Abstract
The importance of epigenetic regulation for maintenance of embryonic stem cell (ESC) pluripotency or for initiation of differentiation is widely accepted. However, the molecular mechanisms are poorly understood. We recently reported that a hypoxic microenvironment induces ESC differentiation. In the present study, we found that hypoxia-responsive histone deacetylase 6 (HDAC6) performs an essential signaling function for myogenic differentiation of ESCs. HDAC6 was downregulated in hypoxic ESCs or during differentiation. A knock-down of HDAC6 in ESCs resulted in induction of myogenic markers, including Pax7. Suppression of HDAC6 increased acetylation of core histones H3 and H4, leading to enhanced binding of RNA polymerase II to the Pax7 promoter. Transplantation of HDAC6 knock-down cells facilitated muscle regeneration in vivo. Importantly, the downregulation of HDAC6 by hypoxia was not mediated by HIF1α or HIF2α, master transcription regulators under hypoxia, but by induction of microRNA-26a that directly targeted the 3′-untranslated region (3′-UTR) of HDAC6. A point mutation of the microRNA-26a-binding sequence in the HDAC6 3′-UTR diminished the luciferase reporter activity. Taken together, these results suggest that environmental cues of differentiation modulate the epigenetic machinery and guide stem cells to commit to a specific lineage.
INTRODUCTION
Hypoxia, a state of low oxygen, influences various pathophysiological events. Oxygen gradient and hypoxia are widespread in developing embryonic tissues that play a crucial role in vascular development (1,2). Hypoxia activates gene expression to promote cell survival, migration, invasion, metastasis and angiogenesis during tumor formation (2–4). In particular, a hypoxic microenvironment plays an important role in determining the stem cell fate, e.g. proliferation, differentiation and maintenance of stem cells (5). We previously reported that hypoxia stimulates embryonic stem cell (ESC) differentiation into the vascular lineage via hypoxia-inducible factor 1 (HIF-1) (6). Our interest is to explore the mechanisms via which a tissue microenvironment, especially hypoxia, regulates this stem cell feature.
Histone deacetylase 6 (HDAC6) is a member of the HDAC family of enzymes which remove an acetyl group from histones, leading to chromatin condensation and repression of gene transcription (7,8). HDACs can also regulate acetylation of non-histone proteins (7). HDAC6 can deacetylase histones, α-tubulin, cortactin and Hsp90 (9–11). HDAC6 modulates Hsp90 function and facilitates degradation and clearance of misfolded proteins (11). HDAC6 is a known regulator of cell motility via control of the tubulin as well as the actin network and supports endothelial-cell migration as well as angiogenesis (10,12). Indeed, overexpression of HDAC6 promotes tumorigenesis, invasive metastatic features and improves cancer survival (9). However, the function of HDAC6 in stem cell differentiation remains largely unknown.
Pluripotent ESCs can differentiate into many cell types (13). Understanding of the regulatory mechanisms of lineage commitment and thus selection of a particular gene expression pattern might allow researchers to manipulate the fate of stem cells for stem cell-based regenerative therapies. Epigenetic control by histone acetylation is known to be important for stem cell fate; however, the effects of histone acetylation on ESC fate remain a controversial topic. Enhanced acetylation of histones is crucial for maintaining ESC pluripotency (14,15). HDAC inhibition by general HDAC inhibitors like Trichostatin A (TSA) improves cell reprogramming and pluripotency (16,17). Valproic acid (VPA) also stimulates the induction of pluripotent stem cells, particularly, VPA enables reprogramming of fibroblasts by means of two factors, Oct4 and Sox2, without the oncogenes c-Myc or Klf4 (18,19). In contrast, TSA or VPA can facilitate differentiation of stem cells into specific lineages, such as neuronal, osteogenic and hepatic cells (20–22). Therefore, uncovering the epigenetic mechanisms that determine stem cell fate is a priority.
MicroRNAs (miRNAs) are small noncoding RNAs (20–22 nucleotides) that act as potent regulators of gene expression (23,24). Mature miRNAs recognize their target protein-coding messenger RNAs (mRNAs) to inhibit mRNA translation or mRNA degradation, via base pairing with complementary sequences within the 3′-untranslated region (3′-UTR) (23,24). Over 10,000 miRNAs have been documented in the ‘MicroRNA Registry’ since the miRNA was first discovered in 1993 (25). With respect to miRNA functions, they play pivotal roles in the coordination of a wide variety of processes such as cell differentiation, proliferation, death and organ development (26–28).
In the present study, we assessed the mRNA level of HDAC family genes under conditions of hypoxia to elucidate the epigenetic control by oxygen during ESC differentiation. We identified HDAC6 as a potential ESC differentiation regulator because its expression is tightly regulated by hypoxia. We found that a knock-down of HDAC6 in ESCs induces commitment to the myogenic lineage where Pax7, a transcription factor essential for skeletal muscle development, is significantly turned on. HDAC6 knock-down ESCs facilitate muscle regeneration in a mouse model of skeletal muscle injury. HDAC6 suppression by hypoxia is not mediated by HIF1α or HIF2α, but by induction of microRNA. Thus, our study demonstrated a previously unknown role of microRNA-26a in HDAC6 downregulation. We reported the regulatory mechanism involving microRNA-26a and HDAC6 during stem cell differentiation in response to environmental cues. These molecules may be therapeutic targets for salvaging damaged muscle tissue.
MATERIALS AND METHODS
Cell culture and in vitro differentiation
Undifferentiated E14 mouse embryonic stem cells (mESCs) and the C57BL/6-bacground mESCs (C57-mESCs, accession no. SCRC-1002; ATCC) were cultured on Mitomycin C (Sigma–Aldrich)-treated mouse embryonic fibroblast (MEF) feeder layer in Dulbecco's modified Eagle's medium (DMEM; GIBCO, Grand Island, NY, USA) with 20% Fetal Bovine Serum (FBS); Hyclone, 1% penicillin/streptomycin (GIBCO), 0.1 mM β-mercaptoethanol (Sigma), 1% non-essential amino acids (GIBCO), 2 mM l-glutamine and 1000 U/ml leukemia inhibitory factor (LIF; Millipore). We regarded as ‘Passage 1’ when we got the E14- or C57-ESCs from ATCC, and we performed all experiment in this manuscript using cells between passages 8–14. For hypoxic culture conditions, cells were incubated in a Hypoxia Chamber (Forma Scientific) maintaining low oxygen tension (1% O2, 5% CO2 and balanced with N2). Forma Hypoxia Chamber (anaerobic system) is more strict control of hypoxia with a closed hypoxia workstation.
Embryoid bodies (EBs) were formed using a hanging drop method (one droplet containing 500 cells/20 μl) in the absence of LIF and feeder cells. To induce spontaneous differentiation, ESCs was cultured on 0.3% gelatin-coated plate in DMEM/10% FBS. For in vitro skeletal muscle-lineage differentiation, ESCs or EBs were plated onto 0.3% gelatin-coated culture wear in DMEM/10% FBS for 1 day, and further incubated followed by replacement with specific media; SkIM (Skeletal muscle Inducing Media) with some modifications (29) [high-glucose DMEM (GIBCO), 10% FBS (Hyclone), 5% Horse serum (Sigma), 1% penicillin/streptomycin (GIBCO), 1 mM non-essential amino acids (GIBCO), 0.1 mM β-mercaptoethanol (Sigma), recombinant mouse VEGF (100 ng/ml: R&D system), transferrin (200 μg/ml: Sigma), bFGF (5 ng/ml: Invitrogen)].
Cardiotoxin-muscle injury model
All animal experiments performed under approval from the Institutional Animal Care and Use Committee of Seoul National University Hospital. For Rotarod test and tissue analysis, male C57BL/6 (8 weeks old) mice were anesthetized, 50 μl of cardiotoxin (10 μM, Sigma) was injected into both right and left tibialis anterior (TA) muscles of each mouse to induce muscle injury (30). One day later, EBs formed with shMock-ESCs or shHDAC6-ESCs (total cell numbers corresponds to 5 × 104/ 50 μl phosphate buffered saline (PBS) buffer) were injected into TA muscle of each group. As sham control, PBS was injected to TA muscle. EBs were labeled with 2 μg/ml of Celltracker CM-DiI (Invitrogen) for 30 min before EB formation by hanging drop to trace differentiation. The TA muscles were harvested and performed immunofluorescence staining to evaluate the differentiation into skeletal muscle cells in injured muscles.
Rotarod test
We tested the ability of mice to remain on rotating rod (Panlab Rota-Rods LE 8200, Harvard Apparatus) (30). We measured the latency time it takes the mouse to fall off the rod rotating under continuous acceleration (from 5 to 40 rpm) as a measurement of competence in motor function. Each mouse was given four trials, measured the each latency time on the rod, and calculated the average latency time. Mice were allowed to rest for at least 5 min between each trial. Before the cardiotoxin muscle injury, the mice were habituated to stay on the stationary drum for 3 min and then to run on the rotating rod up to three times.
Plasmid construction and transfection
For HDAC6 knock-down, we used MISSION™ TRC shRNA Target Set (TRCN00000010413) or the control sh-plamsid (MISSION Non-Target shRNA Control SHC002) (Sigma). C57 ESCs was transfected with Metafetamin (Biotex) and selected with puromycin treatment (10 μg/ml: Sigma). HDAC6 overxpression vector was purchased from Open biosystmes. miR-22 precursor DNA (pre-miR-22) containing 95 bp stem–loop sequence and 348 bp native flank sequence to both upstream and downstream of the stem–loop (NC_000077.6) (31), and miR-26a precursor DNA (pre-miR-26a) containing 77 bp stem–loop sequence and 100 bp native flank sequence to both upstream and downstream of the stem loop (32) were synthesized by PCR and cloned into pcDNA3.1 (Invtrogen). The synthetic miR-22 and miR-26a mimic-oligomers and miRNA-negative control (miR-NC) were obtained from Ambion. AntagomiR against miR-26a was obtained from Invitrogen. Transfection was performed using Metafetamin (Biotex) according to the manufacturer's protocol. Primer information was detailed in Supplementary Table S1.
miRNA target validation by luciferase assay
A 337-bp PCR fragment of HDAC6 3′UTR was cloned into the NotI site, the downstream of the Renilla luciferase gene in psiCHECK™-2 (Promega). Site-directed mutagenesis of miR-26a binding site in HDAC6 3′UTR was achieved by QuickChange II Site-Directed Mutagenesis System (Staratagene) followed by sequence verification. For reporter assays, C57 ESCs cultured in the absence of LIF and feeder cells was transfected with various combinations of effector plasmids. Luciferase assays were performed using the Dual Luciferase Assay System kit (Promega) with a GloMax luminometer (Promega) according to the manufacturer's instruction. The psiCHECK™-2 Vector also contains a constitutively expressed firefly luciferase gene, which served as an internal control to normalize transfection efficiency. Primer information was detailed in Supplementary Table S1.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed with the ChIP assay kit (Millipore) according to the manufacturer's protocol. Chromatin was immunoprecipitated with antibodies against control rabbit IgG, acetyl-H3, acetyl-H4 or RNA polymerase II (Millipore). ChIP (2 μl) or 1% input DNA was used to quantify PCR with primers. PCR primers are listed in Supplementary Table S1. After performing real-time PCR, the values were calculated using the formula 2.5 × 2[CT(input)−CT(AbIP)] (33).
RT-PCR, real-time PCR and microRNA real-time PCR analysis
Total RNA was isolated using QIAshredder and RNeasy mini kit and RNeasy plus Mini kit (Qiagen Inc.). Up to 1 μg of RNA was converted into cDNA according to the PrimeScript™ first strand cDNA Synthesis Kit (Takara). Real-time PCR was performed using the SYBR Green PCR Master Mix (Roche) with specific primers in Supplementary Table S2: HIF1α, HIF2α, Oct4, Nanog, lineage markers (6). Real-time samples were run on an ABI PRISM-7500 sequence detection system (Applied Biosystems). 18S rRNA was simultaneously run as a control and used for normalization. In addition, end-point PCR for HDAC family and Pax7 was performed. GAPDH was used for normalization. For microRNA quantitative real-time PCR, miRNA isolation and single-stranded cDNA from RNA samples were prepared using the miRNeasy mini kit amd the miScript reverse transcription kit (Qiagen Inc.) following the manufacturer's recommendations. Real-time PCR was performed using miScript SYBR green PCR kit (Qiagen Inc.) with specific primers (Supplementary Table S2). miRNAs were quantified with U6 small RNA serving as the normalization control.
Western blot assay
Cells were harvested and lysed in lysis buffer containing protease inhibitors (Roche). Total protein (10–30 μg) was immunoblotted with specific primary antibodies; HDAC6 (Abcam), Pax7 (DSHB), HIF1α (34), HIF2α (Novus), Acetyl-H3, Acetyl-H4 (Millipore). α-Tubulin (Calbiochem) was used as an internal control. Quantification of band intensity was analyzed using TINA 2.0 (RayTest) or ImageJ (NIH) and normalized to the intensity of α-tubulin.
Immunofluorescence staining
Differentiated cells from ESCs or C2C12 mouse skeletal myoblasts on on μ-Dish35mm high(ibidi) were fixed with 4% PFA, blocked with blocking buffer (0.5% goat serum, 0.1% Triton-x100/1% BSA–PBS), and labeled with anti-Pax7 (R&D) followed by fluorescent dye conjugated secondary antibody (Invitrogen). To analysis tissues of CTX injured mice, the TA muscle after cell transplantation was excised, rinsed with PBS and frozen in liquid nitrogen. Histological sections (4–8 μm-thick) were prepared from snap-frozen tissue samples, fixed with acetone, blocked in 1% BSA and incubated with anti-laminin α2 (4H8-2, Alexis Biochemicals) followed by fluorescent dye conjugated secondary antibody (Invitrogen). For quantification of regenerating myofiber, at least five randomly selected fields from transverse-sectioned slides from four different mice were analyzed. The nuclei were stained with DAPI (Molecular Probe) and mounted using Fluorescent Mounting Medium (DAKO®). The fluorescent images were obtained using a confocal microscope (Carl Zeiss LSM710).
Morphometric analysis
Paraffin sections (4–6 μm-thick) of mouse TA muscles were stained with hematoxylin & eosin (H&E) using standard protocols. The microscopic images were obtained with an Olympus TH4-200 microscope. Myofiber cross-sectional area (CSA; μm2) was measured on H&E-stained cross-sections using the ImageJ software (http://imagej.nih.gov/ij/). Ten randomly selected fields from transverse-sectioned slides from four different mice were analyzed (35,36).
Statistical analysis
The results are expressed as means ± standard deviations (SD). The differences between the groups were compared by the unpaired t-test or one-way analysis of variance (ANOVA). P values ≤0.05 were considered statistically significant. All statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, USA).
RESULTS
HDAC6 downregulation and differentiation of ESCs into the mesoderm lineage
We recently reported that hypoxia stimulates stem cell differentiation (6). Thus, we investigated the potential role of HDACs in hypoxia-induced differentiation of stem cells (Supplementary Figure S1). Among the HDAC groups, we focused on HDAC1-11 (HDAC inhibitor-sensitive groups) (7,8) given their possible clinical applications. Then, we selected HDACs responding in parallel expression patterns under both hypoxic conditions and in differentiating embryoid bodies (EBs). Among 11 HDACs, HDAC1, HDAC2 and HDAC6 were downregulated by hypoxic conditions in E14 ESCs (Supplementary Figure S1a and S1b). After differentiation as EB formation, only HDAC2 and HDAC6 mRNA were downregulated (Supplementary Figure S1c). The function of HDAC2 in stem cells has been reported (21,37), whereas the function of HDAC6 in stem cells remains unclear; therefore, we focused on HDAC6.
We confirmed HDAC6 expression in C57 ESCs (Figure 1A and B). Both HDAC6 mRNA and protein were downregulated by hypoxia or by EB formation in these cells, suggesting the possible involvement of HDAC6 in stem cell differentiation. We generated HDAC6 knock-down ESCs using shHDAC6 to study stem cell differentiation more thoroughly. HDAC6 mRNA and protein were markedly decreased after transfection of the cells with shHDAC6 (shHD6), compared to shMock control (Figure 1C and D). shHDAC6 showed selective inhibition of HDAC6 among HDAC1-11 (Figure 1C).
Figure 1.
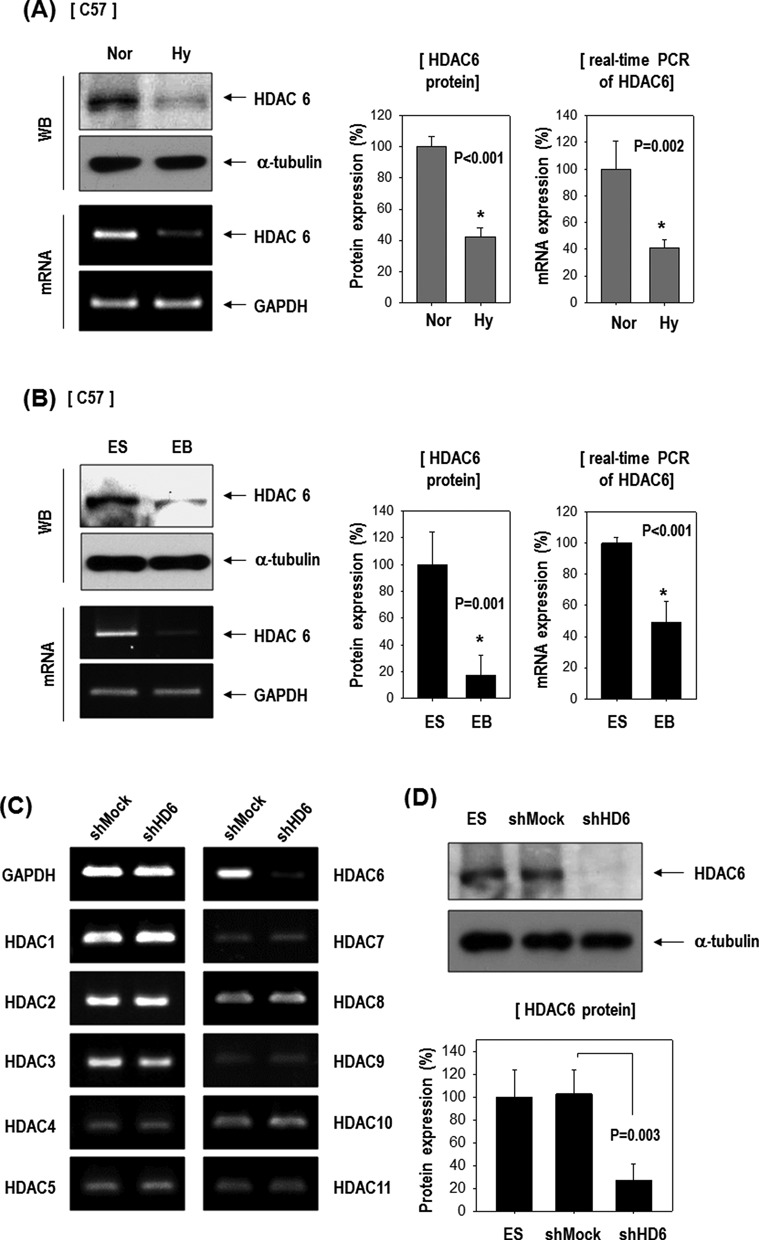
HDAC6 decreased in ESCs in response to hypoxia or during differentiation as EB formation and its knock-down stimulated genes of mesoderm- or myogenic-lineage. (A) HDAC6 protein and mRNA were reduced by 16 h hypoxia in C57 ESCs (n = 3). (B) HDAC6 protein and mRNA were reduced in EBs compared with ES (n = 3). (C and D) Generation of stable HDAC6 knock-down cells. HDAC6 mRNA (C) and protein (D) were markedly reduced by transfection of shHDAC6 (shHD6) compared with the non-target shRNA control (shMock) (n = 3). Especially, shHDAC6 transfection selectively suppressed HDAC6 among eleven HDAC family genes. (E) ESCs were cultured on a gelatin-coated plate in DMEM/10% FBS for 5 days, and real-time PCR was performed (n = 3). (F) Among mesoderm-lineage, myogenic-marker genes (Pax3, Pax7, MyoD) were markedly up-regulated in shHDAC6-cells upon differentiation, whereas vascular-marker genes (PECAM, SMA, VE-cadherin) were not (n = 4).
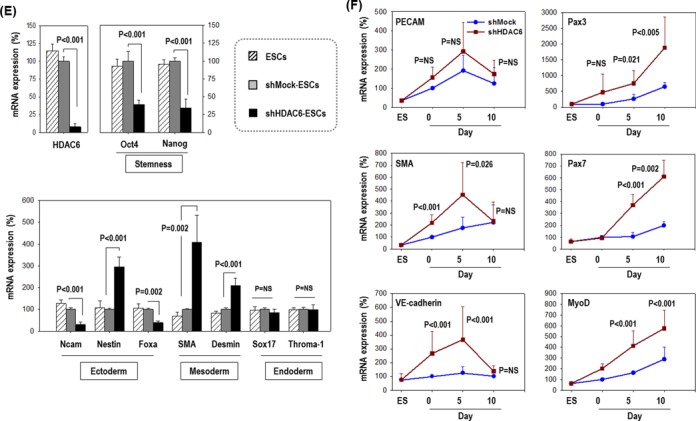
Next, we tested whether depletion of HDAC6 affects ESC commitment to a specific lineage. Pluripotency markers Oct4 and Nanog were significantly downregulated in shHDAC6-ESCs compared to shMock-ESCs, indicating that the HDAC6 knock-down facilitated ESC differentiation (Figure 1E). shHDAC6 significantly increased expression of mesoderm markers (SMA, Desmin) but did not significantly affect endoderm marker genes (Sox17, Throma-1) or ambiguously regulated ectoderm markers (Ncam, Nestin and Foxa).
Because we were interested in vascular and/or myogenic differentiation among mesoderm lineages, we next analyzed the expression of vascular and myogenic marker genes (Figure 1F). Cells were cultured under spontaneous differentiation conditions (DMEM/10% FBS) for up to 10 days, and collected at various time points. Myogenic marker genes (Pax3, Pax7 and MyoD) were markedly upregulated in shHDAC6-ESCs upon differentiation, whereas vascular marker genes (PECAM, SMA and VE-cadherin) were not. Our results suggest that HDAC6 reduction may promote ESC differentiation and the commitment toward the myogenic rather than the vascular lineage (or others).
Myogenic lineage commitment of HDAC6 knock-down ESCs
Given the above results, we analyzed and confirmed Pax7 expression in various clones of shMock- or shHDAC6-ESCs to rule out interclone variation. Pax7 is essential for muscle progenitor cell specification and is the most widely cited marker of embryonic muscle progenitors (38,39). Impressively, Pax7 mRNA and protein were upregulated in all 5 shHDAC6-ESC clones; there were clearly opposite expression patterns between Pax7 and HDAC6 (Figure 2A and B).
Figure 2.
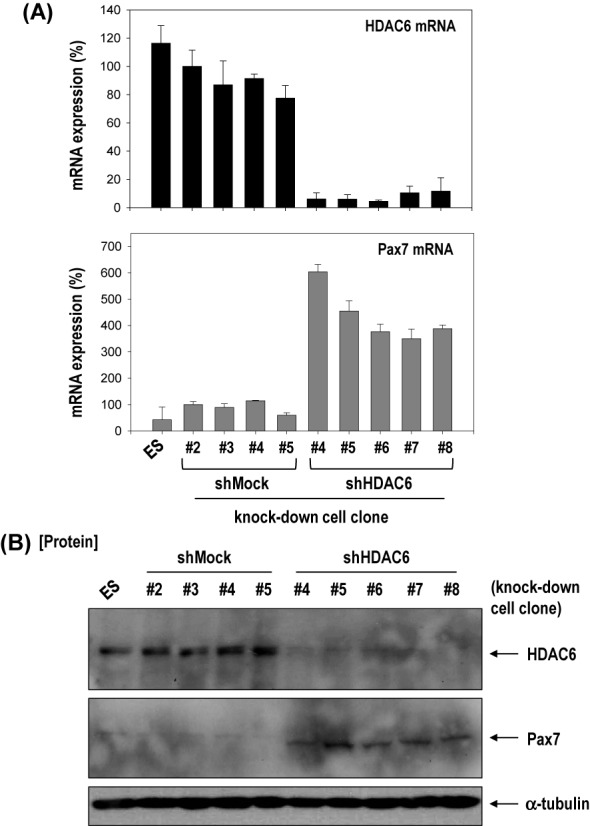
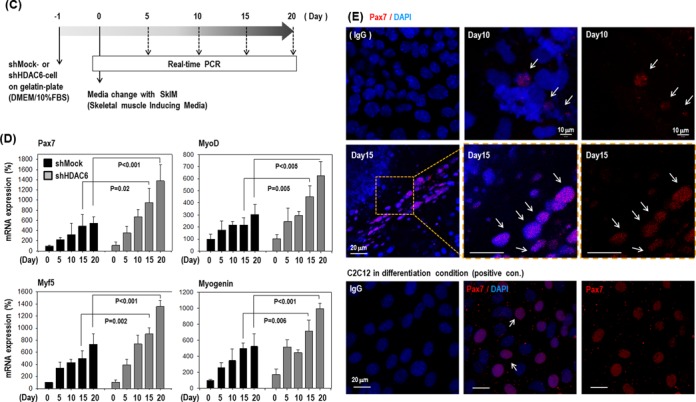
Knock-down of HDAC6 activated differentiation of ECS toward myogenic-lineage. (A and B) Pax7, a marker of muscle progenitor, was remarkably increased at mRNA and protein levels in all five clones with HDAC6 knock-down C57 ESCs compared with all four control clones with shMock transfected. (C) Cells were plated onto a gelatin-coated dish in DMEM/10% FBS for 1 day, and further incubated up to 20 days under SkIM. (D) Pax7 and myogenic factors (Myf5, MyoD, Myogenin) increased after induction of differentiation to muscle in shMock-control cells, which was accentuated at greater degree in shHDAC6-cells (n = 3). (E) Immunofluorescence staining of shHDAC6-ESCs for Pax7 (red)/ nuclei (blue) on day10 or day 15 in SkIM. Positive control for Pax7 staining was myoblast cell line, C2C12 (×200).
As shown in Figure 1F, myogenic marker genes were upregulated in shHDAC6 cells under spontaneous differentiation conditions; therefore, we further analyzed the specific myogenic differentiation potency of shHDAC6-ESCs in comparison with control shMock-ESCs (Figure 2C–E). The cells were cultured in SkIM for specific muscle differentiation (29). As shown in Figure 2D, Pax7 and muscle regulatory factors (Myf5, MyoD and Myogenin) (38,39) were significantly more upregulated during differentiation of shHDAC6-ESCs compared to control shMock-ESCs (Figure 2D). Interestingly, HDAC6 knock-down alone could drive differentiation into the myogenic lineage in the absence of SkIM. Furthermore, additive upregulation of myogenic genes was observed in shHDAC6-ESCs in SkIM during the differentiation (Supplementary Figure S2). In immunofluorescence staining, Pax7 was very rarely and barely detectable in shMock cells (data not shown), whereas strong nuclear staining was observed in shHDAC6 cells (Figure 2E) as in the myoblast cell line C2C12, indicating that shHDAC6 cells efficiently differentiated into the myogenic lineage.
Epigenetic activation of the Pax7 promoter by the knock-down of HDAC6
Because the mRNA and protein expression of muscle progenitor marker Pax7 is remarkably increased in shHDAC6-ESCs (Figure 3A), we tested whether HDAC6 epigenetically represses the Pax7 promoter. We first examined whether HDAC6 knock-down increases acetylation of histones, changes chromatin structure, and stimulates recruitment of transcriptional regulatory proteins to the Pax7 promoter (Figure 3B). We performed ChIP assay with antibodies recognizing acetylation of core histones H3 and H4. The acetylated histones H3 and H4 on the Pax7 promoter was significantly increased in shHDAC6-ESCs (Figure 3C). The increased histone acetylation is a known indicator of an active or more open chromatin structure, which facilitates binding of RNA polymerase II to these regions (40,41). Consistent with enhanced acetylation of histones H3 and H4, the binding of RNA polymerase II (Pol II) to the promoter region of Pax7, especially in region 1, was also enhanced in shHDAC6 cells (Figure 3D). In addition, we carried out western blot analysis of acetylated histones H3 and H4 (Figure 3E). Acetylation of histone H4 was remarkably increased in shHDAC6 cells, and this effect was significantly reversed by HDAC6 overexpression in shHDAC6 cells. Acetylation of histone H3 was also affected by either the HDAC6 knock-down or HDAC6 overexpression but this effect was weaker in comparison with histone H4. These results are consistent with our ChIP data. These findings suggest that the HDAC6 knock-down increased acetylation of histones H3 and H4 and changed chromatin structure, and that the more accessible chromatin structure may facilitate Pax7 transcription in shHDAC6 cells.
Figure 3.
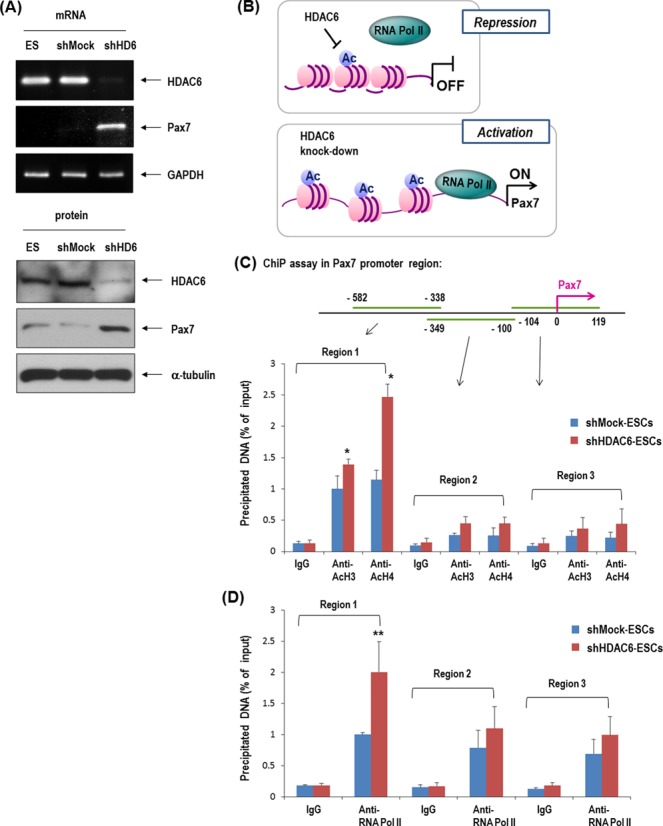
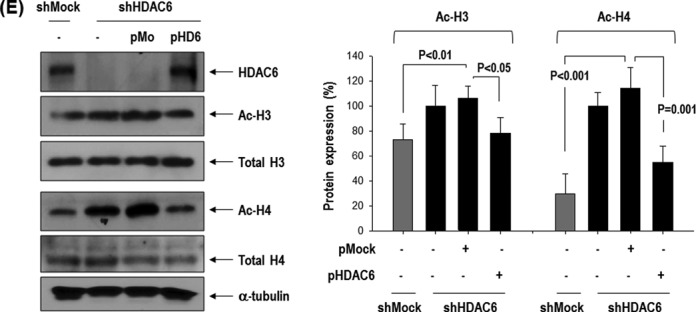
Knock-down of HDAC6 in ESCs induced epigenetic activation of Pax7 promoter. (A) Pax7 mRNA and protein remarkably increased in HDAC6 knock-down C57 ESCs. (B) Depiction of the regulation of histone acetylation and the recruitment of RNA polymerase II (Pol II) to the Pax7 promoter. The increase in the acetylation of H3 and H4 is a known indicator of active chromatin structure (40). (C and D) Quantification of ChIP assay with real-time PCR. Binding of acetylated histones H3 and H4 on Pax7 promoter was greater in HDAC6 knock-down than in control shMock. The binding of RNA polymerase II to the Pax7 promoter was also greater in shHDAC6 cells than in control ones (n = 4, *P < 0.001, **P < 0.05). (E) Western blot of acetylated histones H3 and H4 that were increased in shHDAC6 cells but were decreased by overexpression of HDAC6 in shHDAC6 cells. Quantification graph of western blot (n = 3).
HDAC6 knock-down improves efficacy of ESCs to regenerate skeletal muscle
shHDAC6-ESCs showed efficient commitment to the myogenic lineage in vitro. We tested whether the HDAC6 knock-down in ESCs facilitates muscle differentiation of ESCs in vivo, thereby leading to enhanced muscle regeneration using a mouse model of skeletal muscle injury (Figure 4). In order to monitor the time course of muscle regeneration, we performed Rotarod test after muscle injury by cardiotoxin (CTX). We injured tibialis anterior (TA) muscles of both legs by injecting CTX and transplanted ESCs with or without the knock-down of HDAC6 (CTX+shHD6 versus CTX+shMock) 1 day after CTX injection. We monitored locomotive recovery every week up to week 8 by recording time to fall off the rotating rod (Figure 4A–C). Consistent with the superior myogenic differentiation potential, mice transplanted with HDAC6 knock-down cells (CTX+shHD6 cells) showed significantly better motor function than did mice transplanted with shMock-cells (CTX+shMock cells) since 2 weeks after injury (Figure 4C). Such superiority of the therapeutic efficacy of shHDAC6-ESCs to control shMock-ESCs continued until week 8 (Figure 4B). In the histological analysis (Figure 4D), we observed regenerating myofibers that had a centrally located nucleus, and originated from transplanted ESCs marked by DiI labeling. We also observed laminin α2 immunofluorescence because muscle fiber in skeletal muscles is surrounded by the basal lamina whose major components are laminins (42). The basal lamina surrounding skeletal muscle fibers was strongly stained by laminin α2 antibody (green) in the shHDAC6 group. The regenerating myofibers with DiI fluorescence (red) were more frequently found in the shHDAC6 group than in the shMock group (Figure 4D). Furthermore, myofibers positive for DiI in the shHDAC6 group were bigger than those in the shMock group. Cross sectional area of regenerated myofibers in the shHDAC6 group were significantly bigger than those in the shMock group (Figure 4E). These results are suggestive of successful engraftment, differentiation into the muscle lineage, and improved Rotarod performance in the shHDAC6 group.
Figure 4.
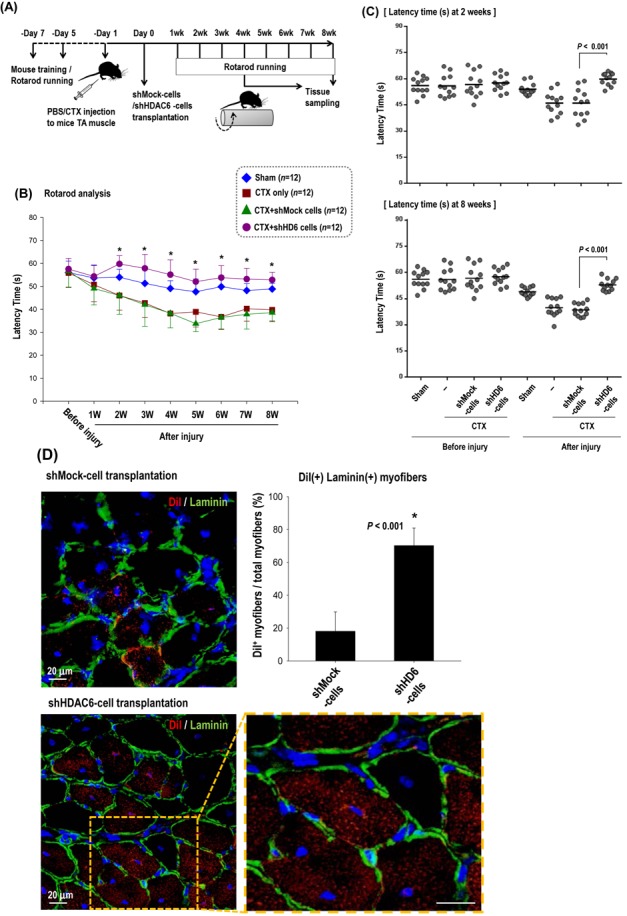
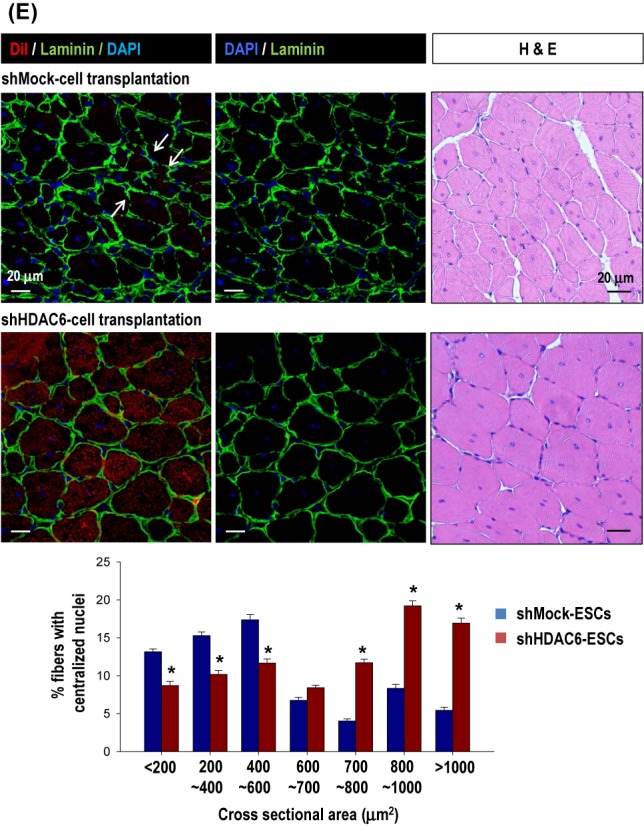
Knock-down of HDAC6 in ESCs facilitated muscle differentiation in vivo mouse model. (A–C) Rotarod test for mice with skeletal muscle injury. One day before transplantation, cardiotoxin (CTX) was injected into the tibialis anterior (TA) muscle (both legs) to induce muscle injury. We monitored the muscle power weekly for up to 8 weeks in four groups: sham control with PBS injection (sham, n = 12), injury control with CTX injection (CTX only, n = 12), injury with cell control (CTX+shMock, n = 12), injury with shHDAC6 cell transplantation (CTX+shHD6, n = 12). The measurement of the latency time for mice to fall off rotating rod at every week for up to 8 weeks (B) and at 2 weeks and 8 weeks (C) after cell transplantation were shown. (*P < 0.001, shMock-cells versus shHDAC6-cells). (D) Cross-section of TA muscle was immunostained for laminin α2 (green)/nuclei (blue) 4 weeks after transplantation of shHDAC6-ESCs that were pre-labeled with DiI. Magnification: ×400. Most of the red cells positive for DiI-fluorescence (red) were the regenerating myofibers with center nucleus. (E) Cross-sectional area of the regenerating myofibers in the injured TA muscles at 8 weeks after treatment with shMock-ESC or shHDAC6-ESC. Myofiber cross sectional area (CSA; μm2) was measured on H&E-stained cross-sections using the ImageJ software. Myofibers positive for DiI in the shHDAC6 group were bigger than those in the shMock group (top). CSA of the regenerating myofibers in the shHDAC6 group were significantly bigger than those in the shMock group (bottom, *P ≤ 0.001. Data are presented as mean ± SEM).
HDAC6 downregulation by hypoxia-induced microRNA-26a
HDAC6 was downregulated by hypoxia; thus, we evaluated the effect of HIF on HDAC6 expression because HIF1α and HIF2α are master transcription factors that control several genes involved in the hypoxic response. We expected that HDAC6 expression would be affected by either HIF1α or HIF2α, but HDAC6 was unaffected (Figure 5A and B). Expression of HIF-responsive genes was increased in HIF1α-and HIF2α-overexpressing cells (Figure 5C), indicating that the expression vectors functioned properly.
Figure 5.
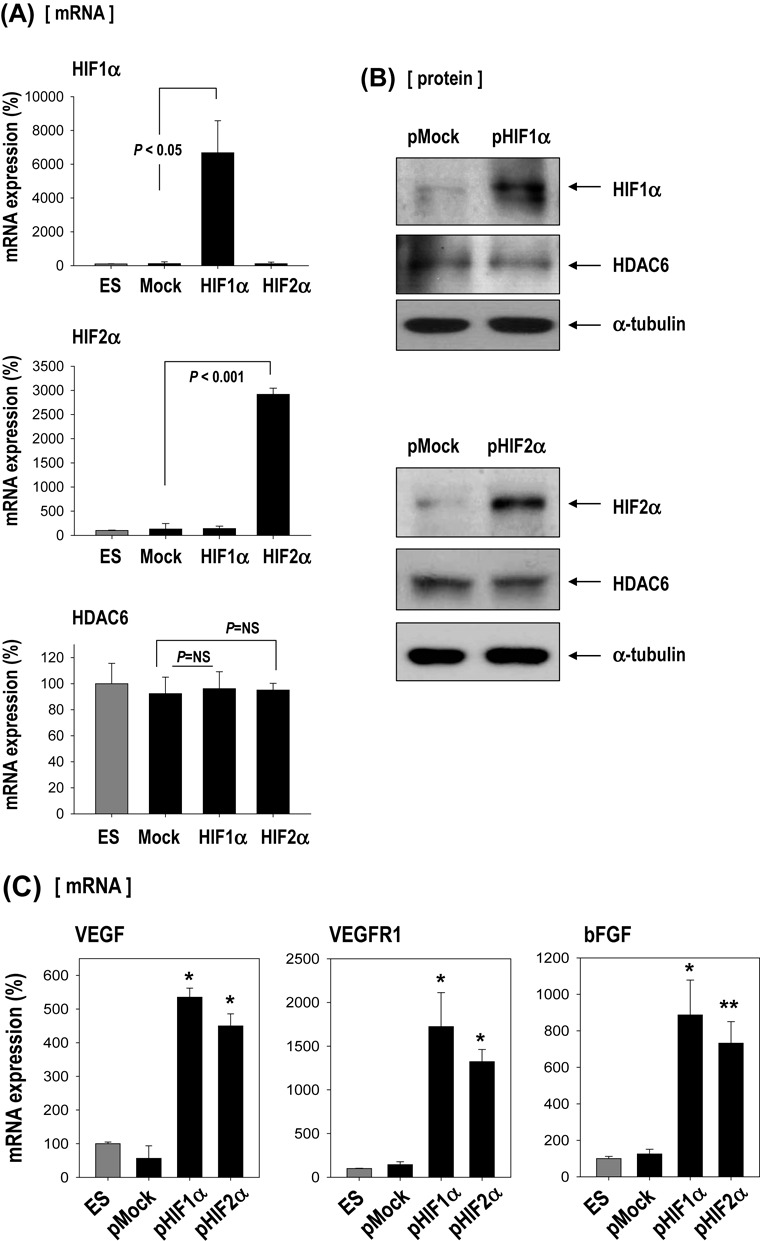
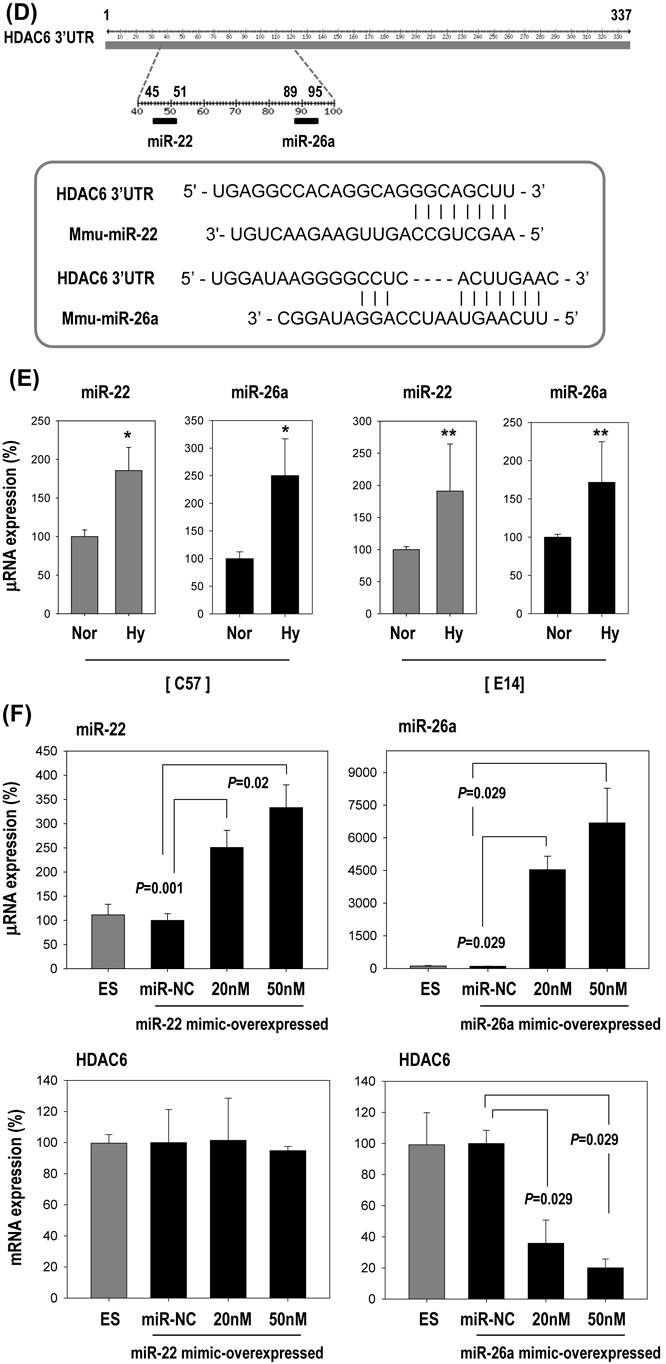
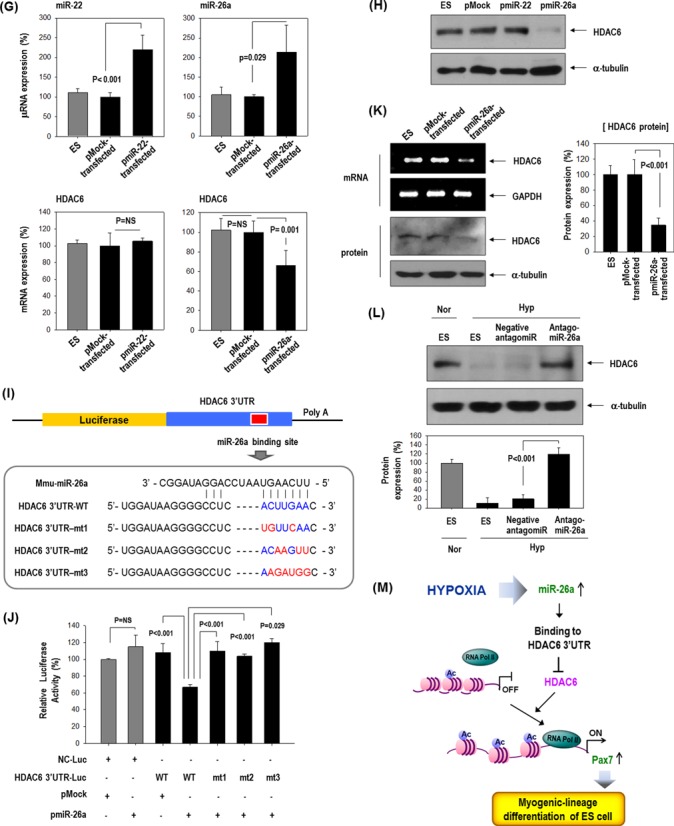
HDAC6 reduction by hypoxia-responsive miR-26a in ESCs. (A–C) Effect of HIF1α or HIF2α over-expression on HDAC6 and other hypoxia-responsive control genes (VEGF, VEGFR1, bFGF). ESCs were transfected with pEGFP-HIF1α or pEGFP-HIF2α (n = 3). Real-time PCR and western blotting for HIF1α, HIF2α and HDAC6. Real-time PCR for control genes (VEGF, VEGFR1, bFGF) suggests that HIF plasmids functioned normally. (n = 3, *P < 0.001, **P < 0.05). (D) Sequence alignment of putative miR-22 and miR-26a targeting site within 3′UTR of HDAC6 (337 bp). (E–G) MicroRNA expression was measured by real-time PCR. (E) Induction of miR-22 and miR-26a by hypoxia in C57 or E14 ESCs (n = 4, *P < 0.001, **P < 0.01). (F) Suppression of HDAC6 only by miR-26a mimic but not by miR-22 in C57 ESCs (n = 3). (miR-NC: miRNA-negative control). (G) Suppression of HDAC6 only by pre-miR-26a plasmid (pmiR-26a) but not by pre-miR-22 plasmid (pmiR-22) in C57 cell (n = 3). (H) Western blot of HDAC6 after transfection of pmiR-22 and pmiR-26a. (I–K) miR-26a directly targeted 3′UTR of HDAC6 gene. (I) Schematic presentation of luciferase reporter constructs showing the predicted structures of each base-paired wild-type (WT) or mutant (mt1, mt2, mt3) 3′UTR of HDAC6. (J) Luciferase activity reflecting HDAC6 expression in C57 ESCs was suppressed by pmiR-26a but not by control pMock. Such inhibitory action of pmiR-26a was abrogated when target sites of 3′UTR were mutated (n = 3). (K) HDAC6 mRNA and protein expression were decreased by transfection of pre-miR-26a (pmiR-26a). Quantification graph of western blot (n = 4). (L) HDAC6 was not suppressed even under hypoxic conditions by transfection with AntagomiR (50 nM) against miR-26a (n = 3). (M) Model for HDAC6 suppression by hypoxia and myogenic differentiation of ESCs.
In search of the factors that suppress HDAC6 expression in response to hypoxia, we analyzed miRNAs that usually suppress target gene expression (23). We hypothesized the existence of HDAC6-suppressive miRNA(s) that are induced by hypoxia. To find the miRNAs that bind to the 3′UTR of HDAC6 gene (HDAC6 3′UTR), we searched for predicted miRNA targets using the bioinformatics target prediction tools TargetScan, microRNA.org and miRBase. We found 2 candidate miRNAs: microRNA-22 (miR-22) and microRNA-26a (miR-26a) (Figure 5D). Both miRNAs were significantly upregulated by hypoxia both in E14 and in C57 ESCs (Figure 5E), and we tested which of the two controlled HDAC6 expression. After transfection with each mimic-oligomer, only miR-26a mimic-overexpression remarkably suppressed HDAC6 whereas miR-22 mimic-overexpression did not (Figure 5F). Likewise, a miR-26a precursor expression vector (pmiR-26a) significantly suppressed HDAC6 expression, whereas a miR-22 precursor expression vector (pmiR-22) had no effect on HDAC6 (Figure 5G and H).
We then tested whether miR-26a can directly target the 3′UTR of HDAC6 gene by using a luciferase reporter assay with wild-type (WT) or mutated (mt) 3′UTR of HDAC6 gene (Figure 5I and J). Overexpression of miR-26a (pmiR-26a) significantly suppressed luciferase activity in 3′UTR-WT of HDAC6 but not in mutated 3′UTR (mt1–mt3) of HDAC6 gene. Control miR (pMock) did not affect the luciferase activity. Expression of the HDAC6 protein was also significantly decreased after transfection of pmiR-26a only (Figure 5K). HDAC6 expression was not suppressed even under hypoxic conditions by transfection of antagomiR against miR-26a (Figure 5L). All these results strongly suggested that HDAC6 may be a direct target of miR-26a.
DISCUSSION
The major finding of our study is that HDAC6 is a regulatory target for the specific lineage commitment of ESCs. In addition, we propose here a novel concept that an environmental factor such as hypoxia regulates the epigenetic machinery and specifies the direction of ESC differentiation. In particular, hypoxia-responsive miR-26a directly targets 3′UTR of HDAC6 gene and suppresses HDAC6 expression, leading to acetylation of histones in the promoter region of Pax7 gene, an essential muscle progenitor marker and to Pax7 activation (Figure 5M).
In our previous study (6), hypoxia induced differentiation of ESCs toward meso-endoderm and vascular-lineage. We hypothesized that the epigenetic machinery like as HDACs may play a role in this process. There are 18 HDACs in mammals that can be grouped into four classes: Class I (HDAC1, -2, -3 and -8), Class II (Class IIa: HDAC4, -5, -7 and -9; Class IIb: HDAC6 and -10), Class III (Sirt1-7) and Class IV (HDAC11) (7). Appropriate HDAC regulation is essential for normal development and is implicated in the pathogenesis of some diseases (43); therefore, the functions of HDACs and their inhibitors have been studied to develop a treatment of such illnesses as cancer, inflammatory diseases, and degenerative disorders (43,44). This epigenetic machinery is also important for stem cell fate. Histone acetylation is crucial for promoting and maintaining ESC pluripotency and reprogramming (14–19). In contrast, other studies suggest that HDAC inhibitors facilitate stem cell differentiation into a specific lineage (20–22). After screening of 11 HDACs, we found that HDAC6 is significantly downregulated both in response to hypoxia and during differentiation as EB formation (Figure 1 and Supplementary Figure S1). Knock-down of HDAC6 in ESCs significantly stimulated stimulates expression of myogenic marker genes including Pax7, which is essential for skeletal muscle development (Figures 1 and 2). Lineage commitment of stem cells via HDAC6 downregulation has not been reported in the literature.
The upstream regulator that suppresses HDAC6 expression in response to hypoxia is not the well-known transcription regulators under conditions of hypoxia such as HIF-1α or -2α but miR-26a (Figure 5). Several miRNAs are reported to be involved in the regulation of stem cell fate (27,28). There are specific miRNA groups: muscle-specific ‘myomiRs’ are regulated during muscle differentiation and muscle activity (27,28), and ‘hypoxamiRs’ are induced by hypoxia (45). MiR-26a, which we encountered here, is not known as either ‘myomiR’ or ‘hypoxamiR’. We demonstrated that miR-26a is upregulated by hypoxia (Figure 5E) and that miR-26a directly targets 3′UTR of HDAC6 and suppresses the latter gene (Figure 5F–J) while increasing Pax7 expression (data not shown). Recent studies have shown the induction of miR-26a during differentiation of myoblast C2C12 cells and point to the necessity of miR-26a for skeletal muscle differentiation in vivo (46,47). Our results clarified the previously unknown role of miR-26a as a sensor of environmental cues that regulates the epigenetic machinery and thereby specifies the direction of stem cell differentiation.
A result that differs slightly from ours was reported recently (10). HDAC6 expression is increased by hypoxia in HUVECs (10), whereas HDAC6 is consistently downregulated by hypoxia in our repeated experiments in both E14 and C57 ESCs (Figure 1 and Supplementary Figure S1). Such discrepant behavior of HDAC6 in response to hypoxia between ESCs and HUVECs was also confirmed in our experiments (Supplementary Figure S3). The reasons for the discrepancy in HDAC6 expression between ESCs and HUVECs under hypoxic conditions are unclear, but we believe that differences in the cell hierarchy may be among the contributing factors; further research is needed to clarify this issue.
Muscle loss occurs as a consequence of a chronic disease, muscle-wasting disorders, or aging, which increases the risk of death (48). Stem cells are a good resource for regenerative medicine; however, the mechanisms that control differentiation into specific lineages have not been fully determined. Commitment of ESCs to the myogenic lineage because of suppression of HDAC6 expression is not only an in vitro finding but is also confirmed in vivo: we observed enhanced muscle regeneration after transplantation of such ESCs into a damaged limb. Therefore, modulation of HDAC6 expression or activity by pharmacological or biological agents may be an option for control of stem cell commitment toward the myogenic lineage in the context of cell therapy of muscle diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Author contributions: S.-W.L. conceived the project, experimental design, analyzed data and wrote manuscript. J.Y. performed experiments, analyzed data and revised manuscript. H.-K.J. and S.-Y.K. performed experiments. W.-J.K. provided consultation, vector construction and assisted in experimental design. J.L. performed animal experiment. E.-J.L. data discussion. H.-S.K. data discussion and revised the manuscript.
Disclosures: None.
Footnotes
These authors contributed equally to the paper as first authors.
FUNDING
Innovative Research Institute for Cell Therapy [A062260]; Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning [2013R1A1A3012024 to S.-W.L.]; Bio & Medical Technology Development Program [2010-0020258] of the NRF funded by the Ministry of Science, ICT & Future Planning of the Korean government. Funding for open access charge: Innovative Research Institute for Cell Therapy [A062260]; Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee S.W., Kim W.J., Choi Y.K., Song H.S., Son M.J., Gelman I.H., Kim Y.J., Kim K.W. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat. Med. 2003;9:900–906. doi: 10.1038/nm889. [DOI] [PubMed] [Google Scholar]
- 2.Semenza G.L. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 3.Lee S.W., Lee Y.M., Bae S.K., Murakami S., Yun Y., Kim K.W. Human hepatitis B virus X protein is a possible mediator of hypoxia-induced angiogenesis in hepatocarcinogenesis. Biochem. Biophys. Res. Commun. 2000;268:456–461. doi: 10.1006/bbrc.2000.2093. [DOI] [PubMed] [Google Scholar]
- 4.Kim M.S., Kwon H.J., Lee Y.M., Baek J.H., Jang J.E., Lee S.W., Moon E.J., Kim H.S., Lee S.K., Chung H.Y., et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat. Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 5.Youn S.W., Lee S.W., Lee J., Jeong H.K., Suh J.W., Yoon C.H., Kang H.J., Kim H.Z., Koh G.Y., Oh B.H., et al. COMP-Ang1 stimulates HIF-1alpha-mediated SDF-1 overexpression and recovers ischemic injury through BM-derived progenitor cell recruitment. Blood. 2011;117:4376–4386. doi: 10.1182/blood-2010-07-295964. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.W., Jeong H.K., Lee J.Y., Yang J., Lee E.J., Kim S.Y., Youn S.W., Lee J., Kim W.J., Kim K.W., et al. Hypoxic priming of mESCs accelerates vascular-lineage differentiation through HIF1-mediated inverse regulation of Oct4 and VEGF. EMBO Mol. Med. 2012;4:924–938. doi: 10.1002/emmm.201101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Ruijter A.J., van Gennip A.H., Caron H.N., Kemp S., van Kuilenburg A.B. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodawadekar S.C., Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–5540. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]
- 9.Aldana-Masangkay G.I., Sakamoto K.M. The role of HDAC6 in cancer. J. Biomed. Biotechnol. 2011;2011:875824. doi: 10.1155/2011/875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaluza D., Kroll J., Gesierich S., Yao T.P., Boon R.A., Hergenreider E., Tjwa M., Rossig L., Seto E., Augustin H.G., et al. Class IIb HDAC6 regulates endothelial cell migration and angiogenesis by deacetylation of cortactin. EMBO J. 2011;30:4142–4156. doi: 10.1038/emboj.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 12.Birdsey G.M., Dryden N.H., Shah A.V., Hannah R., Hall M.D., Haskard D.O., Parsons M., Mason J.C., Zvelebil M., Gottgens B., et al. The transcription factor Erg regulates expression of histone deacetylase 6 and multiple pathways involved in endothelial cell migration and angiogenesis. Blood. 2012;119:894–903. doi: 10.1182/blood-2011-04-350025. [DOI] [PubMed] [Google Scholar]
- 13.Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Li L., Pandey R., Byun J.S., Gardner K., Qin Z., Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan Y., Xue Y., Song C., Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11 493–11 498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durcova-Hills G., Tang F., Doody G., Tooze R., Surani M.A. Reprogramming primordial germ cells into pluripotent stem cells. PLoS One. 2008;3:e3531. doi: 10.1371/journal.pone.0003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hai T., Hao J., Wang L., Jouneau A., Zhou Q. Pluripotency maintenance in mouse somatic cell nuclear transfer embryos and its improvement by treatment with the histone deacetylase inhibitor TSA. Cell. Reprogramm. 2011;13:47–56. doi: 10.1089/cell.2010.0042. [DOI] [PubMed] [Google Scholar]
- 18.Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 19.Moschidou D., Mukherjee S., Blundell M.P., Drews K., Jones G.N., Abdulrazzak H., Nowakowska B., Phoolchund A., Lay K., Ramasamy T.S., et al. Valproic acid confers functional pluripotency to human amniotic fluid stem cells in a transgene-free approach. Mol. Ther. 2012;20:1953–1967. doi: 10.1038/mt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretsovali A., Hadjimichael C., Charmpilas N. Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells Int. 2012;2012:184154. doi: 10.1155/2012/184154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paino F., La Noce M., Tirino V., Naddeo P., Desiderio V., Pirozzi G., De Rosa A., Laino L., Altucci L., Papaccio G. Histone deacetylase inhibition with valproic acid downregulates osteocalcin gene expression in human dental pulp stem cells and osteoblasts: evidence for HDAC2 involvement. Stem Cells. 2014;32:279–289. doi: 10.1002/stem.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong X., Pan R., Zhang H., Yang C., Shao J., Xiang L. Modification of histone acetylation facilitates hepatic differentiation of human bone marrow mesenchymal stem cells. PLoS One. 2013;8:e63405. doi: 10.1371/journal.pone.0063405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Lee H.J. Exceptional stories of microRNAs. Exp. Biol. Med. (Maywood) 2013;238:339–343. doi: 10.1258/ebm.2012.012251. [DOI] [PubMed] [Google Scholar]
- 25.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 27.McCarthy J.J. The MyomiR network in skeletal muscle plasticity. Exerc. Sport Sci. Rev. 2011;39:150–154. doi: 10.1097/JES.0b013e31821c01e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinrich E.M., Dimmeler S. MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ. Res. 2012;110:1014–1022. doi: 10.1161/CIRCRESAHA.111.243394. [DOI] [PubMed] [Google Scholar]
- 29.Mizuno Y., Chang H., Umeda K., Niwa A., Iwasa T., Awaya T., Fukada S., Yamamoto H., Yamanaka S., Nakahata T., et al. Generation of skeletal muscle stem/progenitor cells from murine induced pluripotent stem cells. FASEB J. 2010;24:2245–2253. doi: 10.1096/fj.09-137174. [DOI] [PubMed] [Google Scholar]
- 30.Darabi R., Gehlbach K., Bachoo R.M., Kamath S., Osawa M., Kamm K.E., Kyba M., Perlingeiro R.C. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat. Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- 31.Patel J.B., Appaiah H.N., Burnett R.M., Bhat-Nakshatri P., Wang G., Mehta R., Badve S., Thomson M.J., Hammond S., Steeg P., et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 2011;30:1290–1301. doi: 10.1038/onc.2010.510. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed J.S., Lopez M.A., Boriek A.M. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3beta. J. Biol. Chem. 2010;285:29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo E.J., Chun Y.S., Cho Y.S., Kim J., Lee J.C., Kim M.S., Park J.W. YC-1: a potential anticancer drug targeting hypoxia-inducible factor 1. J. Natl. Cancer Inst. 2003;95:516–525. doi: 10.1093/jnci/95.7.516. [DOI] [PubMed] [Google Scholar]
- 35.Liu N., Williams A.H., Maxeiner J.M., Bezprozvannaya S., Shelton J.M., Richardson J.A., Bassel-Duby R., Olson E.N. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J. Clin. Invest. 2012;122:2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoshino S., Sakamoto K., Vassilopoulos S., Camus S.M., Griffin C.A., Esk C., Torres J.A., Ohkoshi N., Ishii A., Tamaoka A., et al. The CHC22 clathrin-GLUT4 transport pathway contributes to skeletal muscle regeneration. PLoS One. 2013;8:e77787. doi: 10.1371/journal.pone.0077787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter M., Moser M.A., Meunier D., Fischer C., Machat G., Mattes K., Lichtenberger B.M., Brunmeir R., Weissmann S., Murko C., et al. Divergent roles of HDAC1 and HDAC2 in the regulation of epidermal development and tumorigenesis. EMBO J. 2013;32:3176–3191. doi: 10.1038/emboj.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 39.Zammit P.S., Relaix F., Nagata Y., Ruiz A.P., Collins C.A., Partridge T.A., Beauchamp J.R. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Ramirez M., Rocchini C., Ausio J. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 41.Sengupta N., Seto E. Regulation of histone deacetylase activities. J. Cell. Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 42.Patton B.L., Connoll A.M., Martin P.T., Cunningham J.M., Mehta S., Pestronk A., Miner J.H., Sanes J.R. Distribution of ten laminin chains in dystrophic and regenerating muscles. Neuromusc. Disord. : NMD. 1999;9:423–433. doi: 10.1016/s0960-8966(99)00033-4. [DOI] [PubMed] [Google Scholar]
- 43.Haberland M., Montgomery R.L., Olson E.N. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinarello C.A., Fossati G., Mascagni P. Histone deacetylase inhibitors for treating a spectrum of diseases not related to cancer. Mol. Med. 2011;17:333–352. doi: 10.2119/molmed.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loscalzo J. The cellular response to hypoxia: tuning the system with microRNAs. J. Clin. Invest. 2010;120:3815–3817. doi: 10.1172/JCI45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong C.F., Tellam R.L. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 47.Dey B.K., Gagan J., Yan Z., Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26:2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tedesco F.S., Dellavalle A., Diaz-Manera J., Messina G., Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J. Clin. Invest. 2010;120:11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


