Abstract
kis-kid, the auxiliary maintenance system of plasmid R1 and copB, the auxiliary copy number control gene of this plasmid, contribute to increase plasmid replication efficiency in cells with lower than average copy number. It is thought that Kis antitoxin levels decrease in these cells and that this acts as the switch that activates the Kid toxin; activated Kid toxin reduces copB-mRNA levels and this increases RepA levels that increases plasmid copy number. In support of this model we now report that: (i) the Kis antitoxin levels do decrease in cells containing a mini-R1 plasmid carrying a repA mutation that reduces plasmid copy number; (ii) kid-dependent replication rescue is abolished in cells in which the Kis antitoxin levels or the CopB levels are increased. Unexpectedly we found that this coordination significantly increases both the copy number of the repA mutant and of the wt mini-R1 plasmid. This indicates that the coordination between plasmid replication functions and kis-kid system contributes significantly to control plasmid R1 replication.
Keywords: plasmid R1, plasmid replication and copy number control, parD (kis-kid) antitoxin-toxin system, Kis antitoxin, ClpAP protease, transcriptional regulation, CopB repressor, coupling plasmid maintenance systems
1. Introduction
Plasmid R1 is an antibiotic resistance plasmid of enteric bacteria that has contributed important insights into plasmid replication and its control as well as into the regulation and role of auxiliary plasmid maintenance systems [1]. R1 is maintained with a low copy number in the host. Its replication is initiated due to specific interactions of a rate limiting protein, RepA, at oriR1, the origin of replication [2]. The frequency of this process is regulated by the copy number control genes copA and copB. copA, the key regulator gene, codes an unstable antisense RNA, CopA, that inhibits at the posttranscriptional level the synthesis of RepA. CopA RNA targets the polycistronic copB-repA mRNA at copT, its complementary sequence, and inhibits translation of the tap orf which is needed for RepA translation [3,4]. Inactivation of copA leads to uncontrolled amplification of the plasmid or run-away replication [5].
CopB is a tetrameric repressor protein that inhibits transcription of repA from the internal and strong promoter Prep. The levels of this protein, that is transcribed from a constitutive promoter, decrease when plasmid copy number is reduced; at a very low copy number, the tetramer disassembles and the protein lose activity as a repressor; this increases transcription of repA and as consequence the copy number of the plasmid increases. In this way, copB acts as a proper copy number control gene that contributes by rescuing inefficient plasmid replication [6,7,8]. Once the plasmid copy number is restored, CopB levels increase and the protein multimerizes and recovers its repressor activity. Increasing CopB levels in trans favours CopB repressor function but removes its potential to rescue very low plasmid copy number [9]. copB, copA, copT, repA and oriR1, the so called “basic replicon” [10], is the essential maintenance module of the plasmid.
Kis-kid or parD is an auxiliary maintenance module of R1 that is close to the basic replicon [11,12] (see Figure 1). This system contains two genes, kis and kid, encoding respectively two small proteins: an antitoxin, Kis (killer suppressor), and a toxin, Kid (killing determinant). Kid is an RNase that cleaves RNA at sites containing the core sequence 5'-UA(A/C/U)-3; flanking U increase the efficiency of cleavage at these core sequences [13,14,15]. Kis, the antitoxin, is a protein that interacts with Kid and neutralizes its activity. Kis is also a specific repressor of the operon whose efficiency increases in complex with Kid [16,17].
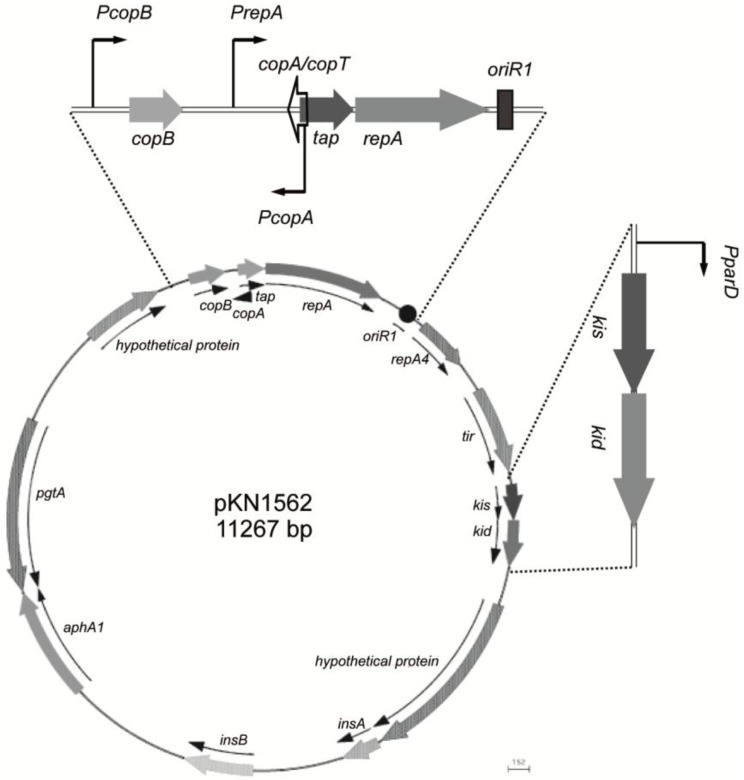
Figure 1.
Localization of maintenance regions in the mini-R1 plasmid pKN1562. Horizontal and vertical projections correspond respectively to the basic replicon module and the kis-kid antitoxin-toxin system. The relative positions of key genes in these modules and the promoters involved in RNA synthesis are also indicated.
kis-kid activity is functionally coupled to the efficiency of R1 replication. The first indication of this coupling was indirect: kis-kid interfered with the isolation of plasmid replication mutants; this was due to the activity of Kid: mutations that inactivated the Kid toxin abolished this interference [18]. Since then, this phenotype, called the “interference” phenotype, has been used as one of the signatures of this coupling. It was later reported that the kis-kid system is activated in low copy mutants of the plasmid and that this partially recovered the plasmid copy number [19]. Two findings were key to explaining, in molecular terms, this new and intriguing signature of the coupling: (i) the identification of the RNase activity of the Kid toxin [13,14,15] and (ii) the finding that copB-repA mRNA contains two sites in the intergenic region of copB-repA mRNA that are efficiently cleaved by Kid [20]; this cleavage reduces the CopB levels, activates the repA promoter and increases plasmid replication efficiency. It has been recently reported that Kid cleaves mRNA of key cell division proteins; in this instance, Kid replication rescue occurs before cell division and effectively enforces plasmid retention by uncoupling plasmid replication and cell division [21]. We reported recently that increasing in trans the Kis antitoxin levels suppressed the “interference” phenotype; this suggested that Kis antitoxin levels could act as the switch connecting the replication and kis-kid toxin-antitoxin maintenance functions [22]. Coupling between replication functions and other maintenance modules has been reported in other plasmids systems: in pSM19035 of Streptococcus pyogenes a global regulator couples plasmid replication, partitioning and toxin-antitoxin modules to achieve high plasmid stability [23]; in the broad host range plasmid RK2, a global regulator controls expression of replication and maintenance systems in different hosts [24]. In the repABC plasmid family of Rhizobiales, transcription of the gene of the replication initiation protein is controlled by proteins of the plasmid partitioning system [25]. In ColE1 plasmid, a multimer resolution system, XerCD couples replication and cell division to achieve plasmid maintenance [26] (see Discussion).
The results reported here support the role of Kis antitoxin as the switch that couples replication functions and the kis-kid maintenance system; they also support the proposal that a Kid-dependent decrease in the CopB levels increases plasmid replication efficiency. In addition, we found that, beyond playing a role as a replication safety device, coupling between plasmid replication and the kis-kid system increases significantly the copy number of the wt plasmid R1. This implies that this coupling plays a significant role as part of the basic mechanisms that control plasmid R1 replication.
2. Results
2.1. Kis Antitoxin Levels Decrease in Cells Containing a pKN1562 Replication Mutant and Increase in a clpP− Background
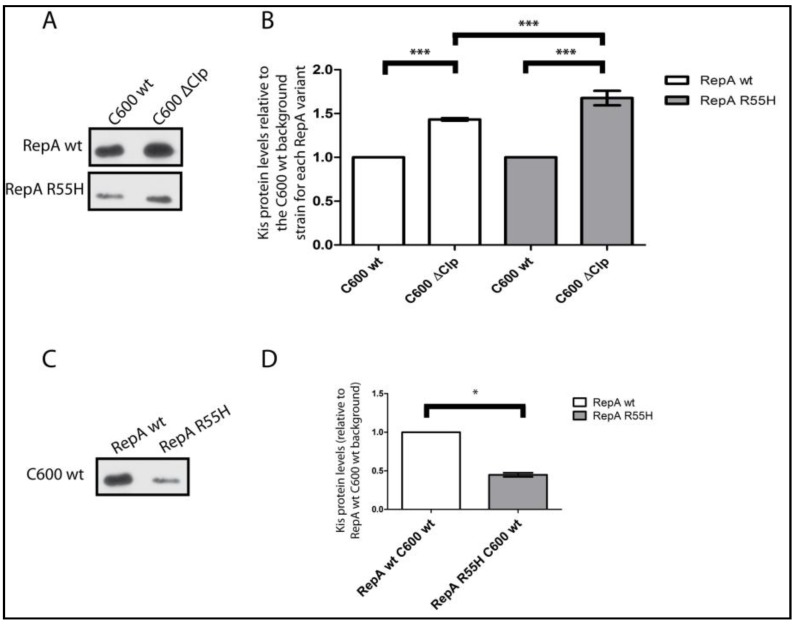
To evaluate the proposal that a reduction in the efficiency of plasmid R1 replication reduced the Kis levels, we compared these levels in cells containing plasmid pKN1562 or its repA55 mutant. This mutation reduces significantly the copy number of the plasmid and activates the modular coupling (see Section 2.2). For these determinations, we tagged kis antitoxin genes of the wt and rep mutant plasmids with a 3×FLAG epitope that is recognized efficiently by specific monoclonal antibodies. Kis antitoxin levels were subsequently evaluated by inmuno-blotting. The data (Figure 2C,D) show that the Kis antitoxin levels decreased in cells containing the plasmid-repA mutant. Consistently with the role of ClpAP as the specific protease cleaving Kis [27], the levels of this protein increased in a strain carrying a deletion of the gene of the ClpP protease; this increase occurred in cells containing the repA-wt miniR1 plasmid pKN1562 or its repA55 mutant (Figure 2A,B).
Figure 2.
Inmunodetection of Kis antitoxin levels. Western blot (A,C) and densitometric analysis (B,D) of Kis levels determined in C600 or in its clpP− derivative (C600−∆clpP). Cells contained either the pKN1562 mini-R1 plasmid coding for RepAwt or its repA55 mutant. The repA55 mutation changes R for H in codon 55 of RepA and this results in a thermosensitive replication protein that, at the permissive temperature, reduces plasmid copy number. An equal amount of total protein extracts were loaded in each lane of the gels and the inmuno-signal of the 3×FLAG labeled Kis was densitometred. Values in B and D represent the average of seven independent densitometries and are corrected for the number of plasmid-containing cells. This percentage was 100% for cells containing the wt plasmid and 78% for cells containing the repA55 plasmid replication mutant. * or *** indicate differences which p-values are = 0.01–0.05 or <0.001 respectively.
2.2. Effects of Increasing the Stability of the Kis Antitoxin on the Modular Coupling
Coupling between replication functions and the kis-kid system reduces drastically the frequency of isolation of plasmid replication mutants (interference phenotype). This phenotype, a signature of the modular coupling, can be abolished by inactivating the gene of the Kid toxin or by overproducing the Kis antitoxin [22]. We now tested if the increased levels of the Kis antitoxin shown in Figure 2 associated to its stabilization in a clpP− strain abolished this phenotype. To this aim we transformed C600 and its clpP− mutant with a preparation of pKN1562 mutagenized with hydroxylamine. The number of pKN1562 thermo-sensitive replication mutants (rep-ts) recovered in this transformation was compared with the number of kanamycin resistant thermo-sensitive mutants (kmr-ts) rescued in the same screening. Note that the rescue of kmr-ts mutants is independent of the presence of a kis-kid system; therefore, they serve as a reference to determine the relative number of rep-ts mutants isolated in different conditions [22]. The analysis indicated that the rep-ts/kmr-ts ratios were 0.22 in the wild-type strain (2/9) and increased to 1.1 in the clpP− strain (10/9). The increase ratio obtained in this strain indicates that stabilization of Kis antitoxin inactivates the interference phenotype, which implies inactivation of the modular coupling. Note that a similar increase ratio has been reproduced in experiments in which the modular coupling was inactivated either by inactivation of kid or by overproduction of Kis [22]. The experimental procedures are detailed in this reference and these procedures are only briefly discussed here (material and methods).
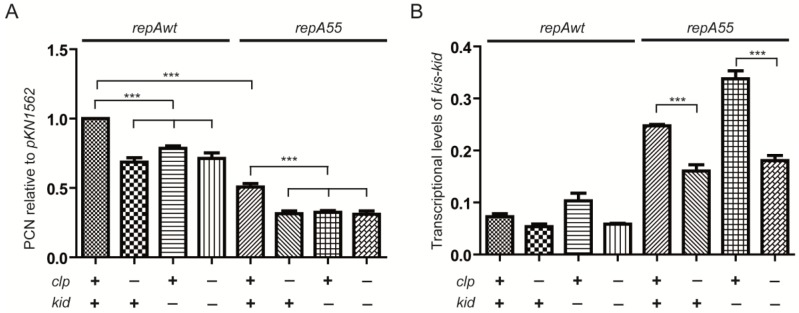
A second signature of the coupling is the replication rescue phenotype, meaning a partial recovery of the efficiency of plasmid replication of repA mutants mediated by kis-kid. Data in Figure 3A show that in a wt background (clp +), the repA55 mutation reduced significantly the copy number of pKN1562. In the isogenic clpP− strain (clp −), this value is further reduced. The difference found is statistically significant and indicates that stabilization of the Kis antitoxin, protein in this last strain abolishes the replication enhancement effect dependent on kis-kid. The results are consistent with the proposal that the clpP− background that stabilizes the Kis antitoxin, inactivates the modular coupling. As a control, it is shown that inactivation of the coupling due to the kid75 mutation, abolishes the replication rescue effect. Important, during this analysis we realized that in the clpP− background, the copy number of the wt plasmid pKN1562 is significantly reduced. Furthermore, the kid75 mutation that inactivates the modular coupling reduces the efficiency of plasmid wt replication to a similar level. This result revealed that the communication between the two maintenance modules form part of the basic mechanisms that control plasmid R1 replication. Results presented in the next two sections further support this conclusion.
Figure 3.
Effects of stabilizing Kis antitoxin (A) on plasmid replication and (B) on kis-kid transcriptional levels. Analyses were done in a wt strain or in an isogenic clpP− background using plasmids KN1562 and its repA55 thermo-sensitive replication mutant; kid + and kid − indicates respectively analysis carried out in the presence of a pKN1562 or of its kid75 mutant pJLV01. (A) Plasmid copy number determinations (PCN) of pKN1562 (kid +) and its kid75 mutant (kid −) in the presence/absence of a functional clpP gene (clp + or clp −). Values were corrected by the number of plasmid free cells and referred to PCN of the lpp chromosomal gene (see Materials and Methods). The PCN value corresponding to pKN1562 in the wt strain (clp +, kid +) was given the reference value of 1; (B) Transcriptional levels of kis-kid per plasmid copy corresponding to the samples analyzed in panel (A). Transcriptional levels of the lpp gene were used as the reference. *** indicates differences which p-values are <0.001.
Coupling between replication and kis-kid modules is also associated with a de-repression of the kis-kid operon. To follow this correlation, we determined the kis-kid mRNA levels in cells containing the wild-type pKN1562 plasmid or its repA55 mutant (Figure 3B). An increase in kis-kid transcription level is most clearly observed in the repA55 mutant and in the presence of a wt clpP gene (clp +). In the absence of ClpP (clp −), Kis repressor levels are stabilized, and accordingly, transcriptional levels of kis-kid are significantly reduced. In cells containing plasmid pKN1562 wt (kid +), kis-kid transcription is maintained at a basal level.
Note that the kid75 mutation inactivates the modular coupling as it inactivates the RNase activity of the toxin; however, the mutations do not interfere with the co-regulatory potential of the toxin and this allows evaluation of kid mutation on kis-kid transcription. We noticed that the presence of the kid75 mutation significantly increases kis-kid transcription levels implying a negative effect of the RNase activity of Kid on the levels of the kis-kid transcript.
2.3. Effects of Overproducing the Kis Antitoxin on the Modular Coupling
Previous results indicate that overproduction of the Kis antitoxin abolished the interference phenotype meaning inactivation of the coupling [22]. We now completed the analysis testing the effect of this overproduction on the replication rescue phenotype and on the transcriptional level of the kis-kid operon.
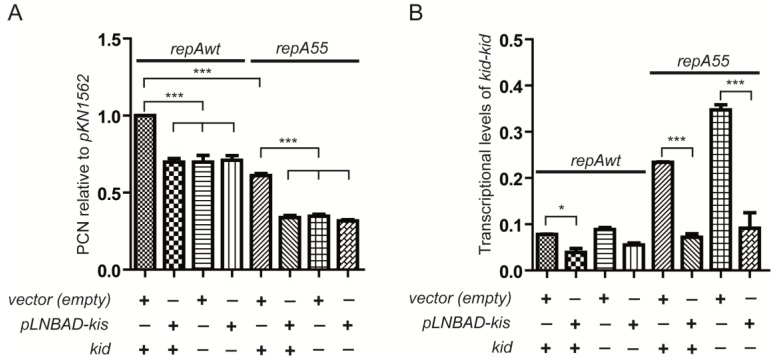
As activation of the modular coupling can also be detected in cells containing the wild-type plasmid pKN1562 (2.1.), we first evaluated the replication rescue phenotype testing the copy number of this plasmid in the presence or absence of excess of Kis antitoxin; this excess was provided in trans by the Kis overproducer pMLM126 in the presence of inducer. Control values were obtained in the presence of the empty vector pLNMBAD. Data show that indeed, the copy number of pKN1562 is substantially reduced in excess of Kis antitoxin (Figure 4A). This result confirms the effect of the coupling on the basal efficiency of replication of pKN1562 and implies that excess of Kis antitoxin inactivates the coupling. Data in Figure 4A further show that the kid75 mutation (pJLV01) abolishes the replication rescue effect, which is consistent with the dependence of this phenotype on an active toxin.
Figure 4.
Effects of overproducing Kis antitoxin on the modular coupling. pLNBAD-kis is a conditional overproducer of the Kis antitoxin and + or − indicates respectively presence or absence of inducer (arabinose). As a control, analyses were carried also in the presence of the empty vector (pLNBAD). Effects on KN1562 wt, its repA55 mutant and on derivatives of these plasmids carrying the kid17 mutation were determined. (A) Copy number determinations. The PCN value corresponding to pKN1562 was given the reference value of 1; (B) kis-kid transcriptional levels. Values were corrected by the number of plasmid free cells and referred to the PCN or the transcriptional level of the lpp chromosomal gene (see Materials and Methods). * or *** indicate differences which p-values are = 0.01–0.05 or <0.001 respectively.
Similar conclusions can be obtained when a pKN1562repA55 mutant is used. As expected the repA55 mutation significantly reduces plasmid copy number. Again the kid75 mutation further reduces the copy number of the repA55 mutant. This reduction is similar to the one observed in excess of Kis meaning that in both cases the modular coupling is lost.
Figure 4B shows the effect of excess of Kis on kis-kid mRNA levels determined in the same conditions analyzed in 4A. The results, best shown in the presence of the repA55 mutation, indicate that this mutation de-represses the kis-kid operon and that de-repression is substantially neutralized in excess of the Kis antitoxin. Although to a lower level, the effect is also seen for the pKN1562 wt plasmid. Note that, as previously shown (Figure 3B), the kis-kid mRNA levels increase in the presence of the kid75 mutation (kid−).
The above results confirm that an excess of Kis abolishes the coupling between plasmid replication and kis-kid maintenance systems both in the wt plasmid and in its repA55 mutant.
2.4. Coupling between Maintenance Modules and Plasmid Stability
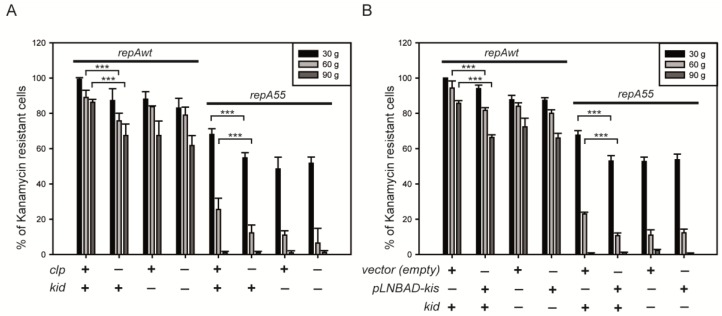
Modular coupling influences plasmid copy number and this has a direct effect on plasmid stability. We evaluated this correlation studying plasmid stability in the presence or absence of a mutation in the host that stabilizes the Kis antitoxin (clp – or clp +) or in the presence or absence of an excess of Kis antitoxin (pLNBAD-kis + or −). As shown previously, both conditions inactivate the modular coupling. In the analysis we used the same strains and constructions used for copy number determinations (see Section 2.2. and Section 2.3.). The results (Figure 5) show that there is a correlation between plasmid copy number and plasmid stability. The effects on plasmid stability are seen more clearly after 60 or 90 generations of propagation in non-selective medium. Conditions that abolish the modular coupling reduce plasmid copy number and this reduces plasmid stability. As predicted, the repA55 mutation that reduces the efficiency of plasmid replication results in all cases in less stable plasmids.
Figure 5.
Effects of abolishing the coupling between maintenance modules on plasmid stability. Plasmid stability was followed determining the percentage of cells retaining the kanamycin resistance marker of pKN1562 and its derivatives after propagation at 30 °C for 30, 60 and 90 generations in rich non-selective media (see Experimental Section). Panel (A) shows the effect of the clpP− background on plasmid stability and Panel (B) shows the effect of overproducing the Kis antitoxin. *** indicate differences which p-values are <0.001.
The results of the stability analysis are consistent with the role of the coupling between maintenance modules in plasmid copy number and are also consistent with the proposal that Kis antitoxin is the switch that connects these modules.
2.5. Excess of CopB Abolishes Replication Rescue
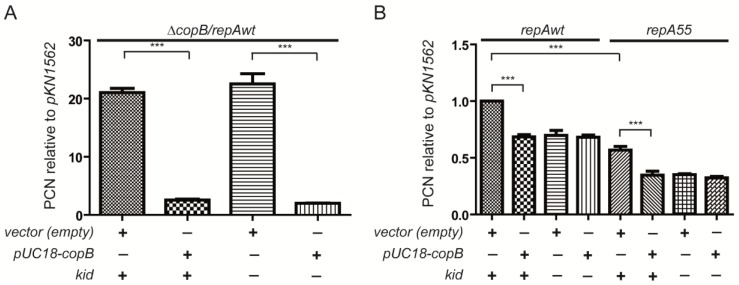
The replication rescue phenotype is dependent on the cleavage of the copB-repA mRNA mediated by the RNase activity of the Kid toxin [20]. We aimed to test the effects of an excess of CopB protein on this phenotype. The analysis was done in the presence or absence of the kid75 mutation both in the pKN1562 wt plasmid and in its repA55 mutant. As an exogenous source of CopB, we used a multi-copy pUC18-copB recombinant that greatly increases the levels of CopB [7]. The functional effect of this excess can be monitored evaluating the complementation of mini-R1 copB deletion mutation that removes the copB promoter and part of the copB gene, thus increasing the plasmid copy number. The results (Figure 6A) show that in the presence of the pUC18-copB recombinant, the copy number of copB deletion mutants of pKN1562 or of its kid75 mutant pJLV01 decreases from values close to 20–25 to a basal level. This clearly indicates that the excess of CopB provided in trans complements the copB mutation. The controls made using the pUC18 vector alone indicates that the complementation observed is a specific effect of CopB.
Figure 6.
Effects of an excess of the CopB protein on the efficiency of replication of mini-R1 plasmids. (A) PCN of copB mutants of pKN1562 (kid +) and of its kid75 mutant (kid −) determined in the presence of and overproducer of CopB (pUC18-copB) or of its empty vector (pUC18); (B) Relative PCN values of the wt pKN1562 plasmid and its repA55 mutant. Values were corrected by the number of plasmid free cells and referred to the PCN of the lpp chromosomal gene (see Materials and Methods). Determinations in the presence (+) or absence (−) of CopB overproducer or of its pUC18 vector were done in the presence of a wt kis-kid system (kid +) or of its kid75 mutant (kid −). In (A) and (B), the PCN value corresponding to pKN1562 in the presence of the empty vector (pUC18) was given the reference value of value 1 (A and B). *** indicate differences which p-values are <0.001.
We then tested the effect of an excess of CopB on the replication efficiency of plasmids pKN1562 or pJLV01, both of them carrying a wt copB gene and therefore a low copy number. Not that pJLV01 carries the kid75 mutation that inactivates the modular coupling. Data in Figure 6B show that, in the presence of the pUC18-copB recombinant, the copy number of pKN1562 is significantly reduced to the value corresponding to pJLV01. This result indicates that the replication rescue is abolished in excess CopB. As expected, the repA55 mutant of pKN1562 has a lower copy number than the wt plasmid; similarly, excess of CopB reduces the plasmid copy number of this repA mutant and this reduction is similar to the one observed in the repA mutant carrying the kid75 mutation.
These data are consistent with the proposal that the replication rescue associated to the modular coupling is dependent on a reduction of the CopB levels due to the action of the RNase activity of Kid toxin. The results confirm again that the modular coupling form part of the basic mechanisms that control plasmid R1 wt replication.
3. Discussion
3.1. Modular Coupling between Maintenance Modules and Efficiency of Plasmid wt Replication
The results of this work consistently support the model for the role of Kis antitoxin as the switch that connects the replication and kis-kid toxin-antitoxin modules of the plasmid. The Kis antitoxin levels are clearly reduced when copy number of the plasmid, that activates the coupling, is reduced. Conversely, it is shown that stabilizing the Kis antitoxin or increasing its levels prevented the modular coupling. This coupling was assessed testing the interference and replication rescue phenotypes as well as studying the transcriptional levels of the operon. Modular coupling and effective replication rescue requires normal levels of CopB. The analyses also show that increasing the CopB levels or inactivating the Kid toxin prevents the modular coupling. Interestingly, we found that the different ways of uncoupling plasmid replication and kis-kid modules consistently reduce plasmid copy number not only of the repA55 mutant but also of the wt plasmid. This implies a more direct involvement of this coupling in control of plasmid replication. The plasmid copy number distribution in individual cells of the culture can reach a significant part of the population, the low level required to trigger the coupling (discussed in [9]). Improving copy number or partitioning at cell division has been pointed out to explain the unexpected high stability of low-copy plasmids [28]. It has been pointed recently that Kid can inhibit cell division and that this toxin achieves plasmid retention by uncoupling plasmid replication and cell division [21]; in addition, the same study proposes that the hok-sok antitoxin-toxin system of this plasmid could eliminate plasmid free cells that could arise due to failures in this process.
3.2. The Pathway: Kis Antitoxin Acts as the Switch Connecting Plasmid Replication and Toxin-Antitoxin Modules
The analysis reported here indicate that the coupling between the basic replicon of R1 and the kis-kid system is initiated when the copy number of the plasmid falls; this reduces the antitoxin level and activates the potential of the Kid toxin to induce the “interference” or the “replication rescue” phenotypes. This last phenotype is dependent on kid and copB; it is activated in cells containing the wt plasmid or its replication mutants and is well defined in molecular terms. The interference phenotype is a signature of the molecular coupling and as the replication rescue is dependent on a wt Kid toxin but it is poorly defined in molecular terms.
3.3. On the Inhibition of the Replication Rescue by Excess of CopB
The replication rescue phenotype is a late event in the pathway described above. It is dependent on Kid and CopB activities. Overproduction of CopB abolishes this phenotype but in principle should not affect the regulation of the kis-kid module as the sequence targeted by this transcriptional repressor is not present in the promoter of the kis-kid operon. In addition CopB overproduction should not affect the “interference” phenotype as CopB does not affect the activity of Kid required for this phenotype. When we compared the plasmid copy number obtained in the presence or absence of an excess of CopB we observed a significant replication enhancement in the presence of normal levels of CopB that was abolished in excess of CopB. This effect is measurable and it can be assigned to the molecular coupling described here.
The modular coupling forms part of the auxiliary machinery that controls plasmid R1 replication via CopB. Its action depends on a reduction of the antitoxin levels that activates the Kid toxin and this can be affected by plasmid copy number but also by the ClpAP protease. Thus the activity of these effectors can influence the role of CopB as auxiliary copy number control protein.
3.4. Coordination of Plasmid Maintenance Functions in Different Systems
Growing information underlines the relevance of connections replication and plasmid maintenance modules and eventually with cell cycle events in different plasmids. In pSM19035, a plasmid of Streptococcus pyogenes, replication, partition and toxin-antitoxin systems act co-ordinately to achieve high plasmid maintenance level with a minimal fitness cost [23]. In the broad host-range plasmid, RK2 replication and partitioning functions act co-ordinately to achieve stable plasmid maintenance in different hosts. Multiple co-ordinately regulated operons contribute to this [24] and the basic mechanisms involved have been recently reviewed [29]. Other singular case of coordination between different functions related to plasmid maintenance has been reported in the XerCD multimer resolution system of plasmid ColE1. Multimers of this multicopy plasmid compromise cell growth and plasmid stability but they can be resolved by the XerCD-mediated site-specific recombination at cer [30]. In addition, multimer formation induces rcd-RNA, a singular component of the system that interacts with and enhances the action of triptophanase thus increasing the concentration of Indol in the cells. This inhibits cell growth and division as well as plasmid replication, thus timing the resolvase to act before cell division can occur [31]. Similarly, in plasmid R1, coordination of the basic replicon functions and the kis-kid system activates the Kid toxin to rescue replication of cells with very low copy number [16]. Activated Kid toxin inhibit cell division, thus effectively achieving plasmid R1 retention by increasing plasmid replication and timing this replication rescue before cell division can occur [21]. Our report adds to this that the kis-kid system plays a more relevant role in control of plasmid copy number than previously suspected.
4. Experimental Section
4.1. Cell Cultures, Strains and Plasmids
Cells were growth at 30 °C in L-Broth (LB) and L-Broth agar (LA) prepared as described [32]. Antibiotics were supplemented according to the resistances carried by the plasmids. The E. coli K12 clpP− mutant SG12050 [33] and its parental strain C600 [34] were used in this work. Plasmid used in this work were: pKN1562, a wild-type (wt) mini-R1 plasmid conferring resistance to kanamycin [10] and carrying the wt kis-kid system [11]; pJLV01, a pKN1562 derivative that carries the point mutation kidD75E that inactivates the RNase function of the Kid toxin but not its co-regulatory activity [22,35,36]; variants of pKN1562 or pJLV01 carrying the repAR55H mutation [22]; pMLM126, a Kis overproducer inducible by arabinose, and pLNBAD its empty vector [36]; finally, we also used a CopB overproducer, pUC18-copB, and its empty vector pUC18 [6].
4.2. Replication Interference Phenotype
Plasmid DNA extractions and transformations were essentially as described [37]. In vitro mutagenesis of plasmids was done with hydroxylamine as previously described [38]. This DNA was extensively dialysed against TE buffer (Tris-HCl 10 mM pH. 8.0, 1 mM EDTA) and used to transform the selected E. coli strains. The ratio of replication thermo-sensitive mutants (rep-ts) to kanamycin resistant thermo-sensitive mutants (kmR-ts) rescued by transformation was used to define the replication interference phenotype. This value is very close to 1 when the coupling is inactivated by a mutation in the kid toxin gene but it is reduced to 0.2–0.3 in the presence of a functional coupling [22].
4.3. Plasmid Stability and Statistical Analysis
The plasmid stability determinations were done in cells containing pKN1562 and its rep and kid mutant derivatives. For this purpose the percentage of colonies carrying the kanamycin resistance marker were determined after cell propagation at 30 °C for 30, 60 and 90 generations in rich solid LA media without kanamycin as previously described [22].
The standard deviation corresponding to the different determinations, were calculated using values obtained at least in three independent experiments. A paired Student’s t-test using GraphPad Prism 5 for Mac software was used. For the statistical analysis of Kis antitoxin levels One way ANOVA test and Wilcoxon non-parametric t-test were used. Values linked by brackets and labelled with * or *** correspond to differences which p-values are <0.05 or <001, respectively.
4.4. Plasmid Copy Number Determinations
R1 mini plasmids copy numbers relative to chromosome were determined by quantitative PCR methodologies (qPCR) as described in [39]. In these determinations, kis and lpp genes were selected as plasmid and chromosome markers, respectively. Specific protocols and primers used in these determinations have been described recently [22]. Plasmid copy number (PCN) per genome was calculated as the ratio between (Size of chromosomal DNA (bp) × Amount of plasmid DNA (ng))/(Size of plasmid DNA (bp) × Amount of genomic DNA bound (ng)). The PCN calculated for pKN1562 was giving the value = 1; the PCNs per genome were normalized to this value. When the analysis was done in the presence of the overproducer of Kis, the primers used for the PCN and transcriptional determinations amplified the 5'region of the Kid toxin and were 5'-GGTCACGCGATTAAAGGC-3' and 5'-GGTCACGCGATTAAAGGC-3'. The PCN values were referred to as the percentage of kanamycin resistant cells evaluated when the sample was taken.
4.5. Quantification of KIS-KID MRNA LEVELS
Transcriptional levels of kis and lpp genes were evaluated by RTqPCR [22]. The kis transcriptional levels were calculated as 2(Ctlpp−Ctkis) where Ct-lpp and Ct-kis are the threshold values corresponding to the PCR amplification of lpp and kis. When the analysis was done in the presence of the Kis overproducer, the primers used for the kis-kid mRNA determinations (see Section 4.4) amplified the 5' region of kid (see Section 4.4). The relative transcriptional levels were referred in all cases to the plasmid copy number, a value that is already corrected from the contribution of plasmid-free cells.
4.6. Determination of Kis Antitoxin Levels by Western Blotting-3XFLAG
Bacterial cultures were growth as indicated above. Equal samples of the cultures were taken at DO600 = 0.5 and the cells were collected and re-suspend in Laemmli buffer. The number of plasmid-containing cells was evaluated in these samples. Antitoxin expression levels were determined as the means of at least seven Western blotting experiments using a pKN1562 version with 3×FLAG-tagged Kis. Kis protein values were referred to the percentage of plasmid-containing cells; the average level determined in the wild-type strain was used as the reference (value = 1). Western blot analysis was done using anti-FLAG (Sigma-Aldrich, St. Louis, MO, USA) diluted 1:500 (2 h 30 min) and anti-mouse antibodies (Sigma-Aldrich, St. Louis, MO, USA) diluted 1:5000 (1 h 30 min) in TBS-Tween containing 3% non-fat milk. The westerns were developed using ECL (Thermo-Scientific, Waltham, MA, USA); band intensity analysis corresponding to samples from five independent cultures, was determined using Quantity One software (Bio-Rad, Berkeley, CA, USA, version 4.6.3).
Acknowledgments
This study was founded by projects CSD2008-00013 and BFU2011-25939 of the Spanish Ministry of Science and Innovation. Discussions related to this work with members of the group are kindly acknowledged. The comments and corrections to the manuscript of Elizabeth Diago-Navarro are kindly acknowledged.
Author Contributions
Juan López Villarejo conceived and performed the evaluation of the kis-kid mRNA levels the copy number determinations and the plasmid stability analysis and also did the relevant constructions needed; Damian Lobato-Marquez was involved in the determinations of the Kis antitoxin levels and did the constructions needed for this analysis; Ramon Díaz-Orejas conceived the first idea of the project, and wrote the manuscript; All the authors were also involved in improving the successive versions of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Nordström K. Plasmid R1—replication and its control. Plasmid. 2006;55:1–26. doi: 10.1016/j.plasmid.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Giraldo R., Diaz R. Differential binding of wild-type and a mutant RepA protein to oriR sequence suggests a model for the initiation of plasmid R1 replication. J. Mol. Biol. 1992;228:787–802. doi: 10.1016/0022-2836(92)90864-G. [DOI] [PubMed] [Google Scholar]
- 3.Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: Genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984;12:71–90. doi: 10.1016/0147-619X(84)90054-4. [DOI] [PubMed] [Google Scholar]
- 4.Nordström K., Wagner E.G. Kinetic aspects of control of plasmid replication by antisense RNA. Trends Biochem. Sci. 1994;19:294–300. doi: 10.1016/0968-0004(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 5.Uhlin B.E., Molin S., Gustafsson P., Nordström K. Plasmids with temperature-dependent copy number for amplification of cloned genes and their products. Gene. 1979;6:91–106. doi: 10.1016/0378-1119(79)90065-9. [DOI] [PubMed] [Google Scholar]
- 6.Riise E., Stougaard P., Bindslev B., Nordström K., Molin S. Molecular cloning and functional characterization of a copy number control gene (copB) of plasmid R1. J. Bacteriol. 1982;151:1136–1145. doi: 10.1128/jb.151.3.1136-1145.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riise E., Molin S. Purification and characterization of the CopB replication control protein, and precise mapping of its target site in the R1 plasmid. Plasmid. 1986;15:163–171. doi: 10.1016/0147-619X(86)90034-X. [DOI] [PubMed] [Google Scholar]
- 8.Light J., Riise E., Molin S. Transcription and its regulation in the basic replicon region of plasmid R1. Mol. Gen. Genet. 1985;198:503–508. doi: 10.1007/BF00332947. [DOI] [PubMed] [Google Scholar]
- 9.Olsson J.A., Paulsson J., Nordström K. Effect of the copB auxiliary replication control system on stability of maintenance of par (+) plasmid R1. J. Bacteriol. 2004;186:207–211. doi: 10.1128/JB.186.1.207-211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molin S., Stougaard P., Uhlin B.E., Gustafsson P., Nordström K. Clustering of genes involved in replication, copy number control, incompatibility, and stable maintenance of the resistance plasmid R1drd-19. J. Bacteriol. 1979;138:70–79. doi: 10.1128/jb.138.1.70-79.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bravo A., de Torrontegui G., Diaz R. Identification of components of a new stability system of plasmid R1, parD, that is close to the origin of replication of this plasmid. Mol. Gen. Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 12.Diago-Navarro E., Hernandez-Arriaga A.M., Lopez-Villarejo J., Muñoz-Gomez A.J., Kamphuis M.B., Boelens R., Lemonnier M., Diaz-Orejas R. parD toxin-antitoxin system of plasmid R1—Basic contributions, biotechnological applications and relationships with closely-related toxin-antitoxin systems. FEBS J. 2010;277:3097–3117. doi: 10.1111/j.1742-4658.2010.07722.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J., Zhang Y., Zhu L., Suzuki M., Inouye M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 2004;279:20678–20684. doi: 10.1074/jbc.M314284200. [DOI] [PubMed] [Google Scholar]
- 14.Muñoz-Gomez A.J., Lemonnier M., Santos-Sierra S., Berzal-Herranz A., Diaz-Orejas R. RNase/anti-RNase activities of the bacterial parD toxin-antitoxin system. J. Bacteriol. 2005;187:3151–3157. doi: 10.1128/JB.187.9.3151-3157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamphuis M.B., Bonvin A.M., Monti M.C., Lemonnier M., Munoz-Gomez A., van den Heuvel R.H., Diaz-Orejas R., Boelens R. Model for RNA binding and the catalytic site of the RNase kid of the bacterial parD toxin-antitoxin system. J. Mol. Biol. 2006;357:115–126. doi: 10.1016/j.jmb.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz-Echevarria M.J., Berzal-Herranz A., Gerdes K., Diaz-Orejas R. The kis and kid genes of the parD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the co-ordinated action of the Kis and Kid proteins. Mol. Microbiol. 1991;5:2685–2693. doi: 10.1111/j.1365-2958.1991.tb01977.x. [DOI] [PubMed] [Google Scholar]
- 17.Monti M.C., Hernandez-Arriaga A.M., Kamphuis M.B., Lopez-Villarejo J., Heck A.J., Boelens R., Diaz-Orejas R., van den Heuvel R.H. Interactions of Kid-Kis toxin-antitoxin complexes with the parD operator-promoter region of plasmid R1 are piloted by the Kis antitoxin and tuned by the stoichiometry of Kid-Kis oligomers. Nucleic Acids Res. 2007;35:1737–1749. doi: 10.1093/nar/gkm073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega S., de Torrontegui G., Diaz R. Isolation and characterization of a conditional replication mutant of the antibiotic resistance factor R1 affected in the gene of the replication protein RepA. Mol. Gen. Genet. 1989;217:111–117. doi: 10.1007/BF00330949. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Echevarria M.J., de-la-Torre M.A., Diaz-Orejas R. A mutation that decreases the efficiency of plasmid R1 replication leads to the activation of parD, a killer stability system of the plasmid. FEMS Microbiol. Lett. 1995;130:129–135. doi: 10.1016/0378-1097(95)00192-8. [DOI] [PubMed] [Google Scholar]
- 20.Pimentel B., Madine M.A., de la Cueva-Mendez G. Kid cleaves specific mRNAs at UUACU sites to rescue the copy number of plasmid R1. EMBO J. 2005;24:3459–3469. doi: 10.1038/sj.emboj.7600815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pimentel B., Nair R., Bermejo-Rodriguez C., Preston M.A., Agu C.A., Wang X., Bernal J.A., Sherratt D.J., de la Cueva-Mendez G. Toxin Kid uncouples DNA replication and cell division to enforce retention of plasmid R1 in Escherichia coli cells. Proc. Natl. Acad. Sci. USA. 2014;111:2734–2739. doi: 10.1073/pnas.1308241111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Villarejo J., Diago-Navarro E., Hernandez-Arriaga A.M., Diaz-Orejas R. Kis antitoxin couples plasmid R1 replication and parD (kis,kid) maintenance modules. Plasmid. 2012;67:118–127. doi: 10.1016/j.plasmid.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Lioy V.S., Pratto F., de la Hoz A.B., Ayora S., Alonso J.C. Plasmid pSM19035, a model to study stable maintenance in firmicutes. Plasmid. 2010;64:1–17. doi: 10.1016/j.plasmid.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Thomas C.M., Smith C.A., Shingler V., Cross M.A., Hussain A.A., Pinkney M. Regulation of replication and maintenance functions of broad host-range plasmid RK2. Basic Life Sci. 1985;30:261–276. doi: 10.1007/978-1-4613-2447-8_21. [DOI] [PubMed] [Google Scholar]
- 25.Cevallos M.A., Cervantes-Rivera R., Gutierrez-Rios R.M. The repABC plasmid family. Plasmid. 2008;60:19–37. doi: 10.1016/j.plasmid.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Field C.M., Summers D.K. Multicopy plasmid stability: Revisiting the dimer catastrophe. J. Theor. Biol. 2011;291:119–127. doi: 10.1016/j.jtbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Diago-Navarro E., Hernandez-Arriaga A.M., Kubik S., Konieczny I., Diaz-Orejas R. Cleavage of the antitoxin of the parD toxin-antitoxin system is determined by the ClpAP protease and is modulated by the relative ratio of the toxin and the antitoxin. Plasmid. 2013;70:78–85. doi: 10.1016/j.plasmid.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Lau B.T., Malkus P., Paulsson J. New quantitative methods for measuring plasmid loss rates reveal unexpected stability. Plasmid. 2013;70:353–361. doi: 10.1016/j.plasmid.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolatka K., Kubik S., Rajewska M., Konieczny I. Replication and partitioning of the broad host-range plasmid RK2. Plasmid. 2010;64:119–134. doi: 10.1016/j.plasmid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Summers D.K., Sherratt D.J. Multimerization of high copy number plasmids causes instability: Co1E1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984;36:1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- 31.Field C.M., Summers D.K. Indole inhibition of ColE1 replication contributes to stable plasmid maintenance. Plasmid. 2012;67:88–94. doi: 10.1016/j.plasmid.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Maniatis T., Fritsch E.F., Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour Laboratory; Cold Spring Harbour, NY, USA: 1982. [Google Scholar]
- 33.Kandror O., Busconi L., Sherman M., Goldberg A.L. Rapid degradation of an abnormal protein in Escherichia coli involves the chaperones GroEl and GroES. J. Biol. Chem. 1994;269:23575–23582. [PubMed] [Google Scholar]
- 34.Appleyard R.K. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diago-Navarro E., Kamphuis M.B., Boelens R., Barendregt A., Heck A.J., van den Heuvel R.H., Diaz-Orejas R. A mutagenic analysis of the RNase mechanism of the bacterial Kid toxin by mass spectrometry. FEBS J. 2009;276:4973–4986. doi: 10.1111/j.1742-4658.2009.07199.x. [DOI] [PubMed] [Google Scholar]
- 36.Lemonnier M., Santos-Sierra S., Pardo-Abarrio C., Diaz-Orejas R. Identification of residues of the Kid toxin involved in autoregulation of the parD system. J. Bacteriol. 2004;186:240–243. doi: 10.1128/JB.186.1.240-243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J., Fristsch E.F., Maniatis T. Molecular Cloning. Cold Spring Harbour Laboratory; Cold Spring Harbour, NY, USA: 1989. [Google Scholar]
- 38.Hashimoto T., Sekiguchi M. Isolation of temperature-sensitive mutants of R plasmid by in vitro mutagenesis with hydroxylamine. J. Bacteriol. 1976;127:1561–1563. doi: 10.1128/jb.127.3.1561-1563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C., Kim J., Shin S.G., Hwang S. Absolute and relative qPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 2006;123:273–280. doi: 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]