Abstract
A rapid method for construction of oligonucleotide arrays on a glass surface, using a novel heterobifunctional reagent, N-(2-trifluoroethanesulfonatoethyl)-N-(methyl)-triethoxysilylpropyl-3-amine (NTMTA), has been described. The heterobifunctional reagent, NTMTA, carries two different thermoreactive groups. The triethoxysilyl group on one end is specific towards silanol functions on the virgin glass surface, while the trifluoroethanesulfonyl (tresyl) group on the other end of the reagent reacts specifically with aminoalkyl- or mercaptoalkyl- functionalized oligonucleotides. Immobilization of oligonucleotides on a glass surface has been realized via two routes. In the first one (A), 5′- aminoalkyl- or mercaptoalkyl-functionalized oligonucleotides were allowed to react with NTMTA to form a oligonucleotide-triethoxysilyl conjugate which, in a subsequent reaction with unmodified (virgin) glass microslide, results in surface-bound oligonucleotides. In the second route (B), the NTMTA reagent reacts first with a glass microslide whereby it generates trifluoroethanesulfonate ester functions on it, which in a subsequent step react with 5′-aminoalkyl or mercaptoalkyl oligonucleotides to generate support-bound oligonucleotides. Subsequently, the oligonucleotide arrays prepared by both routes were analyzed by hybridization experiments with complementary oligonucleotides. The constructed microarrays were successfully used in single and multiple nucleotide mismatch detection by hybridizing these with fluorescein-labeled complementary oligonucleotides. Further more, the proposed method was compared with the existing methods with respect to immobilization efficiency of oligonucleotides.
INTRODUCTION
Last decade has witnessed the emergence of microarray technology, a very powerful and promising tool for gene discovery (1), genome analysis (2), DNA sequencing by hybridization (3), medical diagnostics for genetic diseases (4) and the detection of single nucleotide polymorphisms (5). This also allows undertaking studies relating to nucleic acid–ligand interaction (6) and DNA computing (7). The surface-bound oligonucleotides (microarrays) offer several advantages over the conventional gel-based format. The success of microarrays not only depends on the chemistry used for the immobilization of oligonucleotides but also depends on the good accessibility and functionality of the surface-bound oligonucleotides, density of attachment, thermal stability of the array under experimental conditions and reproducibility of attachment chemistry.
Basically, two approaches are used for fabrication of DNA/oligonucleotide arrays. In one approach, oligonucleotide probes are directly synthesized on the surface at a pre-selected positions (in situ synthesis) (8–13) following conventional and photolithographic techniques such as the Affimetrix on-chip synthesis. This methodology is by far the most efficient method for the construction of high-density oligonucleotide arrays, however, it has practical limitations in terms of flexibility and affordability. The second method, called the deposition method, where pre-fabricated nucleic acids are covalently (12–20) or non-covalently (21) immobilized on solid surfaces (organic or inorganic), offers an excellent flexibility. This latter approach has thus become the most widely used method for creating low- to medium-density DNA microarrays.
Two important factors which influence the quality of microarrays fabricated by the latter method, are (i) the nature of the solid surface used, and (ii) the chemistry employed for fixing of oligonucleotides on solid surfaces. Though a large number of chemical methods have been reported for immobilization of oligonucleotides on pre-functionalized surfaces, there are only two reagents (3-mercaptopropyltriethoxysilane and 3-glycidyloxypropyltrimethoxysilane) avail able that can directly be used for fixing oligonucleotides on virgin glass surface. 3-Mercaptopropyltriethoxysilane results in immobilization of oligonucleotides via disulfide linkage (22), which is a labile linkage and 3-glycidyloxypropyltrimethoxysilane requires longer reaction time (8 h) (23,24). Therefore, a glass-specific reagent, which could fix oligonucleotides on a virgin glass surface rapidly and via a stable linkage, is still elusive. A number of solid surfaces such as polypropylene, polyethylene, nylon, poly(methyl methacrylate) (PMMA), glass, silicon, etc., have been proposed for this purpose. Of these, glass and PMMA appear attractive because these supports can easily be derivatized generating reactive functional groups such as aminoalkyl, mercaptoalkyl, carboxyl, aldehyde, etc. on the surface. Glass, in particular, being an inexpensive material having low intrinsic fluorescence and a relatively homogeneous chemical surface, offers an added advantage in the sense that laser scanners can be used for visualization of fluorescent spots on the surface.
In this communication, we describe a method for construction of oligonucleotide arrays using a novel heterobifunctional reagent, [N-(2-trifluoroethanesulfonatoethyl)-N-(methyl)-triethoxysilylpropyl-3-amine] (NTMTA) (25), on an unmodified (virgin) glass surface. The method is suitable for the construction of oligonucleotide microarrays on a glass surface without employing any additional coupling reagent, thus making the strategy cost-effective, efficient and rapid. The NTMTA reagent has been explored in two ways; in the first one, the NTMTA reagent was reacted with a 5′-aminoalkyl- or mercaptoalkyl-functionalized oligonucleotide through its trifluoroethanesulfonate (tresyl) ester function to generate a oligonucleotide conjugate, which subsequently reacts with the unmodified glass surface to obtain a surface-immobilized oligonucleotide. Whereas, in the second route, the NTMTA reagent was allowed to react first with the unmodified glass surface through its triethoxysilyl function to generate reactive tresyl functions on the surface, which in the subsequent step react with 5′-aminoalkyl or mercaptoalkyl groups bearing oligonucleotides to generate glass surface-bound oligonucleotides. The constructed microarrays were successfully used in detection of nucleotide mismatches by hybridization with fluorescein-labeled complementary oligonucleotides. Further more, the proposed method was compared with some of the existing glass-specific methods.
MATERIALS AND METHODS
General
Reagents and chemicals employed in the present investigation were purified prior to their use. 3-Glycidyloxypropyltri methoxysilane, 3-mercaptopropyltriethoxysilane, 2,2,2-trifluoroethanesulfonyl chloride (tresyl chloride), 3-chloropropyltriethoxysilane, N-methyl-2-aminoethanol and N,N-diisopropylethylamine were procured from Aldrich Chemical Co., St Louis, MO. Fluorescein-DMTrdT-phosphoramidite (dTF) was obtained from Glen Research Inc., Sterling, VA. FAM-phosphoramidite was purchased from Applied Biosystems Inc., Foster City, CA. Modifications at 5′ and 3′ ends in oligonucleotides were incorporated by the methods (27–29) reported from this laboratory. Microslides were scanned under a laser scanner at 570 nm using ScanArray Lite, MicroArray Analysis System, GSI Lumonics. Fluorescence intensity of the spots was determined by QuantArray software.
In the present investigation, the following buffers and conditions were employed for the processing of glass microslides unless and until stated in the experiment itself: (i) reaction buffer: 0.1 M sodium phosphate containing 0.5 M sodium chloride pH 8.0; washing buffer: 0.1 M sodium phosphate containing 0.5 M sodium chloride, pH 7.1; hybridization buffer: 0.1 M sodium phosphate containing 1.0 M sodium chloride pH 7.1.
For immobilization reactions, oligonucleotides were dissolved in reaction buffer to make final concentration to 10 µM and spotted by applying 0.5 µl of this solution per spot. For hybridization experiments, oligonucleotides were dissolved in hybridization buffer in 50 µM concentration and 50 µl of this solution was used in each hybridization reaction.
Preparation of N-(2-hydroxyethyl)-N-(methyl)-triethoxysilylpropyl-3-amine (NHMTA) (III)
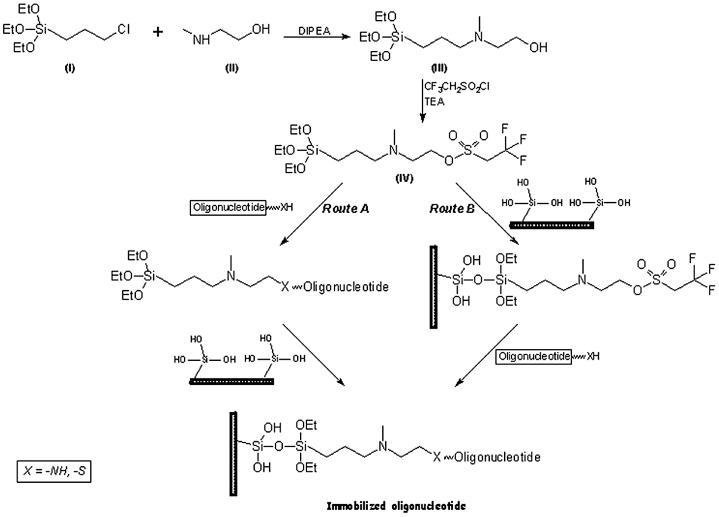
To a solution of anhydrous N-methyl-2-aminoethanol (I) (22.5 g, 0.3 mol) and N,N-diisopropylethylamine (12.9 g, 0.1 mol) in hot toluene (70 ml) was added 3-chloropropyltriethoxysilane (II) (24.05 g, 0.1 mol) in one portion. The mixture was refluxed for 10 h under inert (argon) atmosphere followed by cooling at 4°C for 16 h. The amine-hydrochloride formed in the reaction was removed by filtration under an inert atmosphere through a sintered glass funnel and the residue washed with dry toluene (2 × 25 ml). The combined filtrate and washings were pooled together and concentrated under vacuum. The syrupy crude material was distilled under vacuum to obtain the title compound (III) in 65% yield (18 g). The compound was characterized by 1H-NMR.
1H-NMR, CDCl3 (δ, p.p.m.): 0.58 (t, 2H), 1.14 (m, 9H), 1.30 (m, 4H), 2.27 (s, N-CH3), 2.3–2.6 (m, 7H, -CH2), 3.58 (m, 6H, -OCH2), 3.66 (t, 2H).
Preparation of N-(2-trifluoroethanesulfonatoethyl)-N-(methyl)-triethoxysilyl-propyl-3-amine (NTMTA) (IV)
The title compound was prepared by adding 2,2,2-trifluoroethanesulfonyl (tresyl chloride) (1.1 g, 6 mmol) drop-wise to a mixture of compound III (1.4 g, 5 mmol) and triethylamine (830 µl, 6 mmol) in dichloromethane (25 ml) at 0°C in an ice bath. The reaction was stirred at room temperature for 2 h until complete conversion took place as monitored on TLC. After that, the reaction mixture was cooled to 0°C in an ice-bath and the precipitated amine-hydrochloride removed by filtration under argon. The filtrate was concentrated under vacuum to obtain a syrupy mass of the title compound (IV) in almost quantitative yield. It was then characterized by 1H-NMR and MALDI-TOF.
1H-NMR, CDCl3 (δ, p.p.m.): 0.57 (t, 2H), 1.11 (m, 9H), 1.34 (m, 4H), 2.31 (s, N-CH3), 2.25–2.71 (m, 7H, -CH2), 3.63 (m, 6H, -OCH2), 3.88 (t, 2H, -CH2OSO2). MALDI-TOF (DHBA): 426 (M+H)+.
Oligonucleotide synthesis and purification
Oligonucleotides were synthesized on a 0.2 µmol scale on a Pharmacia Gene Assembler Plus using the standard phosphoramidite approach following the manufacturer’s protocol (26). In order to incorporate reactive nucleophilic groups at the 5′ end of the oligonucleotides, the last coupling reaction in the desired oligomer was performed with either DMTrNH(CH2)5 OP[N(iPr)2]O(CH2)2CN (27) or BzS(CH2)6P[N(iPr)2]O (CH2)2CN (28) to introduce aminoalkyl or mercaptoalkyl group at the 5′ end. The coupling of these reagents was performed in an analogous manner to that of normal nucleoside-phosphoramidites. Labeled oligonucleotides were also synthesized in the same manner by performing the last coupling with FAM-phosphoramidite. Furthermore, an oligomer d(TFTT TTT TTT TTT TTT TTT TT)-OPO3-(CH2)6-SH was synthesized on the thiol polymer support (29) by carrying out the last coupling with fluorescein-dT-phosphoramidite in an analogous manner to that of normal nucleoside-phosphoramidites, except that an extended coupling time (5 min) was employed. A list of oligonucleotide sequences assembled along with their deprotection conditions and yields are given in Table 1. Subsequently, the ammoniacal solutions in all the cases were concentrated in a speed vacuum and the residues dissolved in water (200 µl) in each case and applied on to a desalting column (reverse-phase ODS-silica gel column). The oligonucleotides were eluted with 30% acetonitrile in water, concentrated, purified and stored at 4°C.
Table 1. Oligonucleotides synthesized with their deprotection conditions and yields.
| Sequence no. | Oligonucleotide sequence | Deprotection conditions | Yield (OD at 260 nm) |
|---|---|---|---|
| 1 | H2N-(CH2)5-OPO3-d(CAG AGG TTC TTT GAG TCC TT) | Aq. NH4OH (30%), 16 h, 60°C | 27.4 |
| 2 | HS-(CH2)6-OPO3-d(CAGAGG TTC TTT GAG TCC TT) | Aq. NH4OH (30%) containing 0.1 M DTT, 16 h, 60°C | 24.5 |
| 3 | HS-(CH2)6-OPO3-d(CTC CTG AGG AGA AGG TCT GC) | Aq. NH4OH (30%) containing 0.1 M DTT, 16 h, 60°C | 22.4 |
| 4 | HS-(CH2)6-OPO3-d(CTC CTG AGG CGA AGG TCT GC) | Aq. NH4OH (30%) containing 0.1 M DTT, 16 h, 60°C | 23.6 |
| 5 | H2N-(CH2)5-OPO3-d(CTCCTG AGG AGA AGG TCT GC) | Aq. NH4OH (30%) containing 0.1 M DTT, 16 h, 60°C | 25.8 |
| 6 | H2N-(CH2)5-OPO3-d(CTCCTG AGG CGA AGG TCT GC) | Aq. NH4OH (30%) containing 0.1 M DTT, 16 h, 60°C | 26.3 |
| 7 | FAM-d(AAG GAC TCA AAG AAC CTC TG) | Aq. NH4OH (30%), 16h, 60°C | 24.3 |
| 8 | FAM-d(GCA GAC CTT CTC CTC AGG AG) | Aq. NH4OH (30%), 16 h, 60°C | 25.7 |
| 9 | d(TFTT TTT TTT TTT TTT TTT TT)-OPO3-(CH2)6SH | Aq. NH4OH (30%) containing 0.1 M DTT, 16 h, 60°C | 23.2 |
| 10 | H2N-(CH2)5-OPO3-d(CTC CTG CGG AGA ACG TCT GC) | Aq. NH4OH (30%), 16 h, 60°C | 24.3 |
| 11 | H2N-(CH2)5-OPO3-d(CTC CTG CGG CGA ACG TCT GC) | Aq. NH4OH (30%), 16 h, 60°C | 25.1 |
| 12 | H2N-(CH2)5-OPO3-d(TTT TTT TTT TTT TTT TTT TT) | Aq. NH4OH (30%), 16 h, 60°C | 25.4 |
Determination of optimal time for immobilization of oligomers on the glass surface
In order to determine optimum time required to immobilize aminoalkyl- and mercaptoalkyl-functionalized oligonucleotides on the glass surface, a model experiment was designed to study the immobilization of DMTrO(CH2)5NH2 or DMTrO(CH2)6SH on unmodified controlled pore glass support (500 Å), using NTMTA via two routes.
Route A. O-(4,4′-Dimethoxytrityl)-5-aminopent-1-ol (40.5 mg, 100 µmol) dissolved in ethanol (1.0 ml) was mixed with 1.0 ml of ethanolic solution of NTMTA (42.5 mg, 100 µmol). The reaction was allowed to swirl for 1 h at room temperature in an Eppendorf Mixer. The resulting conjugate (200 µl) was then added to each of the eight centrifuge tubes containing unmodified CPG (∼10 mg), placed in an Eppendorf Thermomixer set at 45°C. The tubes were withdrawn at different time intervals (10, 20, 30, 40, 50, 60, 120, 180 min) and the support, after having been washed with dichloromethane (5 × 1.0 ml) containing triethylamine (0.1%), was subjected to drying under vacuum. The extent of reaction on support in each tube was determined by treating an accurately weighed amount of the support (∼1–2 mg) with 3% trichloroacetic acid in dichloromethane for 5 min. The released DMTr cation was measured spectrophotometrically at 505 nm to determine the loading on the support in µmol/g following the standard method (30). A graph was plotted between the loading on the support (µmol/g) versus time of reaction (min).
Similarly, the extent of reaction as well as optimal time were determined using mercaptoalkyl reagent, DMTrO (CH2)6SH (43.6 mg, 100 µmol), except that the reaction time of 30 min was used for the reaction between mercaptoalkylated ligand and NTMTA reagent.
Route B. In an alternative method, to eight centrifuge tubes containing glass beads (CPG 500A, ∼10 mg in each tube), 100 µl of an ethanolic solution of NTMTA (4.25 mg, 10 µmol) was added to each tube and kept in an Eppendorf Thermomixer set at 45°C. Then the tubes were withdrawn at regular time intervals (10, 20, 30, 40, 50, 60, 120, 180 min) and subjected to washing with ethanol (5 × 1 ml). After drying, to each tube 100µl of an ethanolic solution of O-(4,4′-dimethoxytrityl)-5-aminopent-1-ol (4.05 mg, 10 µmol) was added and the reaction was allowed to proceed for 1 h at room temperature in an Eppendorf Mixer. After that, the contents of the each tube were washed with ethanol (5 × 1 ml) and diethylether (2 × 1 ml) followed by drying under vacuum. The loading on the support in each tube was determined as described above. A graph was drawn between the loading on the support (µmol/g) versus reaction time (min).
Similarly, the above procedure was repeated with mercaptoalkylated reagent, DMTrO(CH2)6SH (4.36 mg, 10 µmol), except a 30 min reaction time was used for the reaction between mercaptoalkylated ligand and support-bound tresyl functions.
Determination of optimal concentration of oligonucleotide spots on glass surface for fluorescent detection
In order to determine the optimal concentration required for the detection of spotted oligonucleotide on the glass surface, a 3′-mercaptoalkyl-modified oligonucleotide sequence d(TFTT TTT TTT TTT TTT TTT TT)-OPO3-(CH2)6SH was serially diluted in five different concentrations (15, 10, 7.5, 5, 2.5 µM) in reaction buffer containing 10% dimethylsulfoxide (DMSO) (v/v). The glass microslide activated with NTMTA (IV) (route B) was spotted with a serially diluted fluorescent oligonucleotide, as mentioned above, in duplicates using a micropipette and kept for 30 min at room temperature in a humid chamber, followed by washings with washing buffer (5 × 10 ml) and dried under vacuum. After that, spots were visualized under laser scanner.
Quantification of immobilized oligonucleotides
In an attempt to quantify the density of immobilized oligonucleotides, an oligomer sequence, d(TFTT TTT TTT TTT TTT TTT TT)-OPO3-(CH2)6SH, diluted to concentrations 0.025, 0.05, 0.10, 0.25, 0.50, 0.75 and 1.00 µM, was spotted on a simple glass microslide in duplicates. After drying the spots on the microslide, it was visualized under a laser scanner and the fluorescence signal intensity corresponding to each spot was measured. Using these values, a graph was plotted between fluorescence intensity and concentration of the spot.
Immobilization of 5′-aminoalkyl- and mercaptoalkyl-oligonucleotides on glass surface
Route A. Oligonucleotide sequence, H2N-(CH2)5-OPO3-d(CAG AGG TTC TTT GAG TCC TT) (0.15 A260 units, 0.85 nmol), dissolved in 100 µl of reaction buffer, was mixed with 85 µl of ethanolic solution of NTMTA (3.6 µg, 8.5 nmol). The reaction mixture was agitated for 60 min at room temperature. Then the solution was concentrated under vacuum to obtain a semi-dried residue of oligonucleotide-triethoxysilyl conjugate, which was re-suspended in double distilled (dd) water (25 µl). The mixture was centrifuged to get rid of excess NTMTA, as undissolved material. The supernatant was concentrated under vacuum to obtain a residue, which was reconstituted in dd water containing 10% DMSO (v/v) to make an oligonucleotide conjugate concentration to 10 µM. The oligonucleotide conjugate was then spotted manually in multiplicates on the glass surface. The spotted microslide was incubated at 45°C in a humid chamber for 40 min followed by washings with washing buffer (5 × 10 ml) and used for the hybridization experiment. The microslide was kept in a hybridization chamber and treated with a solution of the complementary fluorescent oligonucleotide, FAM-d(AAG GAC TCA AAG AAC CTC TG), prepared in hybridization buffer, for 2 h with gentle shaking at 25°C. The glass microslide was then washed several times with hybridization buffer to get rid off excess complementary oligomer. Subsequently, the microslide was dried and examined under a laser scanner.
Similarly, the 5′-mercaptoalkylated oligonucleotide, HS-(CH2)6-OPO3-d(CAG AGG TTC TTT GAG TCC TT), was immobilized on a microslide, except that the reaction between NTMTA and 5′-mercaptoalkylated oligonucleotide was carried out for only 30 min at room temperature.
Route B. In an alternative route, the glass microslide was treated with 2 ml of ethanolic solution of NTMTA (85 µg, 0.2 µmol) by evenly spreading over it, and was kept for 40 min at 45°C in a humid chamber. The microslide was then washed several times with dry ethanol (3 × 15 ml) and dried under vacuum. Subsequently, the activated microslide was subjected to spotting of 10 µM solution of 5′-aminoalkyl and mercaptoalkyl oligonucleotides, H2N-(CH2)5-OPO3-d(CTC CTG AGG AGA AGG TCT GC) and HS-(CH2)6-OPO3-d(CTC CTG AGG AGA AGG TCT GC), dissolved in reaction buffer. The microslide was kept in a humid chamber for 60 min at room temperature. After that, the microslide was kept in 0.1 M Tris buffer pH 8.0 (blocking buffer) for 1 h and then thoroughly washed with washing buffer (5 × 10 ml). Subsequently, the glass microslide was used for the hybridization reaction as described in route A, except that FAM-d(GCA GAC CTT CTC CTC AGG AG) was used as a complementary-labeled oligomer. The spots were visualized under a laser scanner.
Specificity of immobilization chemistry and detection of mismatches
Oligonucleotide sequences H2N-(CH2)5-OPO3-d(CTC CTG AGG AGA AGG TCT GC), H2N-(CH2)5-OPO3-d(CTC CTG AGG CGA AGG TCT GC), H2N-(CH2)5-OPO3-d(CTC CTG CGG AGA ACG TCT GC), H2N-(CH2)5-OPO3-d(CTC CTG CGG CGA ACG TCT GC) and H2N-(CH2)5-OPO3-d(TTT TTT TTT TTT TTT TTT TT) dissolved in reaction buffer, were spotted on an NTMTA activated glass microslide (route B) for 60 min at room temperature in a humid chamber. After blocking and the usual washings, the spots on the microslide were hybridized with a complementary-labeled FAM-d(GCA GAC CTT CTC CTC AGG AG) for 2 h at 25°C. The microslide was washed with hybridization buffer (5 × 10 ml) and subjected to a laser scanner.
Comparison of the proposed method with standard methods
For this purpose, two standard methods [arraying of 5′-aminoalkyl oligonucleotides on epoxide glass surface (23) and 5′-mercaptoalkyl oligonucleotides on 3-mercaptopropyl glass surface (22)] were selected. Appropriately modified oligonucleotides (10 µM) were attached on the modified glass surfaces following the described protocols. Briefly, 5′-aminopentyl-oligomer, H2N-(CH2)5-OPO3-d(CAG AGG TTC TTT GAG TCC TT) (0.2 A260 units, 1.14 nmol), dissolved in 0.1 M KOH (115 µl), was spotted (0.5 µl/spot) on the 3-glycidyloxypropyl glass microslide in quadruplicate and kept at 37°C for 8 h. Thereafter, the microslide was washed with 0.1 M phosphate buffer containing 0.5 M sodium chloride, pH 7.1 (5 × 10 ml). For immobilization of mercaptoalkylated oligonucleotides, 5′-mercaptohexyl oligonucleotide, HS-(CH2)6-OPO3-d(CAG AGG TTC TTT GAG TCC TT) (0.2 A260 units, 1.14 nmol), dissolved in 0.03 M sodium acetate buffer (115 µl) pH 4.3, was reacted with 3-mercaptopropyltriethoxysilane (1.4 µg, 5.7 nmol) for 1 h at room temperature. Then the silanized oligonucleotide was spotted on the glass slides manually (0.5 µl/spot) in quadruplicate and the slide kept in a humid chamber for 15 min at room temperature and then dried at 50°C for 5 min. The slide was then dipped into a boiling water bath for 30 s to remove non-covalently bound oligonucleotide and dried under vacuum. Similarly, an oligonucleotide sequence, H2N-(CH2)5-OPO3-d(CAG AGG TTC TTT GAG TCC TT), was immobilized on a glass slide using the NTMTA method (route A) as discussed above. The three microslides were then kept in a hybridization chamber and treated with a complementary-labeled oligonucleotide, FAM-d(AAG GAC TCA AAG AAC CTC TG), dissolved in hybridization buffer, for 2 h at 25°C. The glass microslides were washed with hybridization buffer (5 × 10 ml) to remove excess complementary oligomer. Subsequently, the microslides were dried and examined under a laser scanner.
RESULTS AND DISCUSSION
A number of hetero- and homobifunctional reagents have been reported to modify peptides and proteins and immobilization of biomolecules on a variety of polymer supports. Most commonly used reagents are either based on thermochemical or photochemical reactive groups. Usually, the thermochemical reactive groups containing reagents require the presence of nucleophilic functions (-SH, -NH2) in the ligand molecules or on the polymer surface. A number of methods are currently available where modified oligonucleotides are immobilized on polymer surfaces for the construction of microarrays. Most of them utilize functionalized polymer surfaces for this purpose. Of the various polymer surfaces tried so far, only glass and PMMA stand a good chance, as these materials can easily be functionalized and adapted to laser scanners for visualization of fluorescent spots on the surface. Unfortunately, there are not many glass-specific reagents available for this purpose. The glass-specific reagents known in the literature have some limitations, such as either taking too long a time (1.5–8 h) for fixing oligonucleotides on the glass surface or they result in support-bound oligonucleotides with labile linkage (disulfide). In the present investigation, we have designed a strategy to immobilize oligonucleotides on a virgin glass surface using a novel heterobifunctional reagent. While designing the synthesis strategy for such a reagent, the following points were kept in mind: (i) the preparation of the reagent should be straightforward and involve commonly available reagents and chemicals; (ii) it should be sufficiently stable during storage; and (iii) one end of the reagent should contain an active chemical group specific towards aminoalkyl and mercaptoalkyl functionalities, easy to introduce at the 5′ end of oligonucleotides during synthesis in the machine itself, while the other end of the reagent should posses a chemical group reactive towards the glass surface. Taking all these factors into consideration, we screened a large number of reactive chemical groups specific towards mercaptoalkyl and aminoalkyl functionalities and reached the conclusion that trifluoroethanesulfonyl- (tresyl) could be the right choice owing to its reactive nature, specificity towards mercaptoalkyl and aminoalkyl functionalities and reasonably good stability. The choice of reactive functionality for the other end of the reagent was, of course, limited, and therefore, a well-known glass-specific group, triethoxysilyl, was selected. A heterobifunctional reagent, NTMTA, was designed and synthesized using commonly available reagents and used for the immobilization of 5′-mercaptoalkyl and aminoalkylated oligonucleotides on an unmodified glass microslide.
The chemical synthesis of the proposed heterobifunctional reagent, NTMTA, is depicted in Scheme 1. The preparation was carried out by a two step process, where 3-chloropropyltriethoxysilane (I) was reacted with N-methyl-2-aminoethanol (II) in the presence of N,N-diisopropylethylamine in hot toluene. The ammonium chloride formed in the reaction was filtered under inert atmosphere, washed twice with toluene and the combined filtrate and washings were pooled together and concentrated under vacuum. The syrupy residue obtained was distilled under vacuum to obtain the intermediate, NHMTA (III) in 65% yield, which was subsequently reacted with trifluoroethanesulfonyl chloride (tresyl chloride) to get the desired heterobifunctional reagent, NTMTA (IV), in almost quantitative yield. The NTMTA reagent was characterized by 1H-NMR and MALDI-TOF.
Scheme 1. Preparation of NTMTA reagent and immobilization of oligonucleotides.
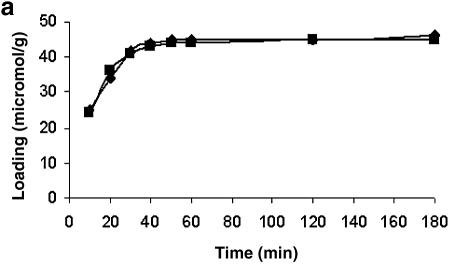
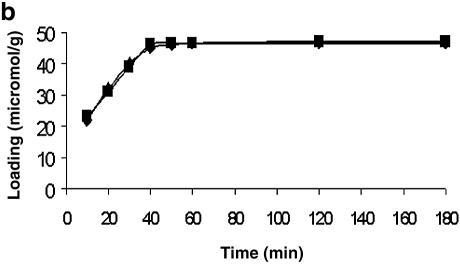
To arrive at the optimum time required to immobilize the 5′ end-modified (mercaptoalkyl or aminoalkyl) oligonucleotides on a glass surface using NTMTA, this was determined indirectly by immobilizing two model compounds, DMTrO (CH2)5NH2 and DMTrO(CH2)-SH, on controlled pore glass (CPG) following both routes. The results are shown in Figure 1a and b, and from these kinetics data, it can be inferred that the immobilization of mercaptoalkyl and aminoalkylated ligands on CPG using NTMTA essentially completes in 40 min at 45°C. Therefore, in all the experiments, the same time period was used for immobilization of oligonucleotides via route A or B.
Figure 1.
Time kinetics to determine the optimal time required to immobilize oligonucleotides on unmodified glass surface via route A (a) and route B (b). Squares, 5′-mercaptoalkylated ligand; diamonds, 5′-aminoalkylated ligand.
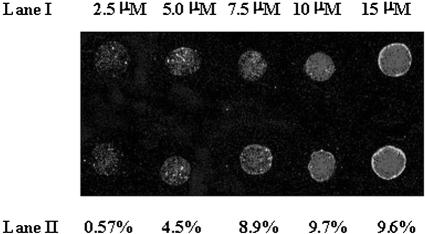
To determine the optimal concentration required for visualization of an oligonucleotide on the glass slide, an activated glass microslide was prepared using NTMTA (route B). On this activated surface, an oligomer, d(TFTT TTT TTT TTT TTT TT-OPO3-(CH2)6-SH), was spotted in five different concentrations (2.5, 5, 7.5, 10, 15 µM). After the usual washings, the spots were visualized under a laser scanner set at 570 nm (Fig. 2, lane I). The immobilization efficiency against each concentration was determined with the help of the standard curve (Fig. 4) and is depicted in lane II of Figure 2. Though the spots corresponding to 2.5–7.5 µM concentration can be visualized easily, a 10 µM concentration was selected for construction of microarrays so as to have good quality spots. Hence, in the rest of the experiments, the same concentration was used for immobilization purposes.
Figure 2.
Threshold concentration of oligonucleotide sequence d(TFTT TTT TTT TTT TTT TTT TT)-OPO3-(CH2)6SH required for visualizing fluorescence under a laser scanner. Lane I, concentration of spots (2.5, 5, 7.5, 10, 15 µM); lane II, immobilization efficiency.
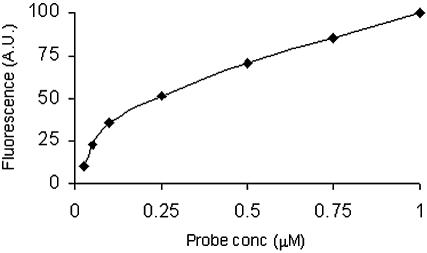
Figure 4.
A correlation sketch between concentration of immobilized probe and fluorescence intensity. Fluorescent oligonucleotide was spotted in 0.025–1.0 µM concentrations. The spotted microslide was scanned under a laser scanner.
Immobilization of oligonucleotides on glass microslides and hybridization studies
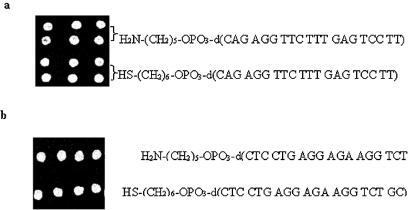
Having arrived at the optimal concentration and time required for immobilization of 5′-modified oligonucleotides on the glass surface, immobilization of oligonucleotides on the glass microslides was carried out using NTMTA following both routes. As NTMTA is a heterobifunctional reagent, it was therefore considered necessary to explore whether this reagent could be used in two ways. In route A, it reacts via its tresyl function with 5′-modified (mercaptoalkyl or aminoalkyl) oligomer to form a oligomer–triethoxysilyl conjugate, which is immobilized in the subsequent reaction on a glass surface via its triethoxysilyl function. The oligomer–triethoxysilyl conjugate was found to be stable over several months at 4°C. In the second route, route B, the NTMTA reagent first reacts with the unmodified glass microslide via its triethoxysilyl function to generate tresyl functions on it, which in the subsequent step react with 5′-modified oligonucleotides to yield surface-bound oligonucleotides. Tresylated microslides were also found to be stable over several months at 4°C. Both the routes offer unique advantages in a sense that the intermediates (oligomer-triethoxysilyl conjugate in route A and tresylated microslides in route B) can be prepared in large quantities and stored for several months safely at 4°C to avoid the hassle of preparing them just before use. Two microarrays prepared following route A and route B were visualized by hybridizing them with the corresponding complementary-labeled oligomers. After thorough washings with washing buffer, the microslides were screened under a laser scanner and the fluorescent spots were visualized, as shown in Figure 3a and b. The fluorescent signals of the spots on microslides prepared via route A and route B were found to be almost of the same intensities. Furthermore, spot morphology was also found to be almost identical.
Figure 3.
(a) Immobilization of oligonucleotides via route A. H2N-(CH2)5-OPO3-d(CAG AGG TTC TTT GAG TCC TT) (entry 1, Table 1) and HS-(CH2)6-OPO3-d(CAG AGG TTC TTT GAG TCC TT) (entry 2, Table 1) visualized after hybridization with FAM-d(AAG GAC TCA AAG AAC CTC TG) (entry 7, Table 1). (b) Immobilization of oligomers via route B. H2N-(CH2)5-OPO3-d(CTC CTG AGG AGA AGG TCT GC) (entry 5, Table 1) and HS-(CH2)6-OPO3-d(CTC CTG AGG AGA AGG TCT GC) (entry 3, Table 1) visualized after hybridization with FAM-d(GCA GAC CTT CTC CTC AGG AG) (entry 8, Table 1).
Quantification of immobilized oligonucleotides
Quantification of immobilized oligonucleotides was carried out by a graphical method. Here, an oligomer, d(TFTT TTT TTT TTT TTT TTT TT-OPO3-(CH2)6SH), diluted in seven different concentrations (0.025, 0.050, 0.10, 0.25, 0.50, 0.75, 1 µM), as detailed in Materials and Methods, was spotted on a glass slide in duplicate and the fluorescence intensity of each spot was determined. The fluorescence intensity of the spots was plotted against its concentration (Fig. 4). An almost linear correlation was observed between the concentration of the spotted oligonucleotide and its fluorescence intensity. The same curve was then used to quantify all the microarrays prepared using the proposed method.
Specificity of immobilization chemistry and detection of mismatches
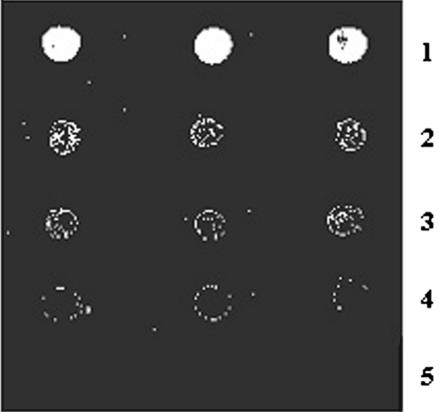
In order to demonstrate the specificity of immobilization chemistry and to detect mismatches in the immobilized oligonucleotides, five oligonucleotide sequences having zero, one, two and three mismatches (shown as bold and underlined), H2N-(CH2)5-OPO3-d(CTC CTG AGG AGA AGG TCT GC), H2N-(CH2)5-OPO3-d(CTC CTG AGG CGA AGG TCT GC), H2N-(CH2)5-OPO3-d(CTC CTG CGG AGA ACG TCT GC), H2N-(CH2)5-OPO3-d(CTC CTG CGG CGA ACG TCT GC) and non-complementary, H2N-(CH2)5-OPO3-d(TTT TTT TTT TTT TTT TTT TT), were immobilized on a glass microslide through route B, as discussed in Materials and Methods. After blocking the reactive sites, the microslide was placed in a hybridization chamber and treated with a labeled oligomer (complementary to zero mismatch sequence), 5′-FAM-d(GCA GAC CTT CTC CTC AGG AG). After thorough washings with washing buffer, the microslide was scanned under a laser scanner. On the basis of fluorescence intensity, base mismatches were detected and the results are shown in Figure 5. The perfectly matched duplex gave the maximum intensity (lane 1) while the spots having one (lane 2), two (lane 3) and three mismatches (lane 4) showed fluorescence intensities in decreasing order and non-complementary (lane 5) did not give any signal at all. It is evident from the results that the immobilized oligonucleotides are specific towards complementary oligonucleotides and non-specific hybridization does not occur (lane 5). The signal-to-noise ratio was on an average >98, as calculated from the signal obtained from complementary hybridized (lane 1) and non-complementary hybridized oligomers (lane 5) at 10 µM concentration. The low noise background in this system may be due to the fact that the surface is not aminated (positively charged) and therefore, not susceptible to non-specific interactions with oligonucleotides (polyanions).
Figure 5.
Detection of nucleotide mismatches and specificity of immobilization via hybridization with fluorescein-labeled complementary oligonucleotides. Lane 1, H2N-(CH2)5-OPO3-d(CTC CTG AGG AGA AGG TCT GC) (entry 5, Table 1); lane 2, H2N-(CH2)5-OPO3-d(CTC CTG AGG CGA AGG TCT GC) (entry 6, Table 1); lane 3, H2N-(CH2)5-OPO3-d(CTC CTG CGG AGA ACG TCT GC) (entry 10, Table 1); lane 4, H2N-(CH2)5-OPO3-d(CTC CTG CGG CGA ACG TCT GC) (entry 11, Table 1); lane 5, H2N-(CH2)5-OPO3-d(TTT TTT TTT TTT TTT TTT TT) (entry 12, Table 1). Hybridization was carried out using FAM-d(GCA GAC CTT CTC CTC AGG AG) (entry 8, Table 1) followed by laser scanning.
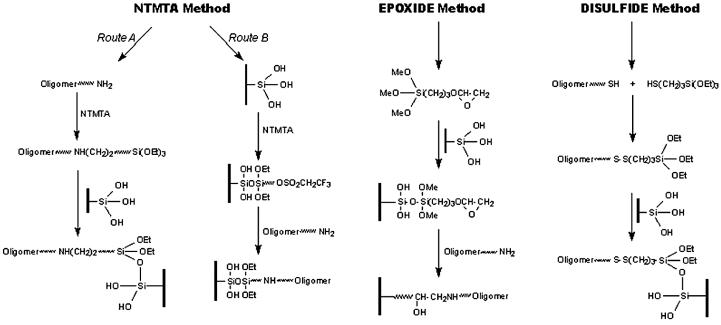
Comparison of the proposed method with the standard methods
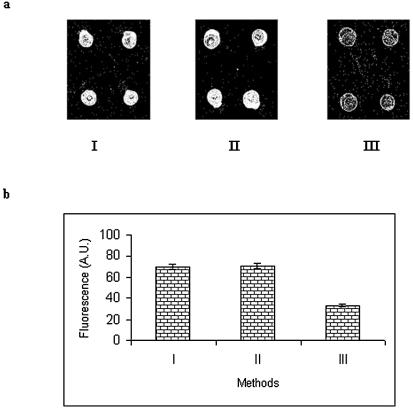
The efficiency of the proposed reagent for immobilization of oligonucleotides was compared with two existing standard methods (Scheme 2). For comparison purposes, an equal concentration (10 µM) of an oligonucleotide was used in all the three methods. However, different reaction times were employed, as reported. The immobilization efficiency in each case was determined after hybridizing the microslides under identical conditions. After hybridization with fluorescent-labeled oligonucleotide, the microslides were visualized under a laser scanner. The results are shown in Figure 6a. Using the fluorescence values from Figure 6a, a histogram was drawn, as shown in Figure 6b. From these figures, it can be deduced that the immobilization efficiency of the proposed method (9.854%) is almost comparable to the epoxide method (9.878%) and much higher than the disulfide method (1.7%). However, the proposed method (40 min) is far superior to the epoxide method (480 min) in terms of immobilization time.
Scheme 2. Comparison of proposed method with the standard methods.
Figure 6.
(a) Morphology of spots obtained after hybridization with labeled complementary oligonucleotide. I, NTMTA method; II, epoxide method; III, disulfide method. (b) Comparison of immobilization efficiency. I, NTMTA method; II, epoxide method; III, disulfide method.
Thermal stability of microarrays constructed using the proposed method
The thermal stability of the constructed arrays using the proposed reagent was evaluated following the PCR conditions. The constructed microarray was subjected to three sets of temperature conditions, as used in a standard PCR. The microarray was first subjected to 94°C for 30 s (denaturing step) in 0.1 M citrate buffer containing 0.5 M sodium chloride pH 7.1, followed by cooling to 54°C for 30 s (annealing step), and then heating to 75°C for 30 s (extension step). This cycle was repeated five times. The virgin microarray (without heat treatment) and heat-treated microarray were hybridized with the complementary-labeled probe. The hybridization signal of the heat-treated microarray was found to be decreased by only 1.6%, clearly indicating that the immobilized oligonucleotides on the glass surface following the proposed method are quite stable for exploitation in biological research. A histogram (Fig. 7) was obtained in a typical experiment comparing the hybridized signal of heat-treated and pre-treated microarrays.
Figure 7.
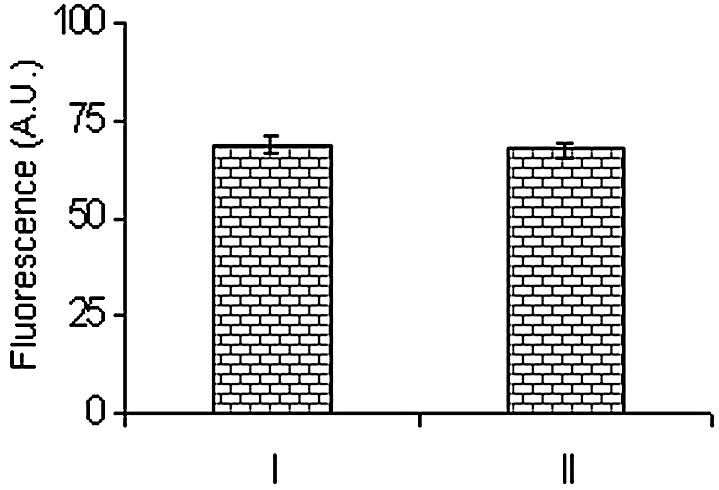
Thermal stability of immobilized oligonucleotides (microarray). Comparison on the basis of fluorescence signals obtained after hybridization of heat-treated and untreated arrays of oligonucleotides. I, untreated microslide; II, heat-treated microslide.
CONCLUSION
A simple and rapid method for the construction of oligonucleotide arrays on a glass surface has been developed employing a novel heterobifunctional reagent. The proposed reagent has been used in two ways. In the first one, 5′-aminoalkyl or mercaptoalkyl oligonucleotide reacts first with the NTMTA reagent, to generate oligonucleotide conjugate, which subsequently reacts with unmodified glass microslides to obtain an oligonucleotide covalently fixed on the surface. The second route involves the reaction of NTMTA with glass microslides to generate an activated surface, where oligonucleotides with 5′-aminoalkyl or 5′-mercaptoalkyl groups were spotted on it to get surface-bound oligonucleotides. The density of immobilized DNA was found to be comparable or better than the existing glass-specific methods and far superior to these methods in terms of reaction time for construction of microarrays and signal-to-noise ratio (>98). Moreover, the covalently immobilized oligonucleotides were stable to varying heating conditions (PCR conditions).
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Professor S. K. Brahmachari, Director, IGIB, Delhi, for his keen interest in the work. We are thankful to Dr G. W. Rembhotkar for his timely help in scanning some of the microarrays. The authors gratefully acknowledge a financial grant from CSIR Task Force Project (NNIOSB).
REFERENCES
- 1.Ramsay G. (1998) DNA-chips: state-of-the-art. Nat. Biotechnol., 16, 40–44. [DOI] [PubMed] [Google Scholar]
- 2.Pollock J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A. Williams,C.F., Jeffery S.S., Botstein,D. and Brown,P.O. (1999) Genome-wide analysis of DNA-copy number changes using cDNA microarrays. Nature Genet., 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 3.Marshall A. and Hodgson,J. (1998) DNA chips: an array of possibilities. Nat. Biotechnol., 16, 27–31. [DOI] [PubMed] [Google Scholar]
- 4.Drobyshev A.N., Mologina,N., Shick,V., Pobedimskaya D., Yershov,G. and Mirzabekov,A. (1997) Sequence analysis by hybridization with oligonucleotide microchip: identification of β-thalessemia mutations. Gene, 188, 45–52. [DOI] [PubMed] [Google Scholar]
- 5.Sosnowskii R.E., Tu,E., Butler,W., O’Connell,J. and Heller,M. (1997) Rapid determination of single base mismatch mutations in DNA hybrids by direct electric field control. Proc. Natl Acad. Sci. USA, 94, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krylov A.S., Zasedateleva,O.A., Prokopenko,D.V., Rouviere-Yaniv,J. and Mirzabekov,A.D. (2001) Massive parallel analysis of the binding specificity of histone-like protein HU to single- and double-stranded DNA with generic oligodeoxyribonucleotide microchips. Nucleic Acids Res., 29, 2654–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong S., Li,C. and Wong,W.H. (2003) ChipInfo: software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res., 31, 3483–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor S.P., Read,J.L., Pirrung,M.C., Stryer,L., Lu,A.T. and Solas,D. (1991) Light directed, spatially addressable parallel chemical synthesis. Science, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- 9.Singh-Gasson S., Green,R.D., Yue,Y., Nelson,C., Blattner,F., Sussman,M.R. and Cerrina,F. (1999) Maskless fabrication of light-director oligonucleotide arrays using a digital micromirror array. Nat. Biotechnol., 17, 974–978. [DOI] [PubMed] [Google Scholar]
- 10.McGall H. and Fidanza,J.A. (2001) Photolithographic synthesis of high density oligonucleotide arrays. Methods Mol. Biol., 170, 71–101. [DOI] [PubMed] [Google Scholar]
- 11.Schena M. (2000) Microarray Biochip Technology. Eaton Publishing, Natick, MA. [Google Scholar]
- 12.Seliger H., Hinz,M. and Happ,E. (2003) Arrays of immobilized oligonucleotides—contributions to nucleic acids technology. Curr. Pharmaceutical Biotechnol., 4, 379–395. [DOI] [PubMed] [Google Scholar]
- 13.Vaijayanthi B., Kumar,P., Ghosh,P.K. and Gupta,K.C. (2003) Recent advances in oligonucleotide synthesis and their applications. Ind. J. Biochem. Biophys., 40, 377–391. [PubMed] [Google Scholar]
- 14.Joos B., Kuster,H. and Cone,R. (1997) Covalent attachment of hybridizable oligonucleotides to glass supports. Anal. Biochem., 247, 96–101. [DOI] [PubMed] [Google Scholar]
- 15.Kumar P. and Gupta,K.C. (2003) A rapid method for the construction of oligonucleotide arrays. Bioconjug. Chem., 14, 507–512. [DOI] [PubMed] [Google Scholar]
- 16.Defrencq E. Hoang,A., Vinet,F. and Dumy,P. (2003) Oxime bond formation for covalent attachment of oligonucleotides on glass support. Biomed. Chem. Lett., 13, 2683–2686. [DOI] [PubMed] [Google Scholar]
- 17.Belosludtsev Y., Iverson,B., Lemeshko,S., Eggers,R., Wiese,R., Lee,S., Powdrill,T. and Hogan,M. (2001) DNA microarrays based on non-covalent oligonucleotide attachment and hybridization in two dimensions. Anal. Biochem., 292, 250–256. [DOI] [PubMed] [Google Scholar]
- 18.Nonglaton G., Benitez,I.O., Guisle,I., Pipelier,M., Leger,J., Dubreuil,D., Tellier,C., Talham,D.R. and Bujoli,B. (2004) New approach to oligonucleotide microarrays using zirconium phosphonate-modified surfaces. J. Am. Chem. Soc., 126, 1497–1502. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X., Nampalli,S., Serino,A.J. and Kumar,S. (2001) Immobilization of oligodeoxyribonucleotides with multiple anchors to microchips. Nucleic Acids Res., 29, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benters R., Niemeyer,C.M., Drutschmann,D., Blohm,D. and Wohrle,D. (2002) DNA microarrays, with PAMAM dendritic linker systems. Nucleic Acids Res., 30, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broude N.E., Woodward,K., Cavallo,R., Cantor,C.R. and Englert,D. (2001) DNA microarrays with stem–loop DNA probes: preparation and applications. Nucleic Acids Res., 29, e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A., Larsson,O., Parodi,D. and Liang,Z. (2000) Silanized nucleic acids: a general platform for DNA immobilization. Nucleic Acids Res., 28, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moller R., Csaki,A., Kohler,A.M. and Fritzsche,W. (2000) DNA probes on chip surfaces studied by scanning force microscopy using specific binding of colloidal gold. Nucleic Acids Res., 28, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamture J.B., Beattie,K.L., Burke,B.E., Eggers,M.D., Ehrlich,D.J., Fowler,R., Hollis,M.A., Kosicki,B.B., Reich,R.K. and Smith,S.R. (1994) Direct detection of nucleic acid hybridization on the surface of a charged coupled device. Nucleic Acids Res., 22, 2121–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar P., Agrawal,S.K., Misra,A. and Gupta,K.C. (2004) A new heterobifunctional reagent for immobilization of biomolecules on glass surface. Biomed. Chem. Lett., 14, 1097–1099. [DOI] [PubMed] [Google Scholar]
- 26.(1989) Pharmacia LKB Gene Assembler Plus Manual. Uppsala, Sweden. [Google Scholar]
- 27.Gaur R.K., Sharma,P. and Gupta,K.C. (1989) A simple method for the introduction of thiol group at 5′-termini of oligonucleotides. Nucleic Acids Res., 17, 4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P., Bhatia,D., Rastogi,R.C. and Gupta,K.C. (1996) Solid phase synthesis and purification of 5′-mercaptoalkylated oligonucleotides. Biomed. Chem. Lett., 6, 683–688. [Google Scholar]
- 29.Gupta K.C., Sharma,P., Sathyanarayana,S. and Kumar,P. (1990) A universal support for the synthesis of 3′-thiol group containing oligonucleotides. Tetrahedron Lett., 31, 247–250. [Google Scholar]
- 30.Gait M.J. (1984) Oligonucleotide Synthesis: A Practical Approach. IRL Press, Oxford. [Google Scholar]