
- 44.Thelen M.P., Venclovas C., Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 45.Onel K., Koff A., Bennett R.L., Unrau P., Holloman W.K. The REC1 gene of Ustilago maydis, which encodes a 3’–>5’ exonuclease, couples DNA repair and completion of DNA synthesis to a mitotic checkpoint. Genetics. 1996;143:165–174. doi: 10.1093/genetics/143.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreau S., Ferguson J.R., Symington L.S. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funabiki H., Hagan I., Uzawa S., Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McEachern M.J., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian L., Moser B.A., Nakamura T.M. Recombination-based telomere maintenance is dependent on Tel1-MRN and Rap1 and inhibited by telomerase, Taz1, and Ku in fission yeast. Mol. Cell. Biol. 2008;28:1443–1455. doi: 10.1128/MCB.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong Z.H., Jiang W.Q., Cesare A.J., Neumann A.A., Wadhwa R., Reddel R.R. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 52.Cesare A.J., Reddel R.R. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 53.Deng Y., Guo X., Ferguson D.O., Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arthur L.M., Gustausson K., Hopfner K.P., Carson C.T., Stracker T.H., Karcher A., Felton D., Weitzman M.D., Tainer J., Carney J.P. Structural and functional analysis of Mre11-3. Nucleic Acids Res. 2004;32:1886–1893. doi: 10.1093/nar/gkh343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dewar J.M., Lydall D. Pif1- and Exo1-dependent nucleases coordinate checkpoint activation following telomere uncapping. EMBO J. 2010;29:4020–4034. doi: 10.1038/emboj.2010.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B., Comai L. Functional interaction between Ku and the werner syndrome protein in DNA end processing. J. Biol. Chem. 2000;275:28349–28352. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- 58.Sanchez-Alonso P., Guzman P. Predicted elements of telomere organization and function in Ustilago maydis. Fungal Genet. Biol. 2008;45(Suppl. 1):S54–S62. doi: 10.1016/j.fgb.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 59.Cesare A.J., Hayashi M.T., Crabbe L., Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol. Cell. 2013;51:141–155. doi: 10.1016/j.molcel.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


